Characterization of Brassica rapa RAP2.4-Related Proteins in Stress Response and as CUL3-Dependent E3 Ligase Substrates
Abstract
1. Introduction
2. Materials and Methods
3. Results
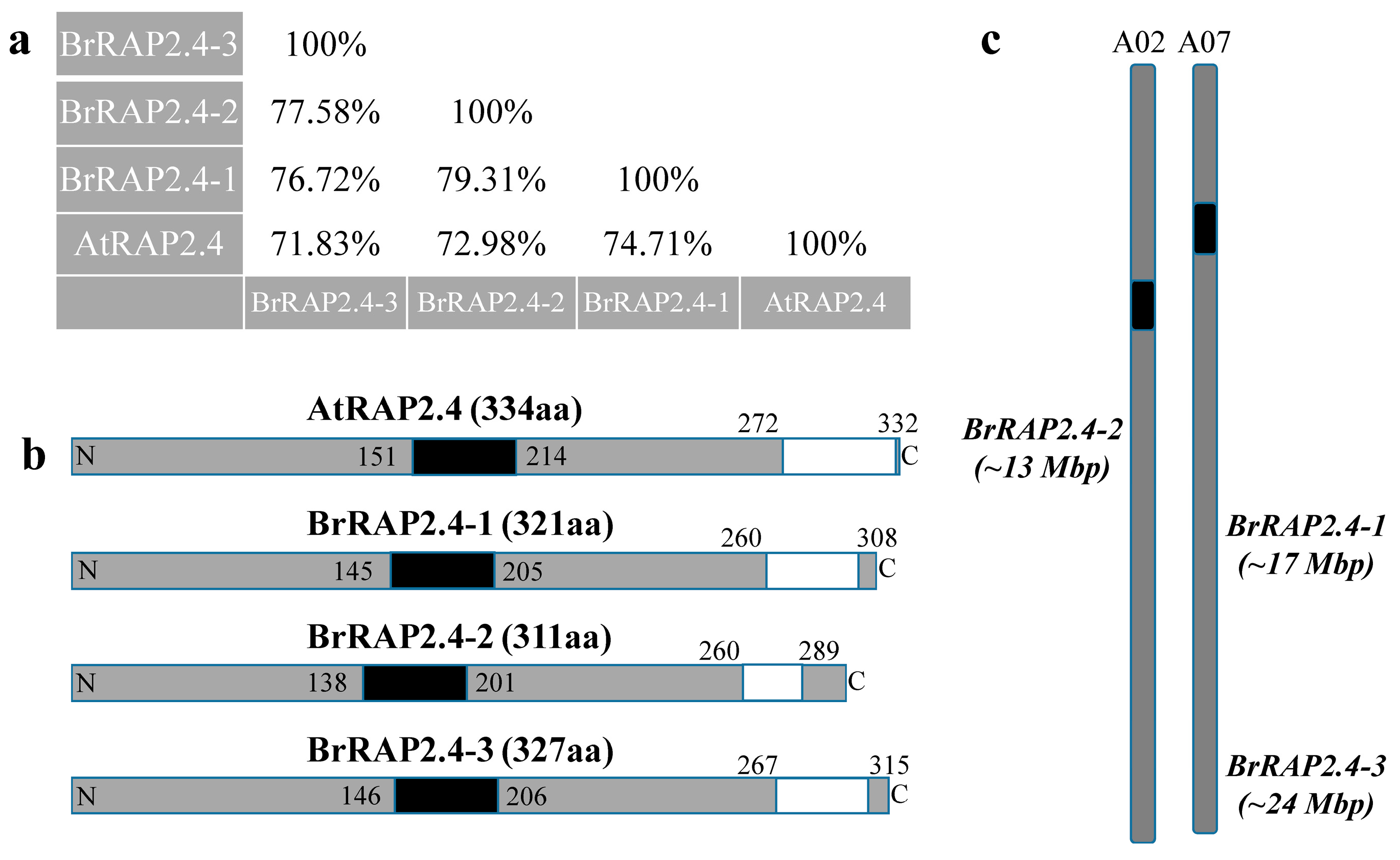
3.1. Brassica Rapa Encodes for Three RAP2.4-Like Genes
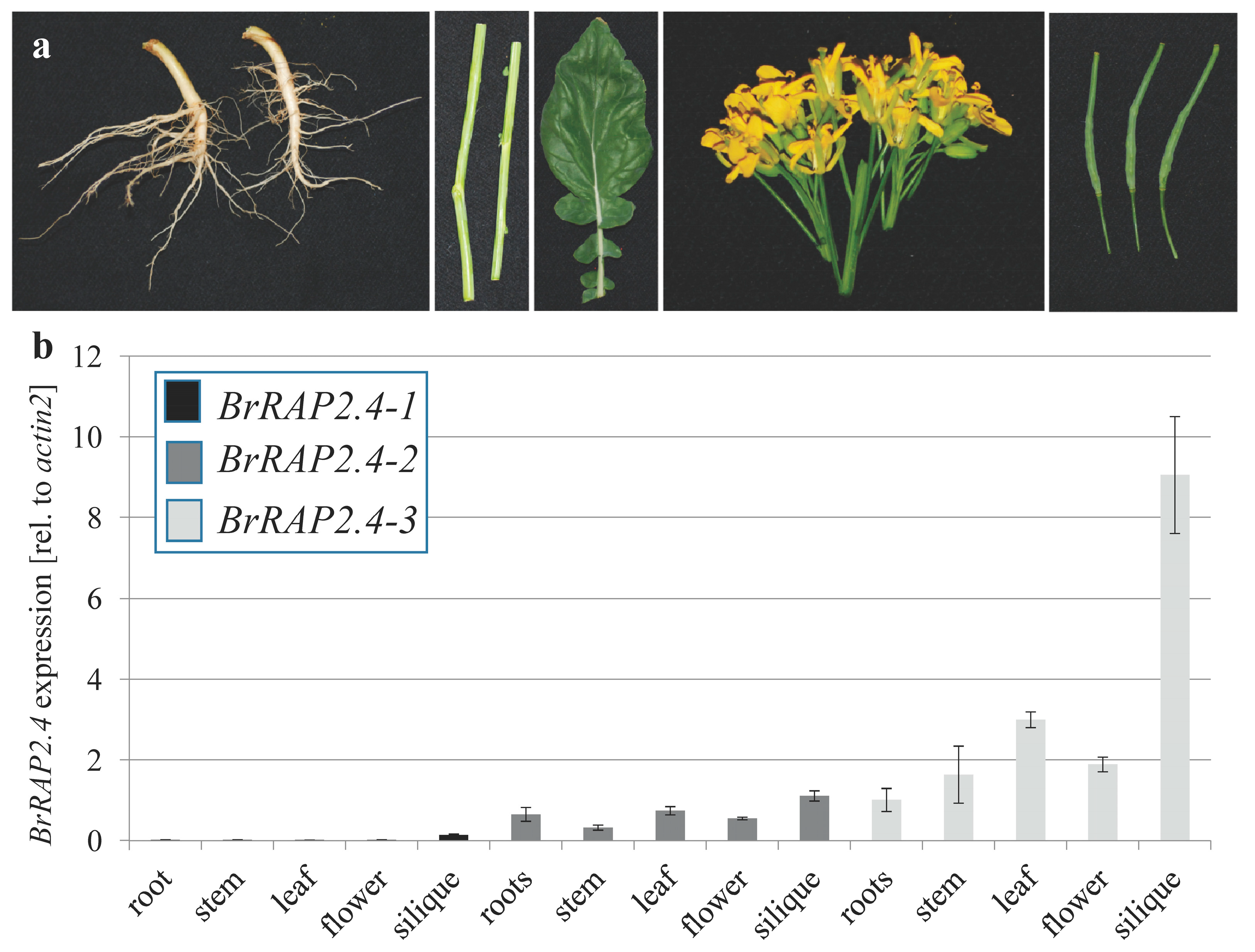
3.2. Tissue Specific Expression of BrRAP2.4-Like Genes
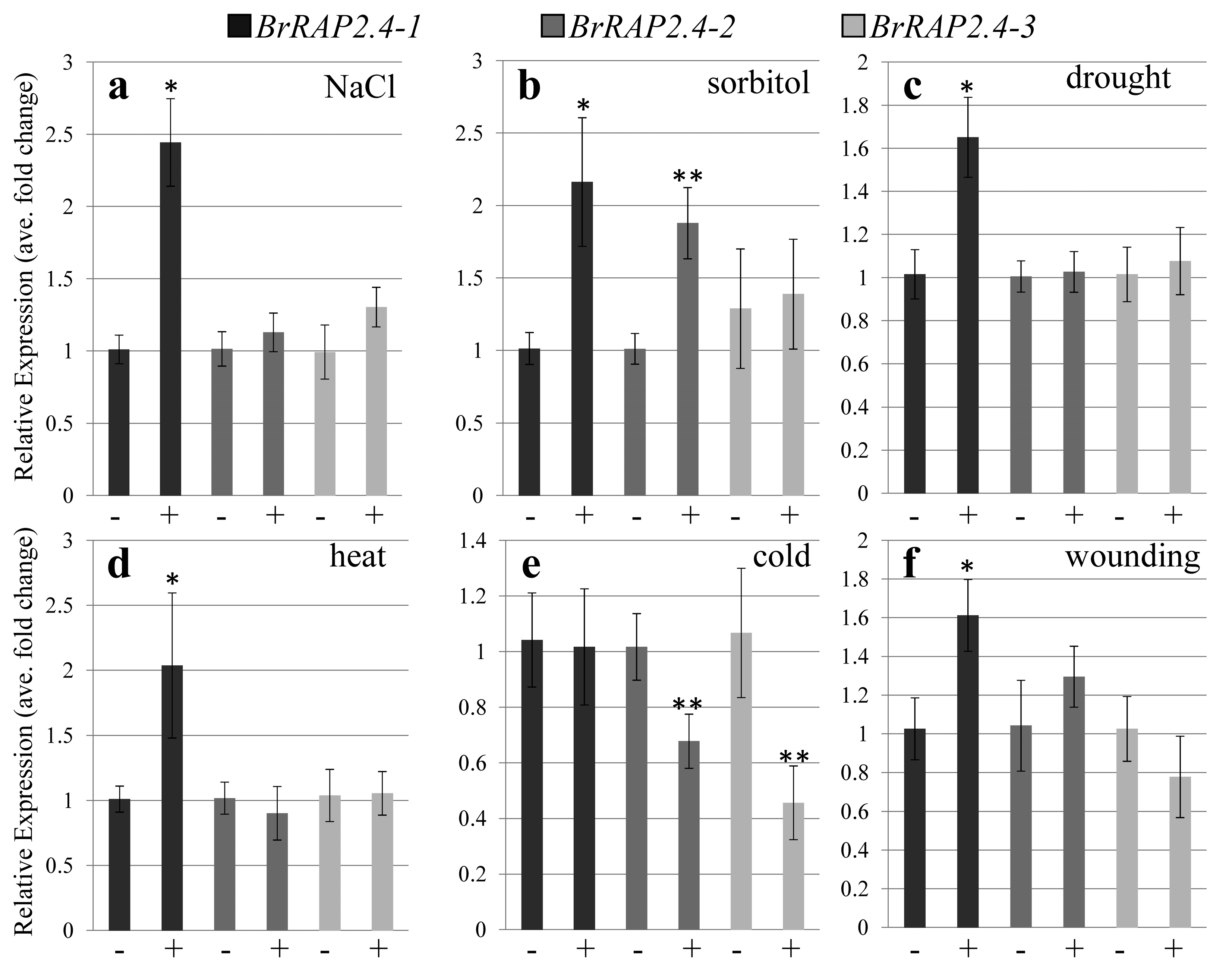
3.3. BrRAP2.4-Like Expression in Response to Abiotic Stress
3.4. Cloning of BrRAP2.4-1 and -3 Showed Differences to Annotated Sequences
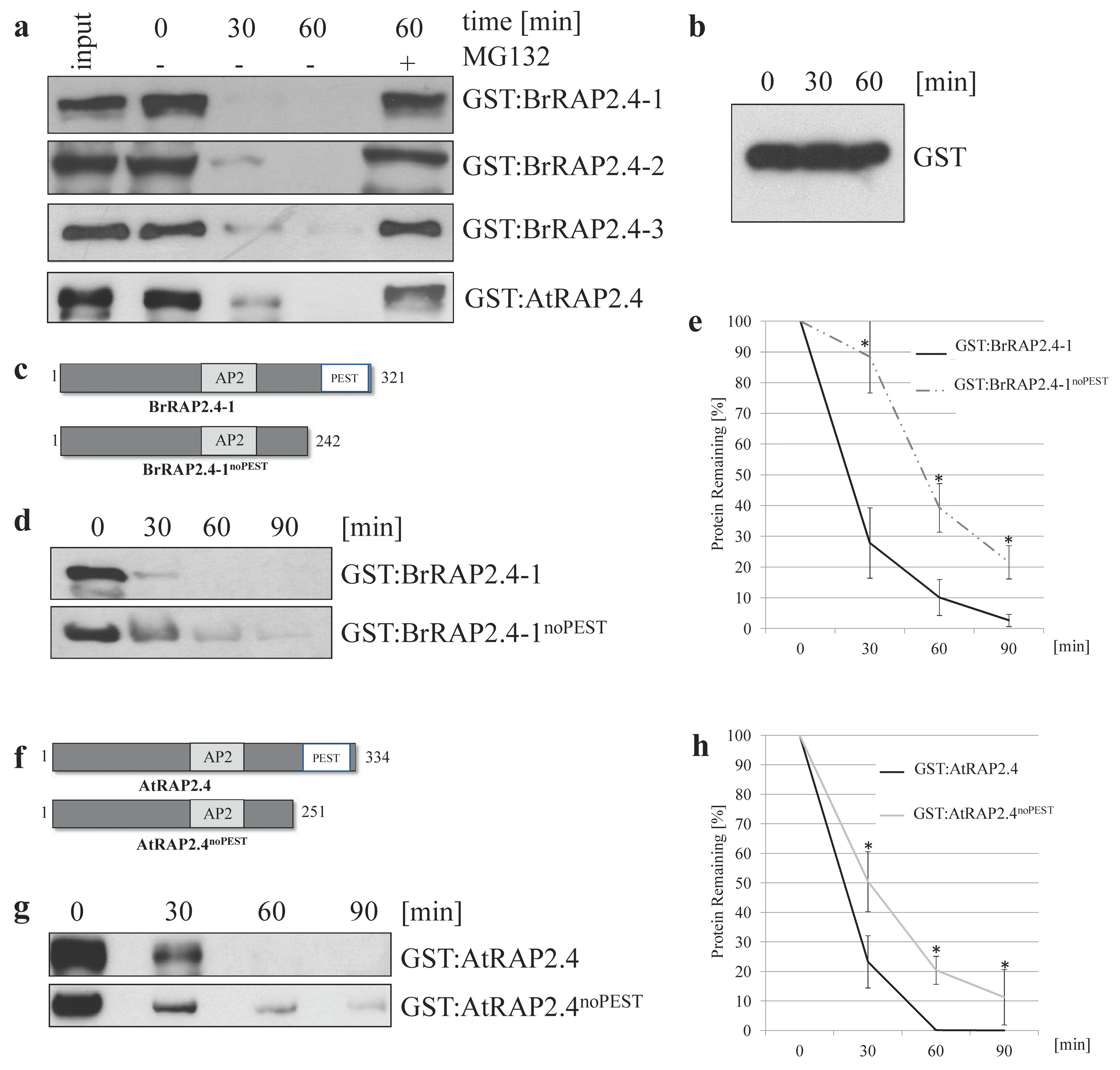
3.5. All BrRAP2.4 Genes Are Instable in a Proteasome-Dependent Manner
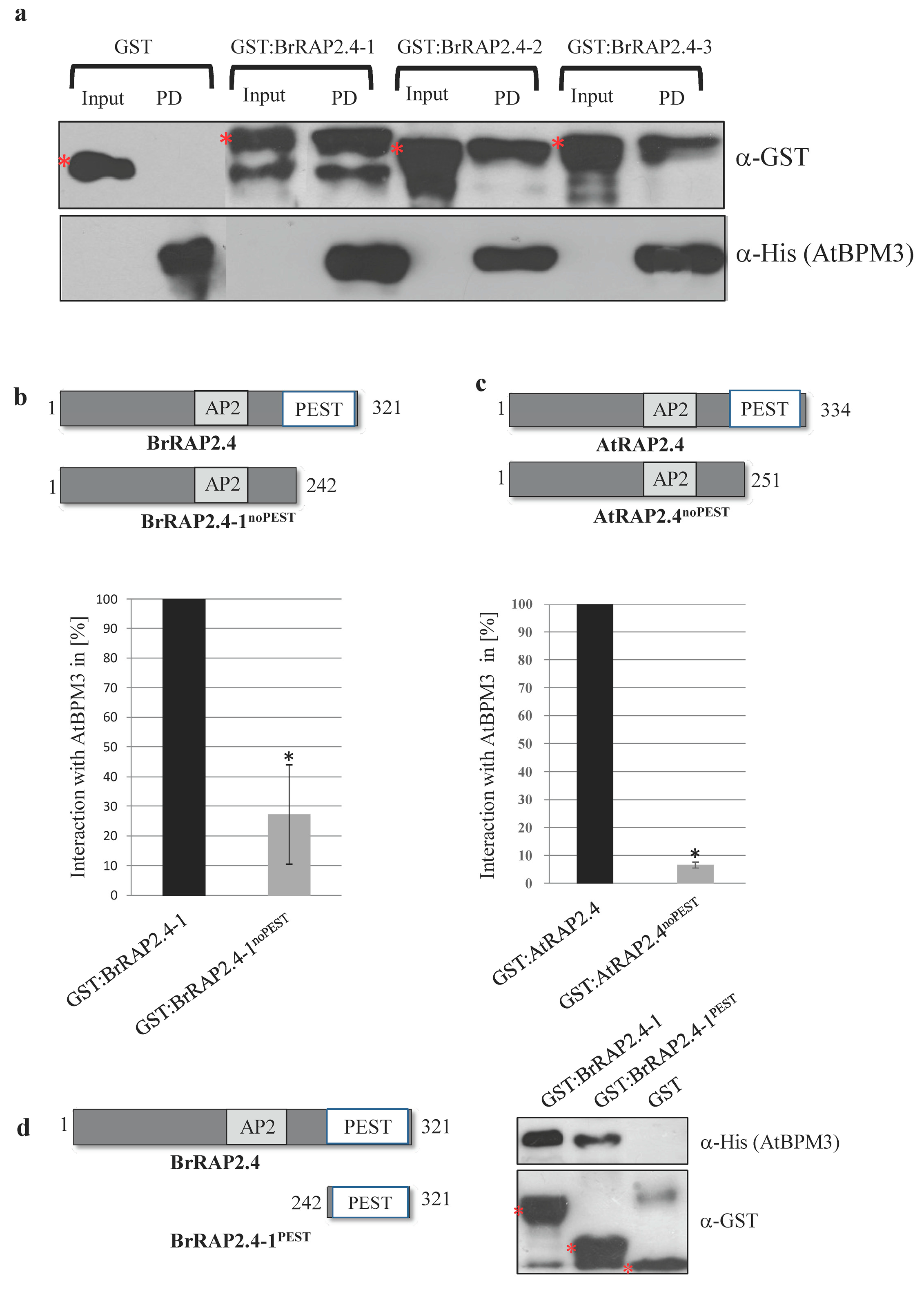
3.6. All BrRAP2.4 Proteins Assemble with Arabidopsis BPM3
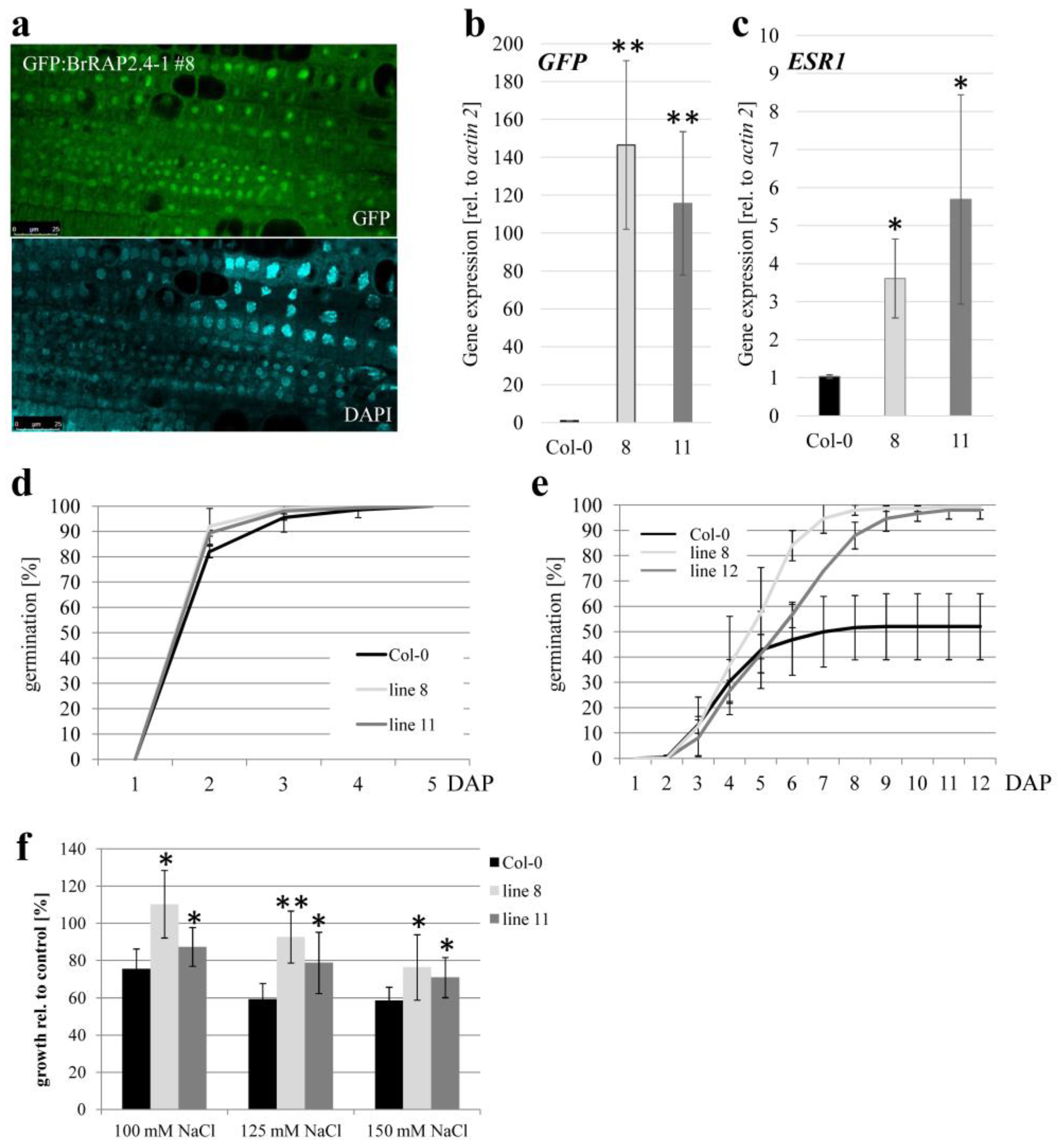
3.7. BrRAP2.4 Proteins Are Located in the Nucleus
3.8. BrRAP2.4-1 Overexpressing Plants Display Increased Salt Tolerance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 2012, 1819, 86–96. [Google Scholar] [CrossRef]
- Xu, Z.S.; Chen, M.; Li, L.C.; Ma, Y.Z. Functions and application of the AP2/ERF transcription factor family in crop improvement. J. Integr. Plant Biol. 2011, 53, 570–585. [Google Scholar] [CrossRef] [PubMed]
- Gutterson, N.; Reuber, T.L. Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr. Opt. Plant Biol. 2004, 7, 465–471. [Google Scholar] [CrossRef] [PubMed]
- Iwase, A.; Mitsuda, N.; Ikeuchi, M.; Ohnuma, M.; Koizuka, C.; Kawamoto, K.; Imamura, J.; Ezura, H.; Sugimoto, K. Arabidopsis WIND1 induces callus formation in rapeseed, tomato, and tobacco. Plant Signal Behav. 2013, 8, e27432. [Google Scholar] [CrossRef] [PubMed]
- Iwase, A.; Mitsuda, N.; Koyama, T.; Hiratsu, K.; Kojima, M.; Arai, T.; Inoue, Y.; Seki, M.; Sakakibara, H.; Sugimoto, K.; et al. The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis. Curr. Biol. Cb 2011, 21, 508–514. [Google Scholar] [CrossRef]
- Rae, L.; Lao, N.T.; Kavanagh, T.A. Regulation of multiple aquaporin genes in Arabidopsis by a pair of recently duplicated DREB transcription factors. Planta 2011, 234, 429–444. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.C.; Park, H.J.; Wang, H.Y. Role of Arabidopsis RAP2.4 in regulating light- and ethylene-mediated developmental processes and drought stress tolerance. Mol. Plant 2008, 1, 42–57. [Google Scholar] [CrossRef] [PubMed]
- Phuong, N.D.; Hoi, P.X. Isolation and characterization of a OsRap2.4A transcription factor and its expression in Arabidopsis for enhancing high salt and drought tolerance. Curr. Sci. India 2015, 108, 51–62. [Google Scholar]
- Figueroa-Yanez, L.; Pereira-Santana, A.; Arroyo-Herrera, A.; Rodriguez-Corona, U.; Sanchez-Teyer, F.; Espadas-Alcocer, J.; Espadas-Gil, F.; Barredo-Pool, F.; Castano, E.; Rodriguez-Zapata, L.C. RAP2.4a Is Transported through the Phloem to Regulate Cold and Heat Tolerance in Papaya Tree (Carica papaya cv. Maradol): Implications for Protection Against Abiotic Stress. PLoS ONE 2016, 11, e0165030. [Google Scholar] [CrossRef] [PubMed]
- Iwase, A.; Ohme-Takagi, M.; Sugimoto, K. WIND1: A key molecular switch for plant cell dedifferentiation. Plant Signal Behav. 2011, 6, 1943–1945. [Google Scholar] [CrossRef] [PubMed]
- Iwase, A.; Mita, K.; Nonaka, S.; Ikeuchi, M.; Koizuka, C.; Ohnuma, M.; Ezura, H.; Imamura, J.; Sugimoto, K. WIND1-based acquisition of regeneration competency in Arabidopsis and rapeseed. J. Plant Res. 2015, 128, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Iwase, A.; Harashima, H.; Ikeuchi, M.; Rymen, B.; Ohnuma, M.; Komaki, S.; Morohashi, K.; Kurata, T.; Nakata, M.; Ohme-Takagi, M.; et al. WIND1 Promotes Shoot Regeneration through Transcriptional Activation of ENHANCER OF SHOOT REGENERATION1 in Arabidopsis. Plant Cell 2017, 29, 54–69. [Google Scholar] [CrossRef] [PubMed]
- Iwase, A.; Mita, K.; Favero, D.S.; Mitsuda, N.; Sasaki, R.; Kobayshi, M.; Takebayashi, Y.; Kojima, M.; Kusano, M.; Oikawa, A.; et al. WIND1 induces dynamic metabolomic reprogramming during regeneration in Brassica napus. Dev. Biol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.B.; Yang, J.W.; Zhang, Z.Y.; Feng, X.F.; Wang, S.M. Two P5CS genes from common bean exhibiting different tolerance to salt stress in transgenic Arabidopsis. J. Genet. 2013, 92, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Cvikrova, M.; Gemperlova, L.; Martincova, O.; Vankova, R. Effect of drought and combined drought and heat stress on polyamine metabolism in proline-over-producing tobacco plants. Plant Phys. Biochem. 2013, 73, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Shelp, B.J.; Zarei, A. Subcellular compartmentation of 4-aminobutyrate (GABA) metabolism in arabidopsis: An update. Plant Signal Behav. 2017, 12, e1322244. [Google Scholar] [CrossRef] [PubMed]
- Scholz, S.S.; Malabarba, J.; Reichelt, M.; Heyer, M.; Ludewig, F.; Mithofer, A. Evidence for GABA-Induced Systemic GABA Accumulation in Arabidopsis upon Wounding. Front. Plant Sci. 2017, 8, 388. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Lee, J.H.; Weber, H.; Tohge, T.; Witt, S.; Roje, S.; Fernie, A.R.; Hellmann, H. Arabidopsis BPM proteins function as substrate adaptors to a cullin3-based E3 ligase to affect fatty acid metabolism in plants. Plant Cell 2013, 25, 2253–2264. [Google Scholar] [CrossRef] [PubMed]
- Weber, H.; Hellmann, H. Arabidopsis thaliana BTB/POZ-MATH proteins interact with members of the ERF/AP2 transcription factor family. Febs J. 2009, 276, 6624–6635. [Google Scholar] [CrossRef]
- Weber, H.; Bernhardt, A.; Dieterle, M.; Hano, P.; Mutlu, A.; Estelle, M.; Genschik, P.; Hellmann, H. Arabidopsis AtCUL3a and AtCUL3b form complexes with members of the BTB/POZ-MATH protein family. Plant Phys. 2005, 137, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, P.; Gusmaroli, G.; Serino, G.; Habashi, J.; Ma, L.; Shen, Y.; Feng, S.; Bostick, M.; Callis, J.; Hellmann, H.; et al. Arabidopsis has two redundant Cullin3 proteins that are essential for embryo development and that interact with RBX1 and BTB proteins to form multisubunit E3 ubiquitin ligase complexes in vivo. Plant Cell 2005, 17, 1180–1195. [Google Scholar] [CrossRef]
- Gingerich, D.J.; Gagne, J.M.; Salter, D.W.; Hellmann, H.; Estelle, M.; Ma, L.; Vierstra, R.D. Cullins 3a and 3b assemble with members of the broad complex/tramtrack/bric-a-brac (BTB) protein family to form essential ubiquitin-protein ligases (E3s) in Arabidopsis. J. Biol. Chem. 2005, 280, 18810–18821. [Google Scholar] [CrossRef]
- Choi, C.M.; Gray, W.M.; Mooney, S.; Hellmann, H. Composition, roles, and regulation of cullin-based ubiquitin e3 ligases. Arab. Book Am. Soc. Plant Biol. 2014, 12, e0175. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Huang, Y.; Hu, Y.; He, X.; Shen, W.; Liu, C.; Ruan, Y. Phylogenetic Analysis of Brassica rapa MATH-Domain Proteins. Curr. Genom. 2013, 14, 214–223. [Google Scholar] [CrossRef]
- Juranic, M.; Dresselhaus, T. Phylogenetic analysis of the expansion of the MATH-BTB gene family in the grasses. Plant Signal Behav. 2014, 9, e28242. [Google Scholar] [CrossRef]
- Gingerich, D.J.; Hanada, K.; Shiu, S.H.; Vierstra, R.D. Large-scale, lineage-specific expansion of a bric-a-brac/tramtrack/broad complex ubiquitin-ligase gene family in rice. Plant Cell 2007, 19, 2329–2348. [Google Scholar] [CrossRef]
- Chen, L.; Bernhardt, A.; Lee, J.; Hellmann, H. Identification of Arabidopsis MYB56 as a novel substrate for CRL3(BPM) E3 ligases. Mol. Plant 2015, 8, 242–250. [Google Scholar] [CrossRef] [PubMed]
- Lechner, E.; Leonhardt, N.; Eisler, H.; Parmentier, Y.; Alioua, M.; Jacquet, H.; Leung, J.; Genschik, P. MATH/BTB CRL3 receptors target the homeodomain-leucine zipper ATHB6 to modulate abscisic acid signaling. Dev. Cell 2011, 21, 1116–1128. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, K.; Ohama, N.; Kidokoro, S.; Mizoi, J.; Takahashi, F.; Todaka, D.; Mogami, J.; Sato, H.; Qin, F.; Kim, J.S.; et al. BPM-CUL3 E3 ligase modulates thermotolerance by facilitating negative regulatory domain-mediated degradation of DREB2A in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, E8528–E8536. [Google Scholar] [CrossRef] [PubMed]
- Estelle, M.A.; Somerville, C. Auxin resistantmutants of Arabidopsis thaliana with altered morphology. Mol. Gen. Gen. 1987, 206, 200–206. [Google Scholar] [CrossRef]
- Curtis, M.D.; Grossniklaus, U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Phys. 2003, 133, 462–469. [Google Scholar] [CrossRef]
- Sparkes, I.A.; Runions, J.; Kearns, A.; Hawes, C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 2006, 1, 2019–2025. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Lu, Q.S.; Paz, J.D.; Pathmanathan, A.; Chiu, R.S.; Tsai, A.Y.; Gazzarrini, S. The C-terminal domain of FUSCA3 negatively regulates mRNA and protein levels, and mediates sensitivity to the hormones abscisic acid and gibberellic acid in Arabidopsis. Plant J. 2010, 64, 100–113. [Google Scholar] [CrossRef]
- Wang, F.; Zhu, D.; Huang, X.; Li, S.; Gong, Y.; Yao, Q.; Fu, X.; Fan, L.M.; Deng, X.W. Biochemical insights on degradation of Arabidopsis DELLA proteins gained from a cell-free assay system. Plant Cell 2009, 21, 2378–2390. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Leuendorf, J.E.; Osorio, S.; Szewczyk, A.; Fernie, A.R.; Hellmann, H. Complex Assembly and Metabolic Profiling of Arabidopsis thaliana Plants Overexpressing Vitamin B-6 Biosynthesis Proteins. Mol. Plant 2010, 3, 890–903. [Google Scholar] [CrossRef]
- Ikeda, M.; Ohme-Takagi, M. TCPs, WUSs, and WINDs: Families of transcription factors that regulate shoot meristem formation, stem cell maintenance, and somatic cell differentiation. Front. Plant Sci. 2014, 5, 427. [Google Scholar] [CrossRef]
- Kato, H.; Sakaki, K.; Mihara, K. Ubiquitin-proteasome-dependent degradation of mammalian ER stearoyl-CoA desaturase. J. Cell Sci. 2006, 119, 2342–2353. [Google Scholar] [CrossRef]
- Fung, T.K.; Siu, W.Y.; Yam, C.H.; Lau, A.; Poon, R.Y. Cyclin F is degraded during G2-M by mechanisms fundamentally different from other cyclins. J. Biol. Chem. 2002, 277, 35140–35149. [Google Scholar] [CrossRef]
- Karsies, A.; Hohn, T.; Leclerc, D. Degradation signals within both terminal domains of the cauliflower mosaic virus capsid protein precursor. Plant J. 2001, 27, 335–343. [Google Scholar] [CrossRef]
- Salama, S.R.; Hendricks, K.B.; Thorner, J. G1 cyclin degradation: The PEST motif of yeast Cln2 is necessary, but not sufficient, for rapid protein turnover. Mol. Cell Biol. 1994, 14, 7953–7966. [Google Scholar] [CrossRef]
- Liu, F.; Dowling, M.; Yang, X.J.; Kao, G.D. Caspase-mediated specific cleavage of human histone deacetylase 4. J. Biol. Chem. 2004, 279, 34537–34546. [Google Scholar] [CrossRef]
- Berset, C.; Griac, P.; Tempel, R.; La Rue, J.; Wittenberg, C.; Lanker, S. Transferable domain in the G(1) cyclin Cln2 sufficient to switch degradation of Sic1 from the E3 ubiquitin ligase SCF(Cdc4) to SCF(Grr1). Mol. Cell Biol. 2002, 22, 4463–4476. [Google Scholar] [CrossRef]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B.; et al. Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef]
- Franchi, G.G.; Piotto, B.; Nepi, M.; Baskin, C.C.; Baskin, J.M.; Pacini, E. Pollen and seed desiccation tolerance in relation to degree of developmental arrest, dispersal, and survival. J. Exp. Bot. 2011, 62, 5267–5281. [Google Scholar] [CrossRef]
- Lastdrager, J.; Hanson, J.; Smeekens, S. Sugar signals and the control of plant growth and development. J. Exp. Bot. 2014, 65, 799–807. [Google Scholar] [CrossRef]
- Broeckx, T.; Hulsmans, S.; Rolland, F. The plant energy sensor: Evolutionary conservation and divergence of SnRK1 structure, regulation, and function. J. Exp. Bot. 2016, 67, 6215–6252. [Google Scholar] [CrossRef]
- Iordachescu, M.; Imai, R. Trehalose biosynthesis in response to abiotic stresses. J. Integr. Plant Biol. 2008, 50, 1223–1229. [Google Scholar] [CrossRef]
- Ma, W.; Kong, Q.; Grix, M.; Mantyla, J.J.; Yang, Y.; Benning, C.; Ohlrogge, J.B. Deletion of a C-terminal intrinsically disordered region of WRINKLED1 affects its stability and enhances oil accumulation in Arabidopsis. Plant J. 2015, 83, 864–874. [Google Scholar] [CrossRef]
- Zhuang, M.; Calabrese, M.F.; Liu, J.; Waddell, M.B.; Nourse, A.; Hammel, M.; Miller, D.J.; Walden, H.; Duda, D.M.; Seyedin, S.N.; et al. Structures of SPOP-substrate complexes: Insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol. Cell 2009, 36, 39–50. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mooney, S.; Al-Saharin, R.; Choi, C.M.; Tucker, K.; Beathard, C.; Hellmann, H.A. Characterization of Brassica rapa RAP2.4-Related Proteins in Stress Response and as CUL3-Dependent E3 Ligase Substrates. Cells 2019, 8, 336. https://doi.org/10.3390/cells8040336
Mooney S, Al-Saharin R, Choi CM, Tucker K, Beathard C, Hellmann HA. Characterization of Brassica rapa RAP2.4-Related Proteins in Stress Response and as CUL3-Dependent E3 Ligase Substrates. Cells. 2019; 8(4):336. https://doi.org/10.3390/cells8040336
Chicago/Turabian StyleMooney, Sutton, Raed Al-Saharin, Christina M. Choi, Kyle Tucker, Chase Beathard, and Hanjo A. Hellmann. 2019. "Characterization of Brassica rapa RAP2.4-Related Proteins in Stress Response and as CUL3-Dependent E3 Ligase Substrates" Cells 8, no. 4: 336. https://doi.org/10.3390/cells8040336
APA StyleMooney, S., Al-Saharin, R., Choi, C. M., Tucker, K., Beathard, C., & Hellmann, H. A. (2019). Characterization of Brassica rapa RAP2.4-Related Proteins in Stress Response and as CUL3-Dependent E3 Ligase Substrates. Cells, 8(4), 336. https://doi.org/10.3390/cells8040336





