Nuclear Progestin Receptor Phosphorylation by Cdk9 Is Required for the Expression of Mmp15, a Protease Indispensable for Ovulation in Medaka
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Tissues
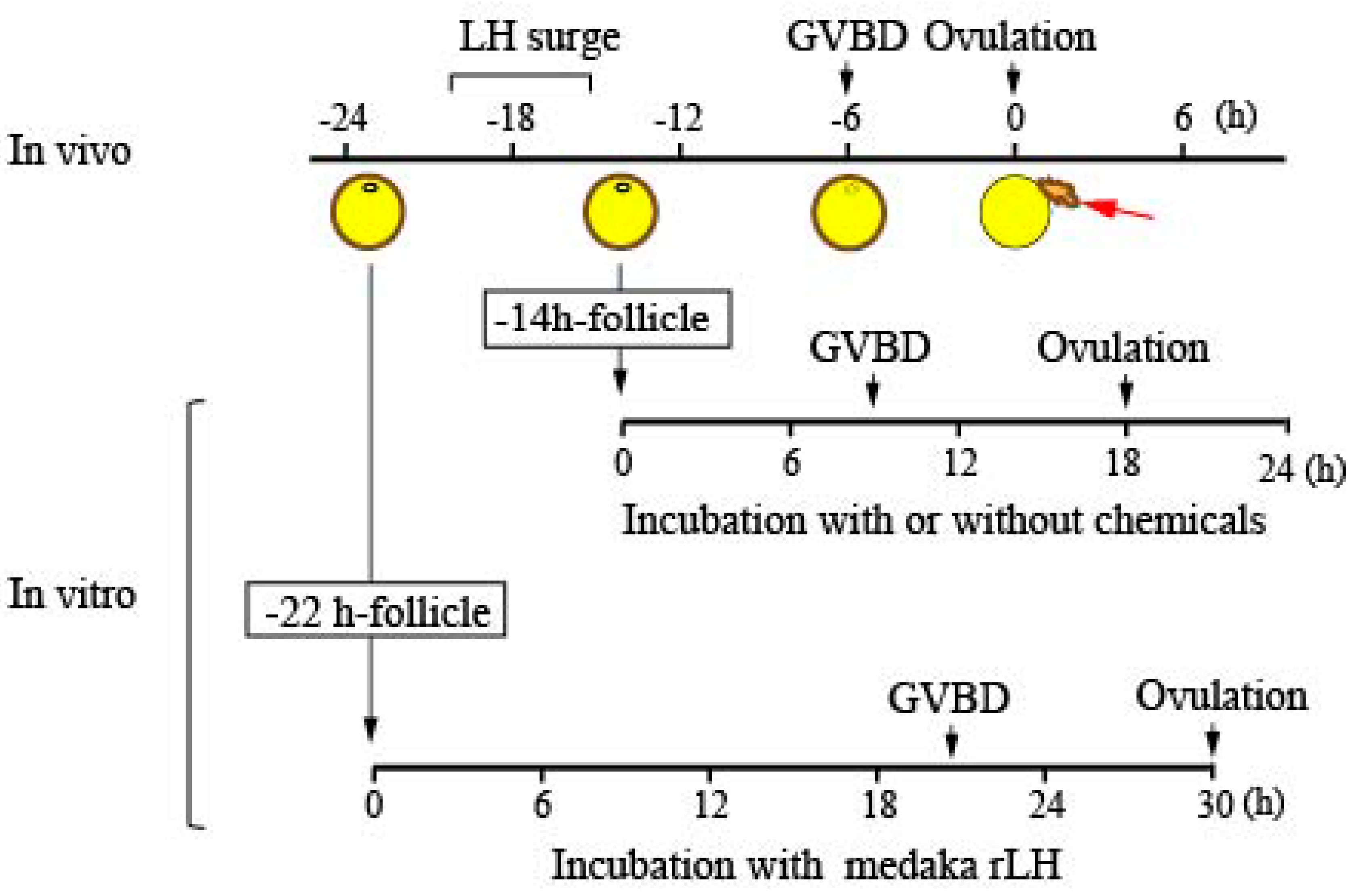
2.2. In Vitro Culture of Isolated Follicles
2.3. cDNA Cloning for ccni
2.4. RNA Isolation, Reverse Transcription (RT), and Real-Time Polymerase Chain Reaction (PCR)
2.5. Preparation of Antigens and Antibodies
2.6. Immunoprecipitation and Western Blot Analysis
2.7. Phosphatase Treatment
2.8. Primary Culture of Medaka Granulosa Cells (GC)
2.9. Digestion of ECM Proteins by Medaka Recombinant Mmp15
2.10. Culturing Cells Stably Expressing Medaka Pgr
2.11. Chromatin Immunoprecipitation (ChIP)
2.12. Detection of Phosphorylated Pgr
2.13. Immunohistochemistry
2.14. Knockout Experiments for cdk9
2.15. Coimmunoprecipitation
2.16. Statistical Analysis
3. Results
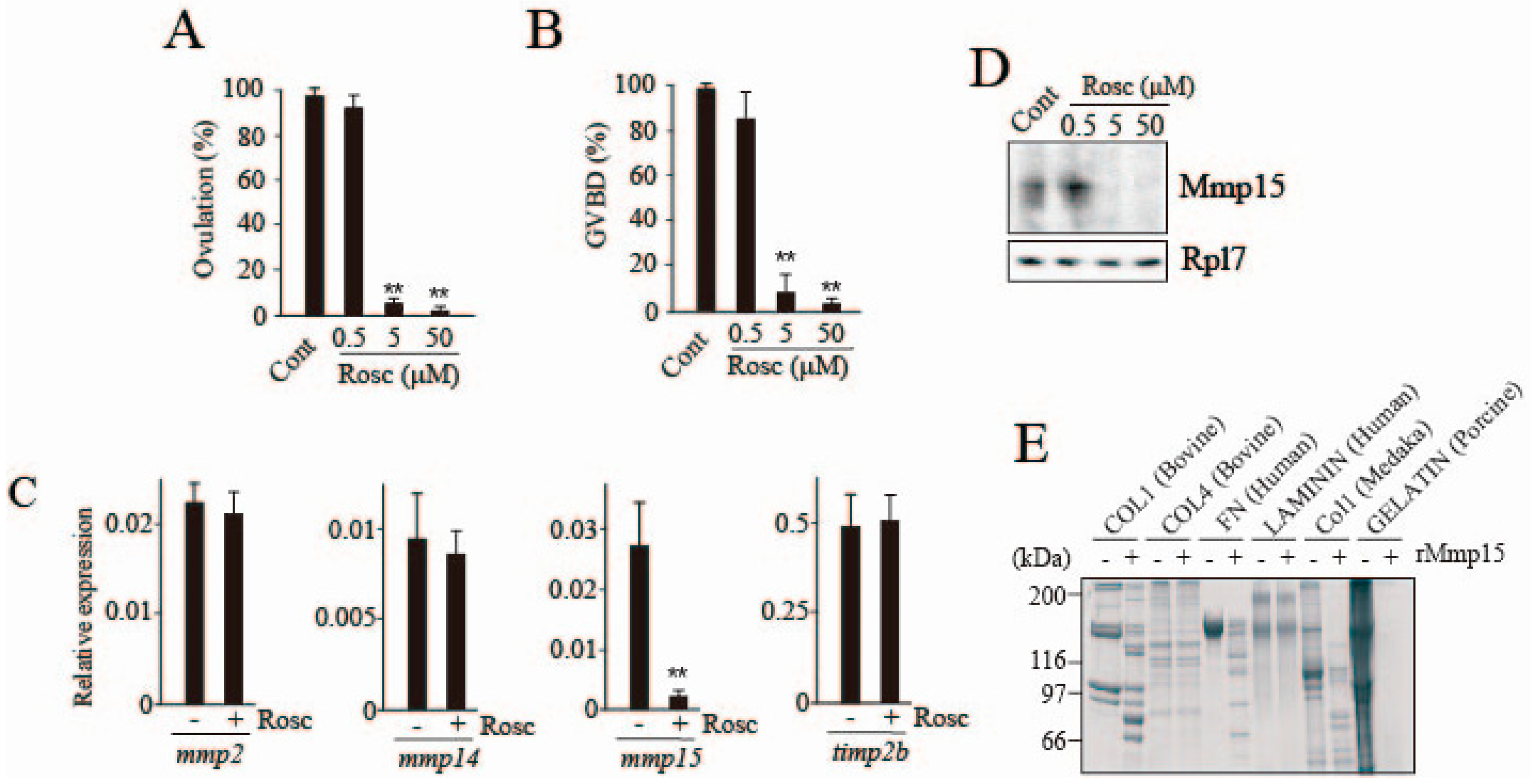
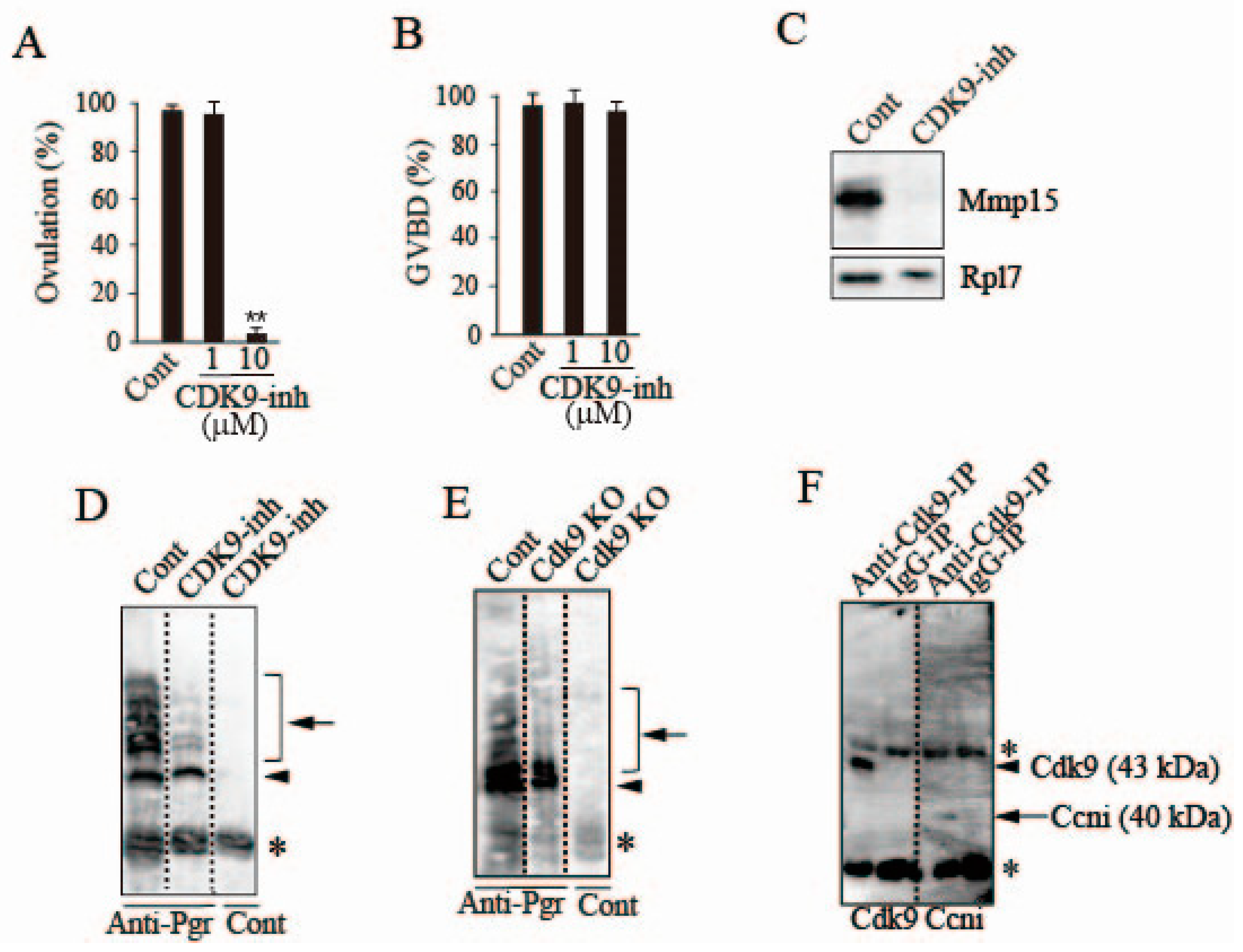
3.1. Inhibitory Effect of Roscovitine on Medaka Follicle Ovulation
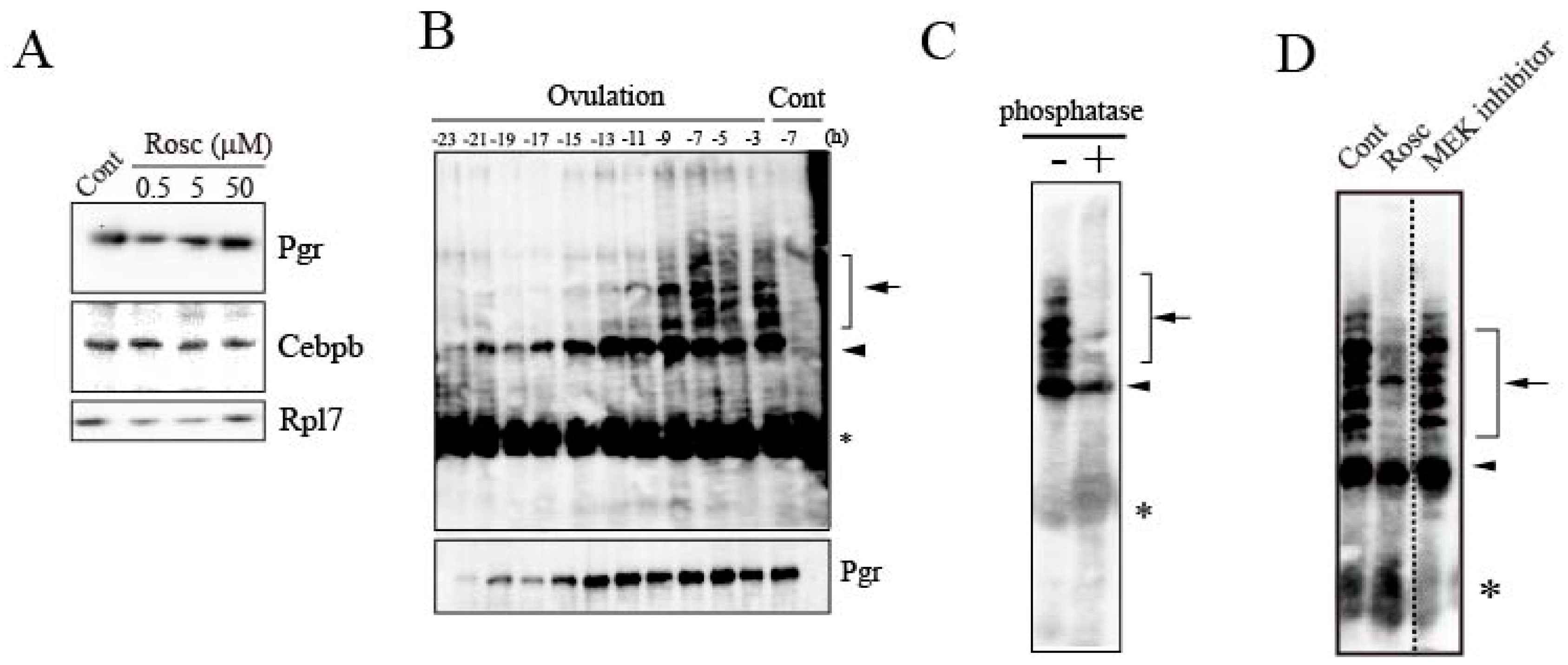
3.2. Effects of Roscovitine on Transcription Factors Involved in mmp15 Gene Expression
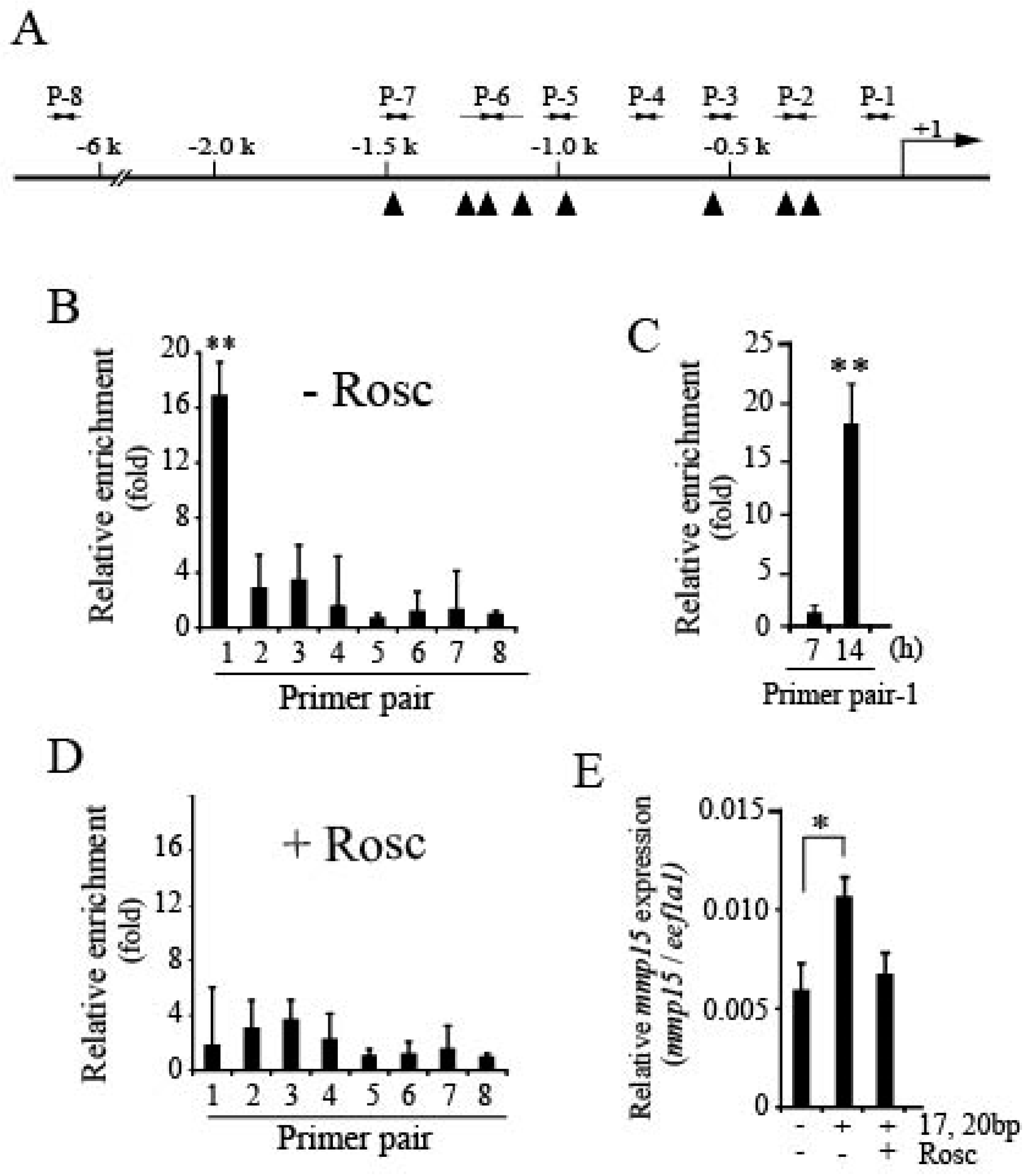
3.3. Loss of Pgr Binding Ability to the Promoter Region of the mmp15 Gene in Preovulatory Follicles under the Effect of Roscovitine Treatment
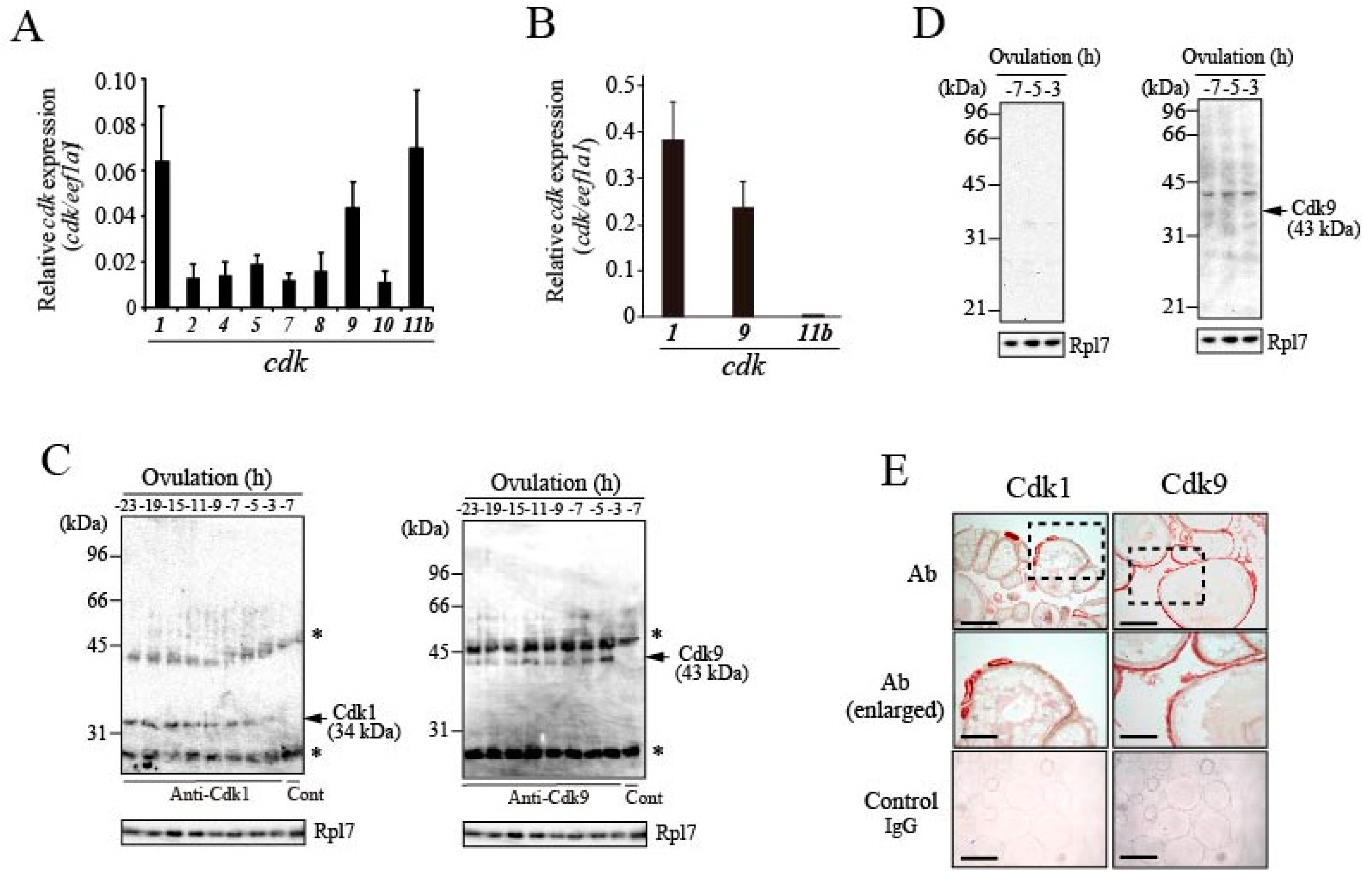
3.4. Expression of CDKs in Preovulatory Follicles of the Medaka Ovary
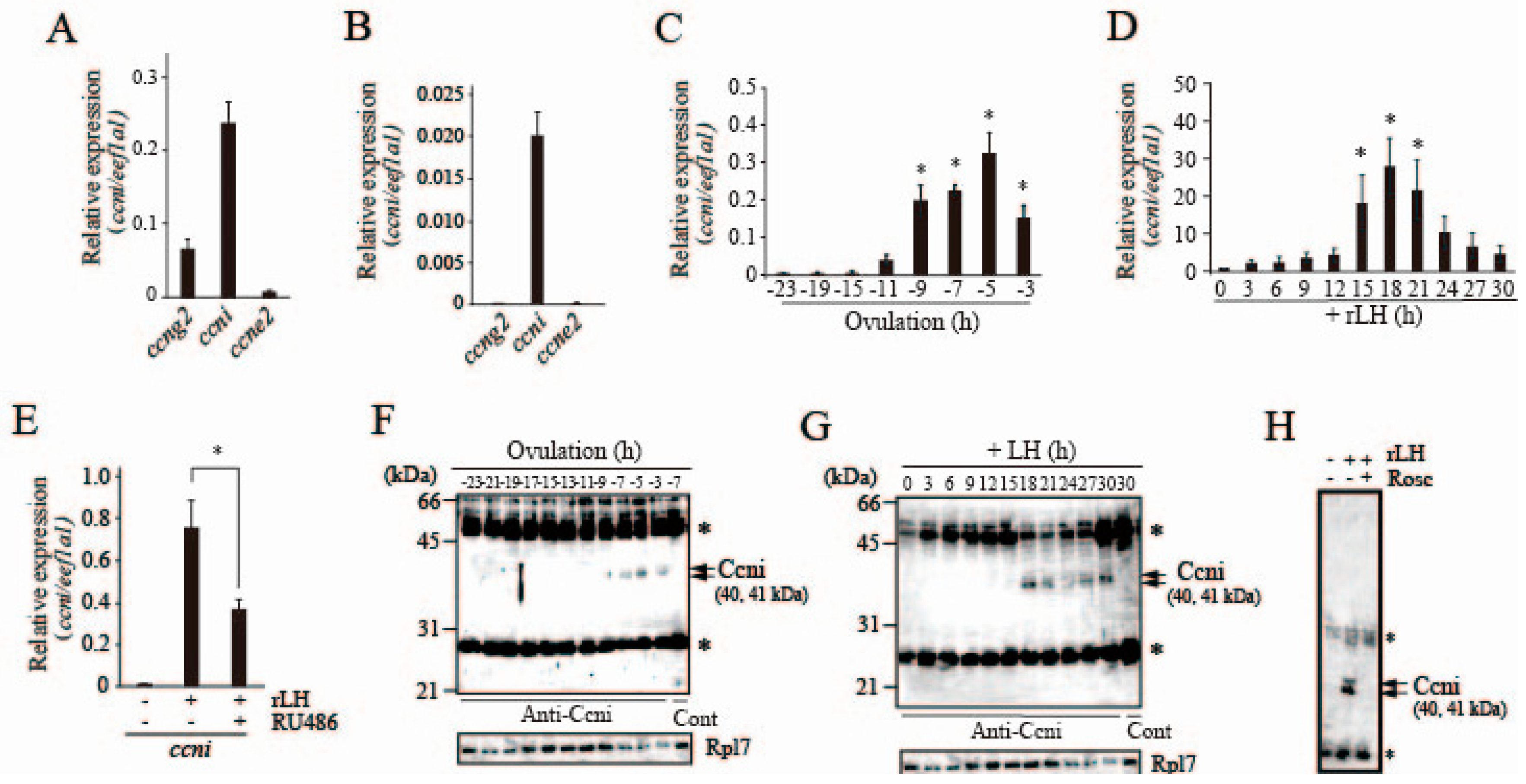
3.5. Expression of Cyclins in Preovulatory Follicles of the Medaka Ovary
3.6. Possible Involvement of Ccni/Cdk9 in Pgr Phosphorylation in the Preovulatory Follicle
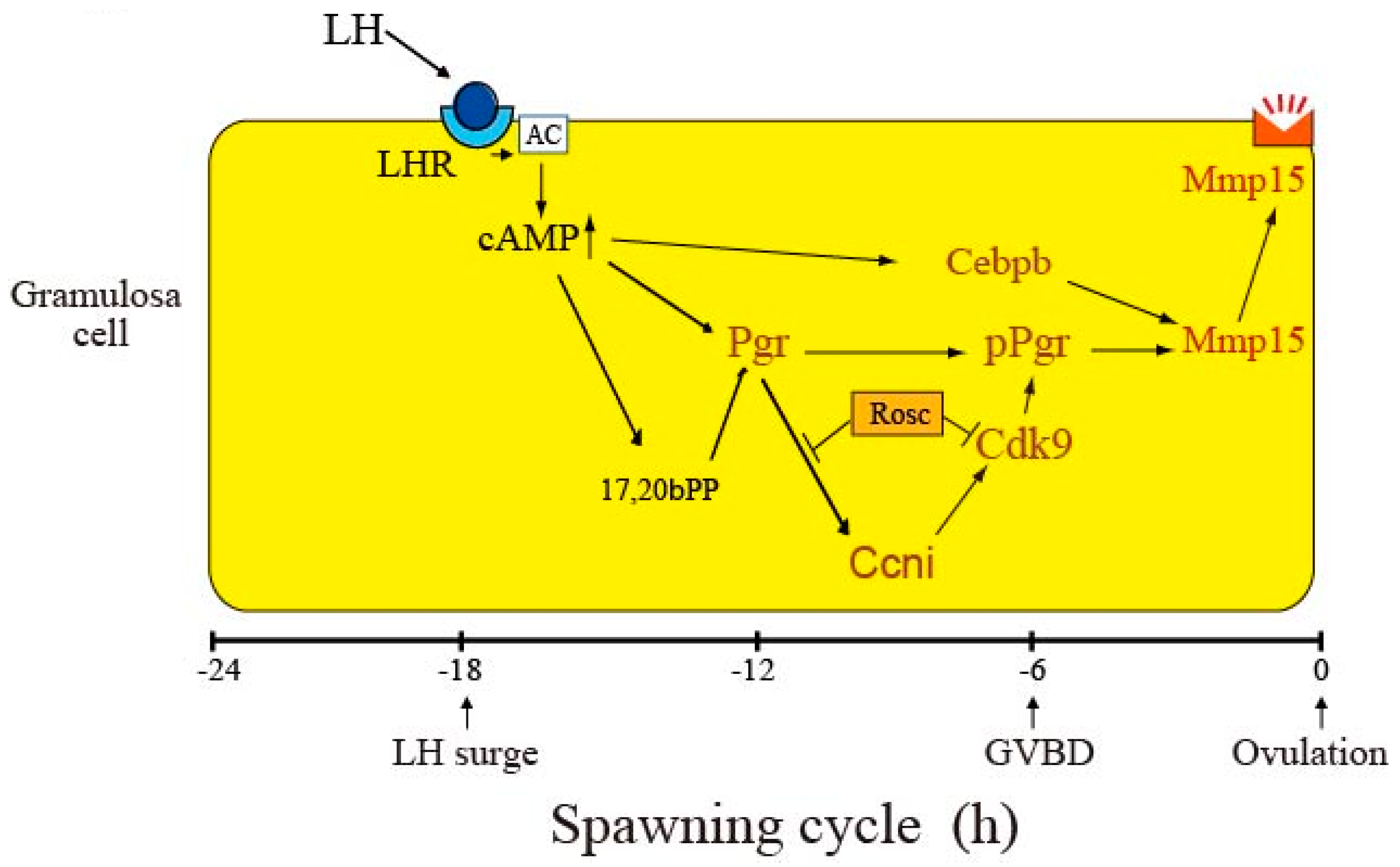
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Espey, L.L.; Richards, J.S. Knobil and Neill’s Physiology of Reproduction, 3rd ed.; Neill, J.D., Ed.; Academic Press: Amsterdam, The Netherlands, 2006; Volume 1, pp. 425–474. ISBN 0-12-515401-1. [Google Scholar]
- Lubzens, E.; Young, G.; Bobe, J.; Cerdà, J. Oogenesis in teleosts: How fish eggs are formed. Gen. Comp. Endocrinol. 2010, 165, 367–389. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhu, B.; Ge, W. Genetic analysis of zebrafish gonadotropin (FSH and LH) functions by TALEN-mediated gene disruption. Mol. Endocrinol. 2015, 29, 76–98. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, A.; Kanda, S.; Abe, T.; Oka, Y. Evolution of the hypothalamic-pituitary-gonadal axis regulation in vertebrates revealed by knockout medaka. Endocrinology 2016, 157, 3994–4002. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.; Fujimori, C.; Hagiwara, A.; Ogiwara, K. Recent advances in the understanding of teleost medaka ovulation: The role of proteases and prostaglandins. Zool. Sci. 2013, 30, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Ogiwara, K.; Takano, N.; Shinohara, M.; Murakami, M.; Takahashi, T. Gelatinase A and membrane-type matrix metalloproteinases 1 and 2 are responsible for follicle rupture during ovulation in the medaka. Proc. Natl. Acad. Sci. USA 2005, 102, 8442–8447. [Google Scholar] [CrossRef] [PubMed]
- Ogiwara, K.; Minagawa, K.; Takano, N.; Kageyama, T.; Takahashi, T. Apparent involvement of plasmin in early-stage of follicle rupture during ovulation in medaka. Biol. Reprod. 2012, 86, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ogiwara, K.; Hagiwara, A.; Rajapakse, S.; Takahashi, T. The role of urokinase plasminogen activator and plasminogen activator inhibitor-1 in follicle rupture during ovulation in the teleost medaka. Biol. Reprod. 2015, 92, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.; Hagiwara, A.; Ogiwara, K. Follicle rupture during ovulation with an emphasis on recent progress in fish models. Reproduction 2018, REP-18-0251. [Google Scholar] [CrossRef] [PubMed]
- Sakai, N.; Iwamatsu, T.; Yamauchi, K.; Nagahama, Y. Development of thesteroidogenic capacity of medaka (Oryzias latipes) ovarian follicles during vitellogenesis and oocyte maturation. Gen. Comp. Endocrinol. 1987, 66, 333–342. [Google Scholar] [CrossRef]
- Fukada, S.; Sakai, N.; Adachi, S.; Nagahama, Y. Steroidogenesis in the ovarian follicle of medaka (Oryzias latipes, a daily spawner) during oocyte maturation. Dev. Growth Differ. 1994, 36, 81–88. [Google Scholar] [CrossRef]
- Nagahama, Y.; Yamashita, M. Regulation of oocyte maturation in fish. Dev. Growth Differ. 2008, 50, S195–S219. [Google Scholar] [CrossRef] [PubMed]
- Ogiwara, K.; Takahashi, T. Involvement of the nuclear progestin receptor in LH-induced expression of membrane type 2-matrix metalloproteinase required for follicle rupture during ovulation in the medaka, Oryzias latipes. Mol. Cell. Endocrinol. 2017, 450, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Ogiwara, K.; Fujimori, C.; Rajapakse, S.; Takahashi, T. Characterization of luteinizing hormone and luteinizing hormone receptor and their indispensable role in the ovulatory process of the medaka. PLoS ONE 2013, 8, e54482. [Google Scholar] [CrossRef] [PubMed]
- Ogiwara, K.; Takahashi, T. Specificity of the medaka enteropeptidase serine protease and its usefulness as a biotechnological tool for fusion-protein cleavage. Proc. Natl. Acad. Sci. USA 2007, 104, 7021–7026. [Google Scholar] [CrossRef] [PubMed]
- Hagiwara, A.; Ogiwara, K.; Katsu, Y.; Takahashi, T. Luteinizing hormone-induced expression of Ptge4b, a prostaglandin E2 receptor indispensable for ovulation of the medaka Oryzias latipes, is regulated by a genomic mechanism involving nuclear protestin receptor. Biol. Reprod. 2014, 90, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ogiwara, K.; Takahashi, T. A dual role for melatonin in medaka ovulation: Ensuring prostaglandin synthesis and actin cytoskeleton rearrangement in follicular cells. Biol. Reprod. 2016, 94, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Ansai, S.; Kinoshita, M. Targeted mutagenesis using CRISPR/Cas system in medaka. Biol. Open 2014, 3, 362–371. [Google Scholar] [CrossRef] [PubMed]
- Horiguchi, M.; Fujimori, C.; Ogiwara, K.; Moriyama, A.; Takahashi, T. Collagen type I α1 chain mRNA is expressed in the follicle cells of the medaka ovary. Zool. Sci. 2008, 937–945. [Google Scholar] [CrossRef] [PubMed]
- Kato, Y.; Ogiwara, K.; Fujimori, C.; Kimura, A.; Takahashi, T. Expression and localization of collagen type IV α1 chain in medaka ovary. Cell Tissue Res. 2010, 340, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Xue, Y.; Yu, G.K.; Arias, C.; Lin, J.; Fong, S.; Faure, M.; Weisburd, B.; Ji, X.; Mercier, A.; et al. Compensatory induction of MYC expression by sustained CDK9 inhibition via a BRD4-dependent mechanism. eLife 2015, 4, e06535. [Google Scholar] [CrossRef] [PubMed]
- Hagen, C.R.; Daniel, A.R.; Dressing, G.E.; Lange, C.A. Role of phosphorylation in progesterone receptor signaling and specificity. Mol. Cell. Endocrinol. 2012, 357, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Hafiz, H.A.; Horwitz, K.B. Post-translational modifications of the progesterone receptors. J. Steroid Biochem. Mol. Biol. 2014, 140, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.; Hagiwara, A.; Ogiwara, K. Prostaglandins in teleost ovulation: A review of the roles with a view to comparison with prostaglandins in mammalian ovulation. Mol. Cell. Endocrinol. 2017, 461, 236–247. [Google Scholar] [CrossRef] [PubMed]
- Qiu, M.; Lange, C.A. MAP kinases couple multiple functions of human progesterone receptors: Degradation, transcriptional synergy, and nuclear association. J. Steroid Biochem. Mol. Biol. 2003, 85, 147–157. [Google Scholar] [CrossRef]
- Lange, C.A.; Shen, T.; Horwitz, K.B. Phosphorylation of human progesterone receptors at serine-294 by mitogen-activated protein kinase signals their degradation by the 26S proteasome. Proc. Natl. Acad. Sci. USA 2000, 97, 1032–1037. [Google Scholar] [CrossRef] [PubMed]
- Qiu, M.; Olsen, A.; Faivere, E.; Horwitz, K.B.; Lange, C.A. Mitogen-activated protein kinase regulates nuclear association of human progesterone receptors. Mol. Endocrinol. 2003, 17, 628–642. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ogiwara, K.; Takahashi, T. Nuclear Progestin Receptor Phosphorylation by Cdk9 Is Required for the Expression of Mmp15, a Protease Indispensable for Ovulation in Medaka. Cells 2019, 8, 215. https://doi.org/10.3390/cells8030215
Ogiwara K, Takahashi T. Nuclear Progestin Receptor Phosphorylation by Cdk9 Is Required for the Expression of Mmp15, a Protease Indispensable for Ovulation in Medaka. Cells. 2019; 8(3):215. https://doi.org/10.3390/cells8030215
Chicago/Turabian StyleOgiwara, Katsueki, and Takayuki Takahashi. 2019. "Nuclear Progestin Receptor Phosphorylation by Cdk9 Is Required for the Expression of Mmp15, a Protease Indispensable for Ovulation in Medaka" Cells 8, no. 3: 215. https://doi.org/10.3390/cells8030215
APA StyleOgiwara, K., & Takahashi, T. (2019). Nuclear Progestin Receptor Phosphorylation by Cdk9 Is Required for the Expression of Mmp15, a Protease Indispensable for Ovulation in Medaka. Cells, 8(3), 215. https://doi.org/10.3390/cells8030215



