Supramolecular Structures of the Dictyostelium Lamin NE81
Abstract
1. Introduction
2. Materials and Methods
2.1. Vector Constructions
2.2. Protein Purifications
2.3. Assembly Studies
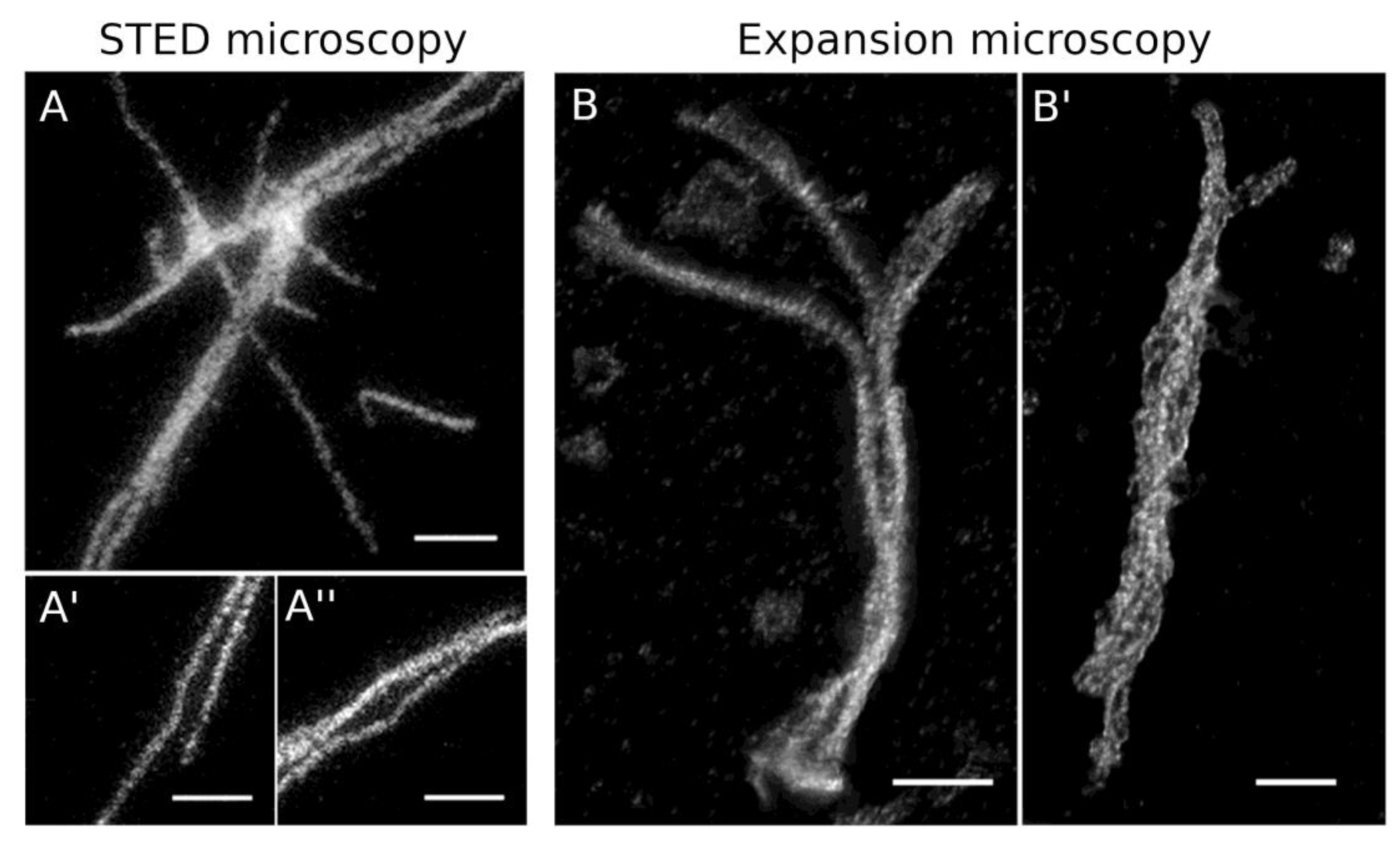
2.4. Light Microscopy
2.5. Transmission Electron Microscopy (TEM)
2.6. Field-Emission Scanning Electron Microscopy (feSEM) of Xenopus Oocyte Nuclear Membranes
2.7. Other Methods
2.8. Antibodies and Conjugates
3. Results
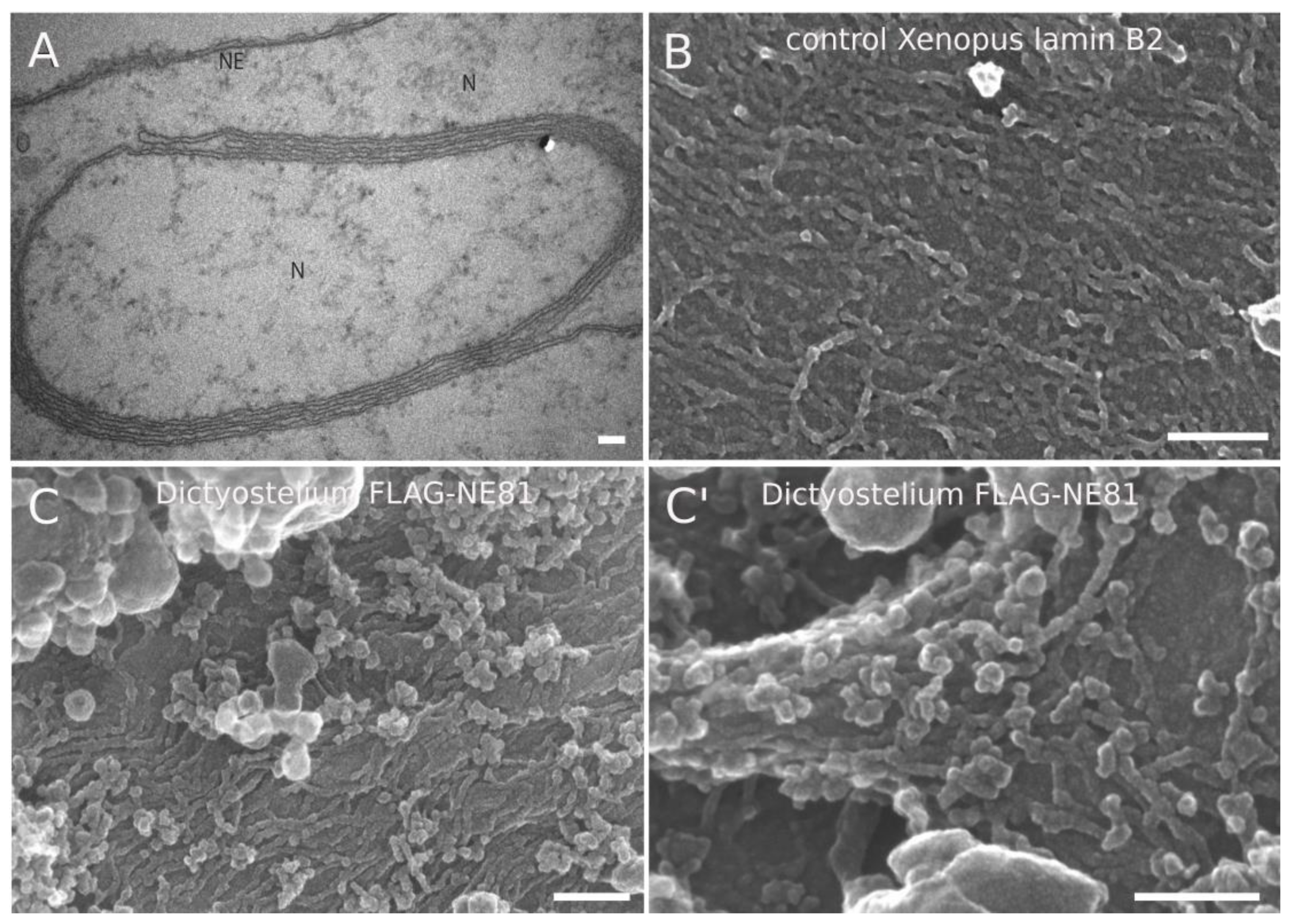
3.1. Field-Emission SEM Analysis of NE81 Assemblies at Xenopus Oocyte Nuclear Membranes
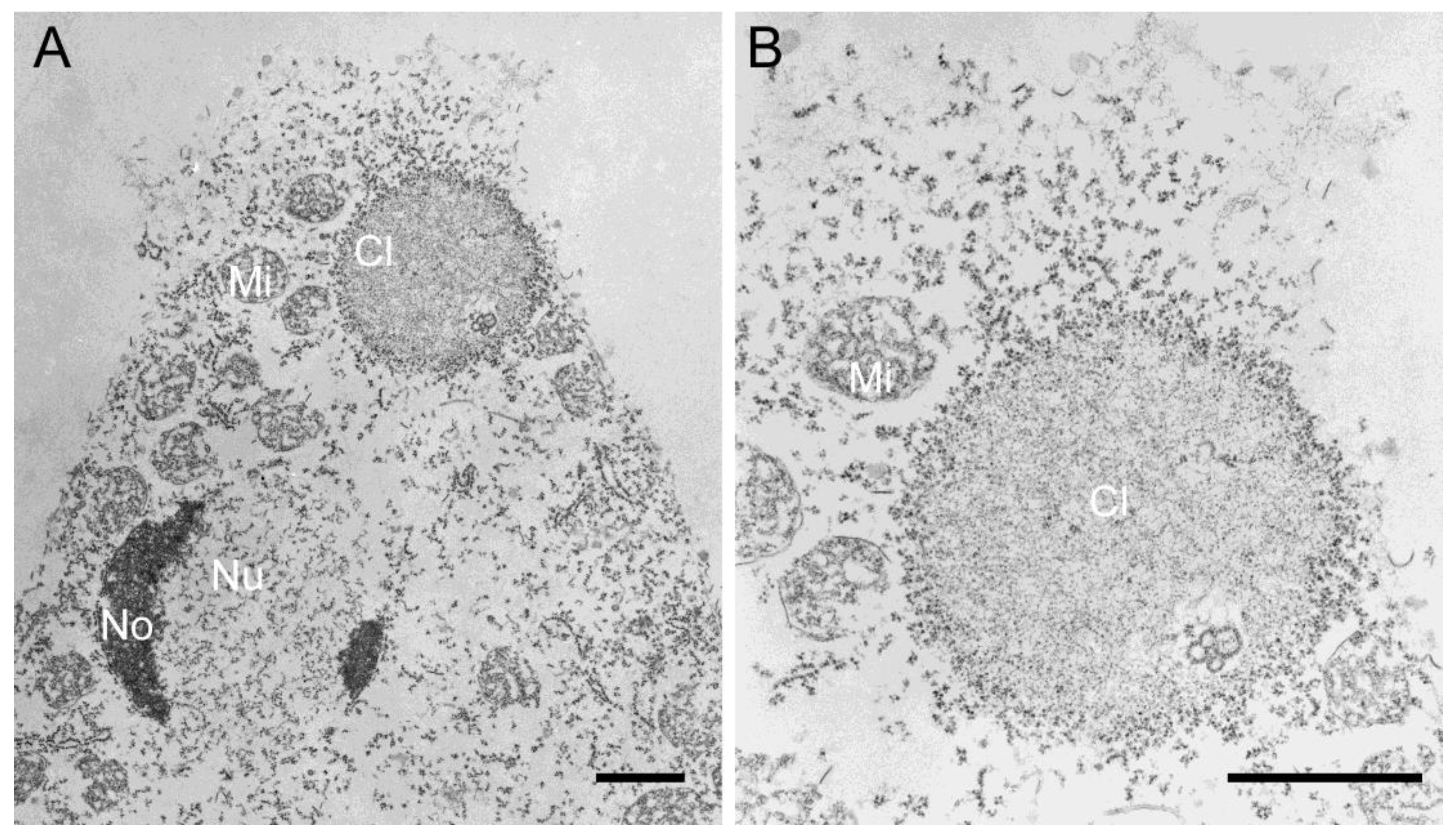
3.2. In Vitro Assembly of NE81 Expressed in Dictyostelium
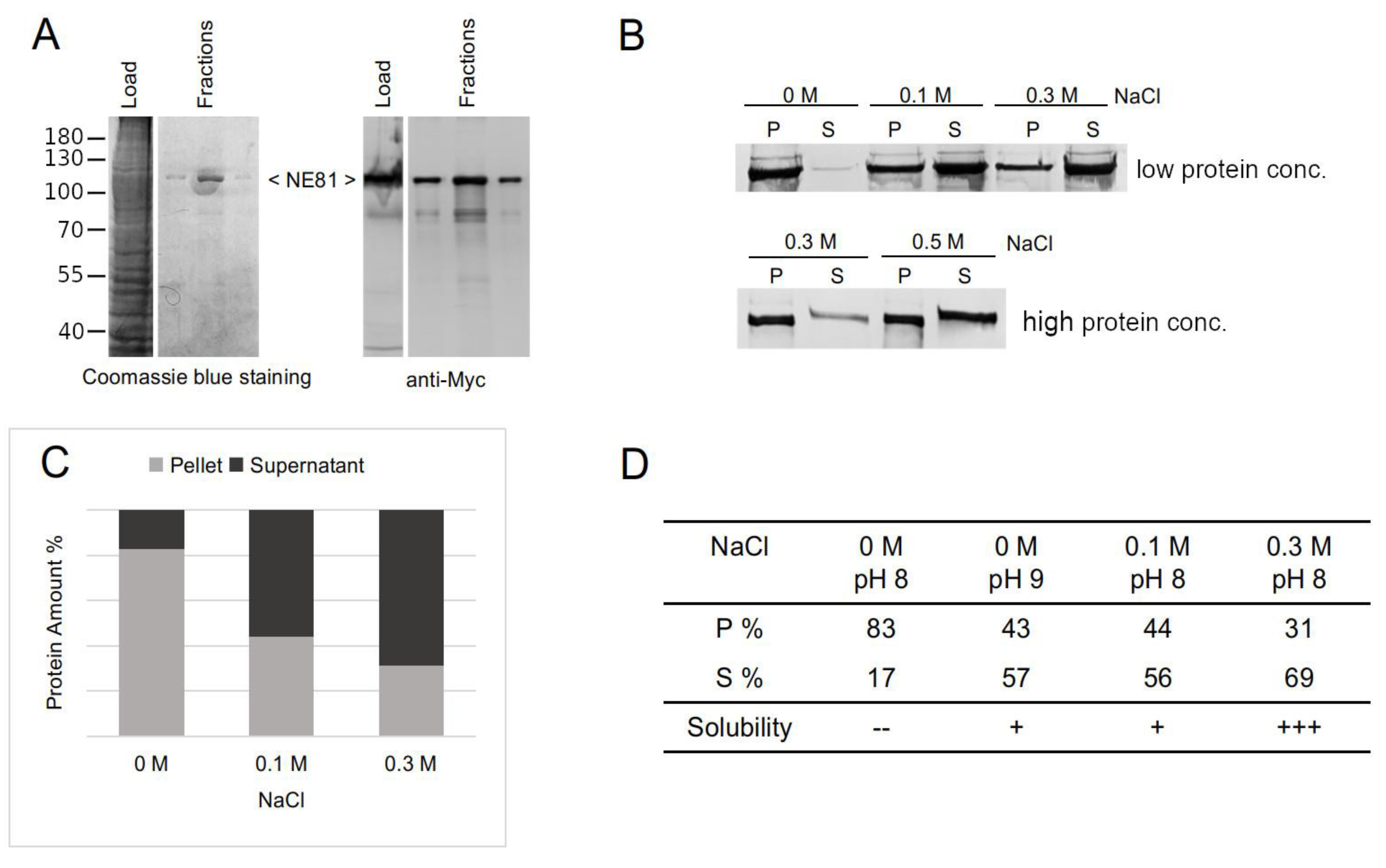
3.2.1. NE81 Lacking CaaX-box and NLS Is a Suitable Source to Study Protein Assembly
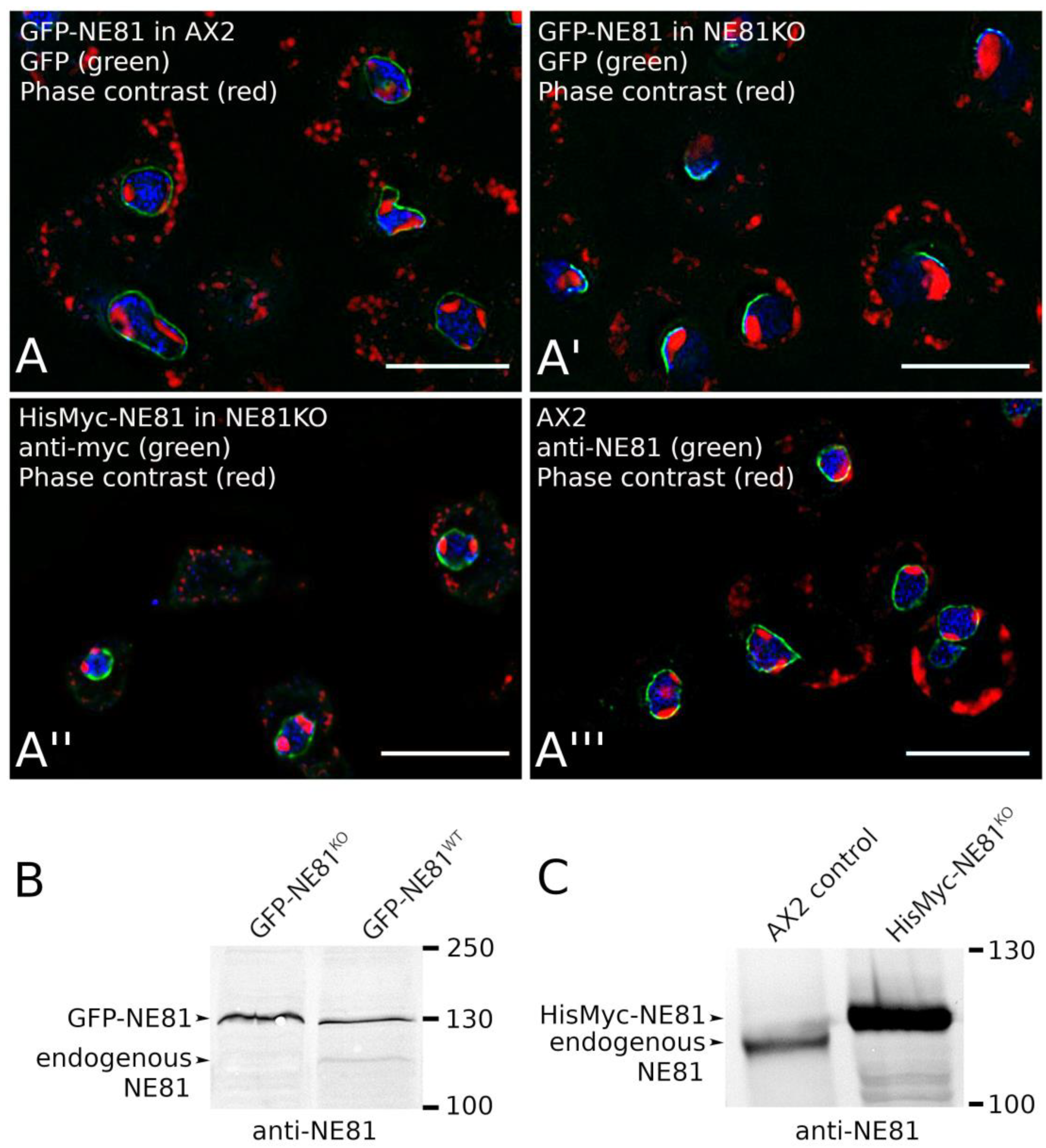
3.2.2. The GFP Tag, But Not the HisMyc-Tag, Interferes with NE81 Protein Assembly
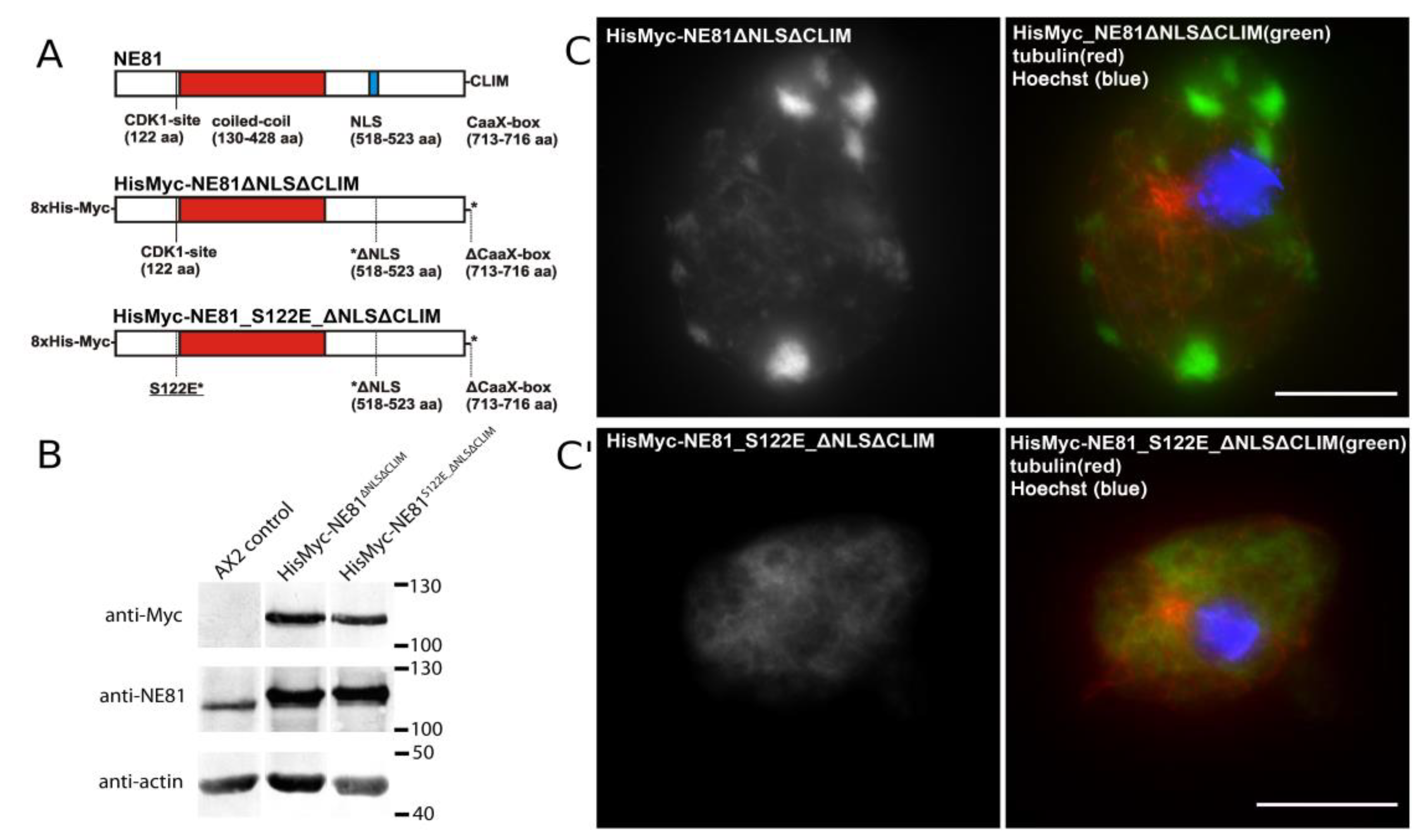
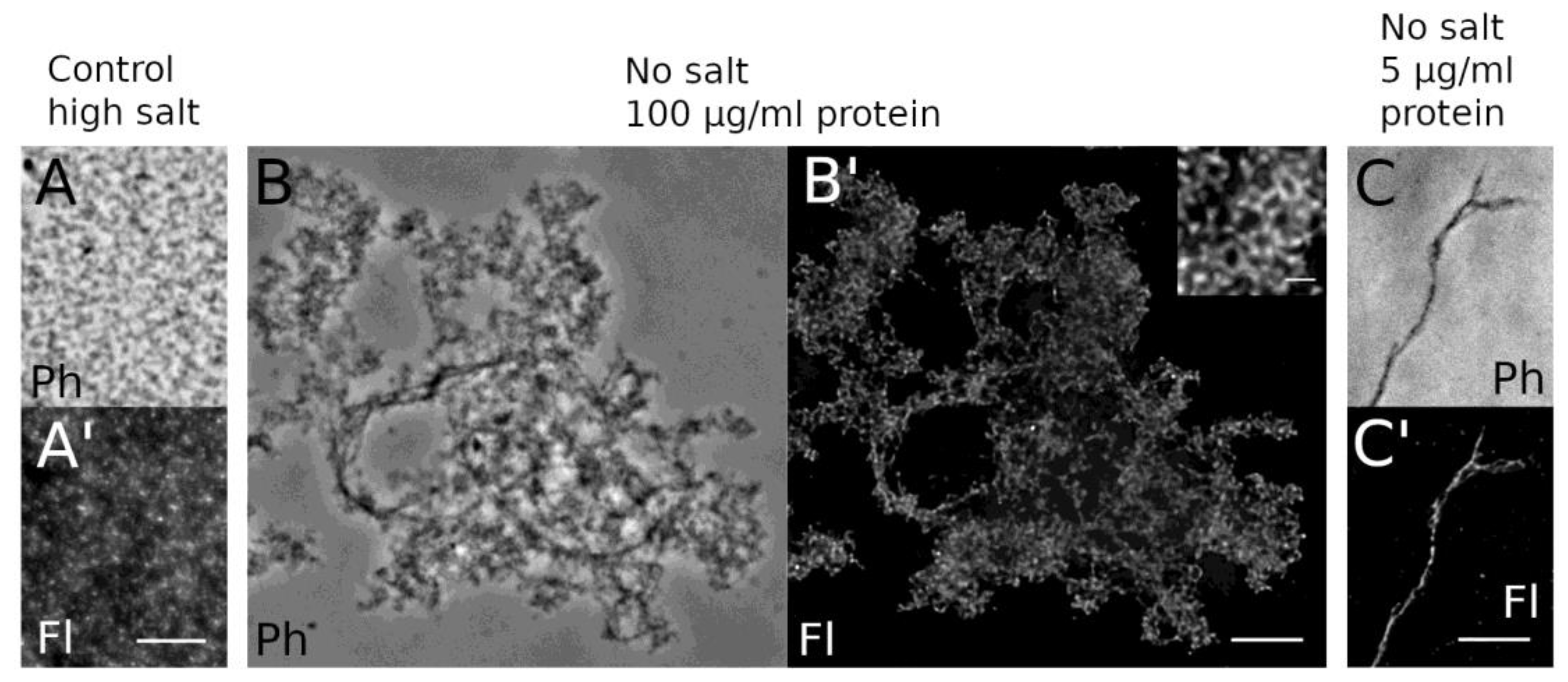
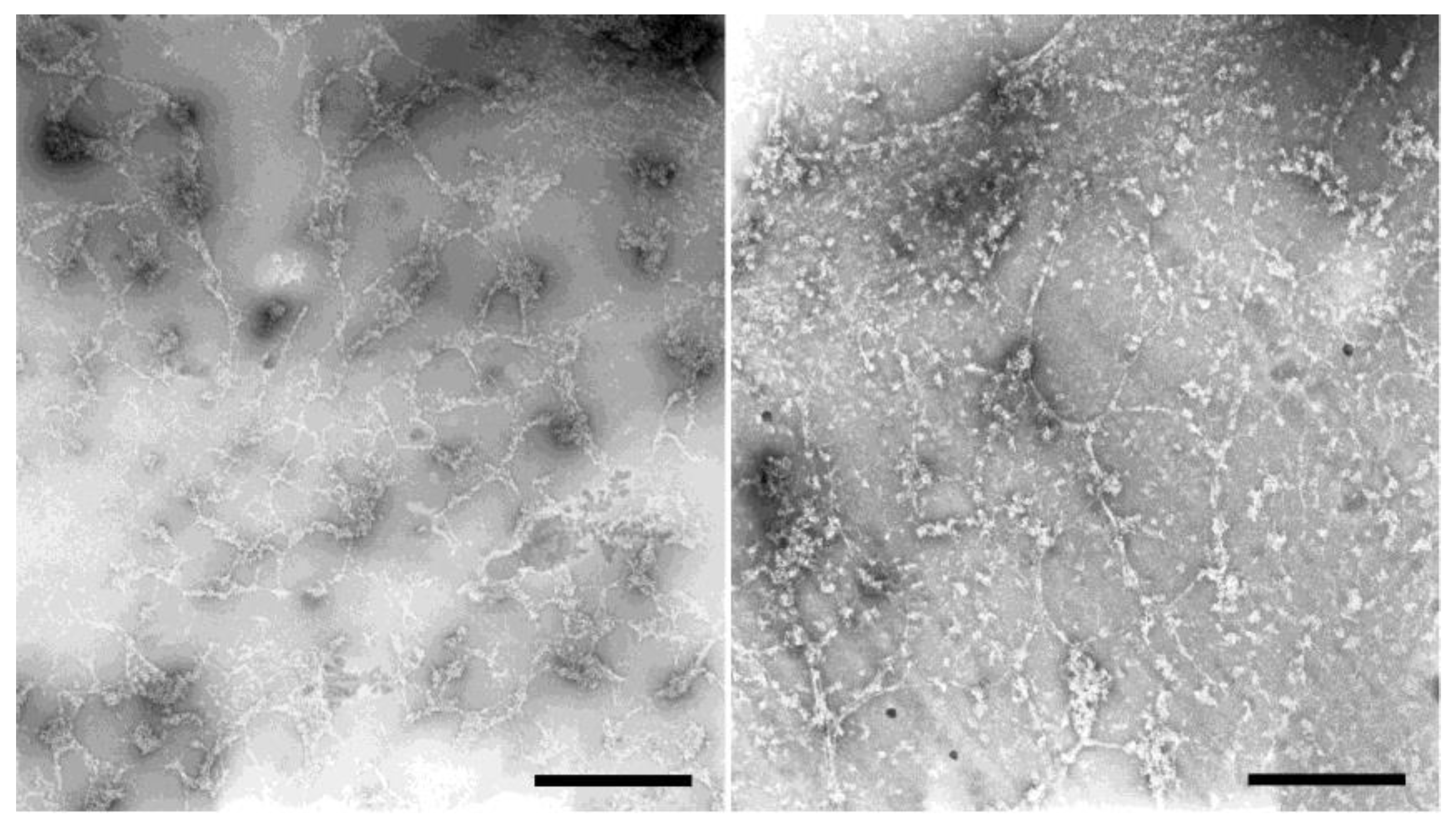
3.2.3. Isolation of HisMyc-NE81ΔNLSΔCLIM and In Vitro Assembly of Filaments
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Herrmann, H.; Bar, H.; Kreplak, L.; Strelkov, S.V.; Aebi, U. Intermediate filaments: From cell architecture to nanomechanics. Nat. Rev. Mol. Cell Biol. 2007, 8, 562–573. [Google Scholar] [CrossRef] [PubMed]
- Devos, D.P.; Gräf, R.; Field, M.C. Evolution of the nucleus. Curr. Opin. Cell Biol. 2014, 28C, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Crisp, M.; Liu, Q.; Roux, K.; Rattner, J.B.; Shanahan, C.; Burke, B.; Stahl, P.D.; Hodzic, D. Coupling of the nucleus and cytoplasm: Role of the LINC complex. J. Cell Biol. 2006, 172, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Van Bortle, K.; Corces, V.G. Spinning the web of cell fate. Cell 2013, 152, 1213–1217. [Google Scholar] [CrossRef]
- Dahl, K.N.; Kahn, S.M.; Wilson, K.L.; Discher, D.E. The nuclear envelope lamina network has elasticity and a compressibility limit suggestive of a molecular shock absorber. J. Cell Sci. 2004, 117, 4779–4786. [Google Scholar] [CrossRef] [PubMed]
- Tatli, M.; Medalia, O. Insight into the functional organization of nuclear lamins in health and disease. Curr. Opin. Cell Biol. 2018, 54, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Camozzi, D.; Capanni, C.; Cenni, V.; Mattioli, E.; Columbaro, M.; Squarzoni, S.; Lattanzi, G. Diverse lamin-dependent mechanisms interact to control chromatin dynamics: Focus on laminopathies. Nucleus 2014, 5, 427–440. [Google Scholar] [CrossRef] [PubMed]
- Discher, D.E.; Smith, L.; Cho, S.; Colasurdo, M.; Garcia, A.J.; Safran, S. Matrix Mechanosensing: From Scaling Concepts in ′Omics Data to Mechanisms in the Nucleus, Regeneration, and Cancer. Annu. Rev. Biophys. 2017, 46, 295–315. [Google Scholar] [CrossRef] [PubMed]
- Isermann, P.; Lammerding, J. Nuclear mechanics and mechanotransduction in health and disease. Curr. Biol. CB 2013, 23, R1113–R1121. [Google Scholar] [CrossRef]
- de Leeuw, R.; Gruenbaum, Y.; Medalia, O. Nuclear Lamins: Thin Filaments with Major Functions. Trends Cell Biol. 2017, 28, 34–45. [Google Scholar] [CrossRef]
- Peter, A.; Stick, R. Evolution of the lamin protein family: What introns can tell. Nucleus 2012, 3, 44–59. [Google Scholar] [CrossRef] [PubMed]
- Krüger, A.; Batsios, P.; Baumann, O.; Luckert, E.; Schwarz, H.; Stick, R.; Meyer, I.; Gräf, R. Characterization of NE81, the first lamin-like nucleoskeleton protein in a unicellular organism. Mol. Biol. Cell 2012, 23, 360–370. [Google Scholar] [CrossRef] [PubMed]
- Batsios, P.; Peter, T.; Baumann, O.; Stick, R.; Meyer, I.; Gräf, R. A lamin in lower eukaryotes? Nucleus 2012, 3, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Kollmar, M. Polyphyly of nuclear lamin genes indicates an early eukaryotic origin of the metazoan-type intermediate filament proteins. Sci. Rep. 2015, 5, 10652. [Google Scholar] [CrossRef] [PubMed]
- Preisner, H.; Habicht, J.; Garg, S.G.; Gould, S.B. Intermediate filament protein evolution and protists. Cytoskeleton 2018. [Google Scholar] [CrossRef]
- Koreny, L.; Field, M.C. Ancient Eukaryotic Origin and Evolutionary Plasticity of Nuclear Lamina. Genome Biol. Evol. 2016, 8, 2663–2671. [Google Scholar] [CrossRef]
- Gräf, R.; Batsios, P.; Meyer, I. Evolution of centrosomes and the nuclear lamina: Amoebozoan assets. Eur. J. Cell Biol. 2015, 94, 249–256. [Google Scholar] [CrossRef]
- Batsios, P.; Ren, X.; Baumann, O.; Larochelle, D.A.; Gräf, R. Src1 is a Protein of the Inner Nuclear Membrane Interacting with the Dictyostelium Lamin NE81. Cells 2016, 5, 13. [Google Scholar] [CrossRef]
- Adam, S.A.; Sengupta, K.; Goldman, R.D. Regulation of nuclear lamin polymerization by importin alpha. J. Biol. Chem. 2008, 283, 8462–8468. [Google Scholar] [CrossRef]
- Schulz, I.; Erle, A.; Gräf, R.; Krüger, A.; Lohmeier, H.; Putzler, S.; Samereier, M.; Weidenthaler, S. Identification and cell cycle-dependent localization of nine novel, genuine centrosomal components in Dictyostelium discoideum. Cell Motil. Cytoskelet. 2009, 66, 915–928. [Google Scholar] [CrossRef]
- Hofemeister, H.; Weber, K.; Stick, R. Association of prenylated proteins with the plasma membrane and the inner nuclear membrane is mediated by the same membrane-targeting motifs. Mol. Biol. Cell 2000, 11, 3233–3246. [Google Scholar] [CrossRef]
- Batsios, P.; Meyer, I.; Gräf, R. Proximity-Dependent Biotin Identification (BioID) in Dictyostelium Amoebae. Methods Enzym. 2016, 569, 23–42. [Google Scholar]
- Gräf, R.; Euteneuer, U.; Ueda, M.; Schliwa, M. Isolation of nucleation-competent centrosomes from Dictyostelium discoideum. Eur. J. Cell Biol. 1998, 76, 167–175. [Google Scholar] [CrossRef]
- Schneider, C.A. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671. [Google Scholar] [CrossRef] [PubMed]
- Batsios, P.; Baumann, O.; Gräf, R.; Meyer, I. Isolation of Dictyostelium nuclei for light and electron microscopy. Methods Mol. Biol. 2013, 983, 283–294. [Google Scholar] [PubMed]
- Kuhnert, O.; Baumann, O.; Meyer, I.; Gräf, R. Functional characterization of CP148, a novel key component for centrosome integrity in Dictyostelium. Cell Mol. Life Sci. 2012, 69, 1875–1888. [Google Scholar] [CrossRef]
- Klauss, A.; König, M.; Hille, C. Upgrade of a Scanning Confocal Microscope to a Single-Beam Path STED Microscope. PLoS ONE 2015, 10, e0130717. [Google Scholar] [CrossRef]
- Klauss, A.; Conrad, F.; Hille, C. Binary phase masks for easy system alignment and basic aberration sensing with spatial light modulators in STED microscopy. Sci. Rep. 2017, 7, 15699. [Google Scholar] [CrossRef]
- Tillberg, P.W.; Chen, F.; Piatkevich, K.D.; Zhao, Y.; Yu, C.C.; English, B.P.; Gao, L.; Martorell, A.; Suk, H.J.; Yoshida, F.; et al. Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nat. Biotechnol. 2016, 34, 987–992. [Google Scholar] [CrossRef]
- Chozinski, T.J.; Halpern, A.R.; Okawa, H.; Kim, H.J.; Tremel, G.J.; Wong, R.O.; Vaughan, J.C. Expansion microscopy with conventional antibodies and fluorescent proteins. Nat. Methods 2016, 13, 485–488. [Google Scholar] [CrossRef]
- Goldberg, M.W.; Huttenlauch, I.; Hutchison, C.J.; Stick, R. Filaments made from A- and B-type lamins differ in structure and organization. J. Cell Sci. 2008, 121, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Sive, H.L.; Grainger, R.M.; Harland, R.M. Defolliculation of Xenopus oocytes. Cold Spring Harb. Protoc. 2010, 2010. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, M.W.; Allen, T.D. High resolution scanning electron microscopy of the nuclear envelope: Demonstration of a new, regular, fibrous lattice attached to the baskets of the nucleoplasmic face of the nuclear pores. J. Cell Biol. 1992, 119, 1429–1440. [Google Scholar] [CrossRef] [PubMed]
- Gräf, R.; Euteneuer, U.; Ho, T.H.; Rehberg, M. Regulated Expression of the Centrosomal Protein DdCP224 Affects Microtubule Dynamics and Reveals Mechanisms for the Control of Supernumerary Centrosome Number. Mol. Biol. Cell 2003, 14, 4067–4074. [Google Scholar] [CrossRef] [PubMed]
- Wehland, J.; Willingham, M.C.; Sandoval, I.V. A rat monoclonal antibody reacting specifically with the tyrosylated form of alpha-tubulin. II. Effects on cell movement, organization of microtubules, and intermediate filaments, and arrangement of Golgi elements. J. Cell Biol. 1983, 97, 1476–1490. [Google Scholar] [CrossRef]
- Evan, G.I.; Lewis, G.K.; Ramsay, G.; Bishop, J.M. Isolation of monoclonal antibodies specific for human c-myc proto- oncogene product. Mol. Cell Biol. 1985, 5, 3610–3616. [Google Scholar] [CrossRef] [PubMed]
- Westphal, M.; Jungbluth, A.; Heidecker, M.; Mühlbauer, B.; Heizer, C.; Schwartz, J.M.; Marriott, G.; Gerisch, G. Microfilament dynamics during cell movement and chemotaxis monitored using a GFP-actin fusion protein. Curr. Biol. 1997, 7, 176–183. [Google Scholar] [CrossRef]
- Goldberg, M.W.; Fiserova, J.; Huttenlauch, I.; Stick, R. A new model for nuclear lamina organization. Biochem. Soc. Trans. 2008, 36, 1339–1343. [Google Scholar] [CrossRef]
- Linde, N.; Stick, R. Intranuclear membranes induced by lipidated proteins are derived from the nuclear envelope. Nucleus 2010, 1, 343–353. [Google Scholar] [CrossRef]
- Foeger, N.; Wiesel, N.; Lotsch, D.; Mucke, N.; Kreplak, L.; Aebi, U.; Gruenbaum, Y.; Herrmann, H. Solubility properties and specific assembly pathways of the B-type lamin from Caenorhabditis elegans. J. Struct. Biol. 2006, 155, 340–350. [Google Scholar] [CrossRef]
- Makarov, A.A.; Rizzotto, A.; Meinke, P.; Schirmer, E.C. Purification of Lamins and Soluble Fragments of NETs. Methods Enzym. 2016, 569, 79–100. [Google Scholar]
- Kreplak, L.; Richter, K.; Aebi, U.; Herrmann, H. Electron microscopy of intermediate filaments: Teaming up with atomic force and confocal laser scanning microscopy. Methods Cell Biol. 2008, 88, 273–297. [Google Scholar] [PubMed]
- Putzler, S.; Meyer, I.; Gräf, R. CP91 is a component of the Dictyostelium centrosome involved in centrosome biogenesis. Eur. J. Cell Biol. 2016, 95, 124–135. [Google Scholar] [CrossRef] [PubMed]
- Aebi, U.; Cohn, J.; Buhle, L.; Gerace, L. The nuclear lamina is a meshwork of intermediate-type filaments. Nature 1986, 323, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Karabinos, A.; Schunemann, J.; Meyer, M.; Aebi, U.; Weber, K. The single nuclear lamin of Caenorhabditis elegans forms in vitro stable intermediate filaments and paracrystals with a reduced axial periodicity. J. Mol. Biol. 2003, 325, 241–247. [Google Scholar] [CrossRef]
- Pitzen, V.; Askarzada, S.; Gräf, R.; Meyer, I. CDK5RAP2 Is an Essential Scaffolding Protein of the Corona of the Dictyostelium Centrosome. Cells 2018, 7. [Google Scholar] [CrossRef]
- Grossman, E.; Dahan, I.; Stick, R.; Goldberg, M.W.; Gruenbaum, Y.; Medalia, O. Filaments assembly of ectopically expressed Caenorhabditis elegans lamin within Xenopus oocytes. J. Struct. Biol. 2012, 177, 113–118. [Google Scholar] [CrossRef]
- Shimi, T.; Kittisopikul, M.; Tran, J.; Goldman, A.E.; Adam, S.A.; Zheng, Y.; Jaqaman, K.; Goldman, R.D. Structural Organization of Nuclear Lamins A, C, B1 and B2 Revealed by Super-Resolution Microscopy. Mol. Biol. Cell 2015, 26, 4075–4086. [Google Scholar] [CrossRef]
- Xie, W.; Chojnowski, A.; Boudier, T.; Lim, J.S.; Ahmed, S.; Ser, Z.; Stewart, C.; Burke, B. A-type Lamins Form Distinct Filamentous Networks with Differential Nuclear Pore Complex Associations. Curr. Biol. 2016, 26, 2651–2658. [Google Scholar] [CrossRef]
- Xie, W.; Burke, B. Nuclear networking. Nucleus 2017, 8, 323–330. [Google Scholar] [CrossRef]
- Bhattacharjee, P.; Dasgupta, D.; Sengupta, K. DCM associated LMNA mutations cause distortions in lamina structure and assembly. Biochim. Biophys. Acta 2017, 1861, 2598–2608. [Google Scholar] [CrossRef] [PubMed]
- Turgay, Y.; Eibauer, M.; Goldman, A.E.; Shimi, T.; Khayat, M.; Ben-Harush, K.; Dubrovsky-Gaupp, A.; Sapra, K.T.; Goldman, R.D.; Medalia, O. The molecular architecture of lamins in somatic cells. Nature 2017, 543, 261–264. [Google Scholar] [CrossRef]
- Turgay, Y.; Medalia, O. The structure of lamin filaments in somatic cells as revealed by cryo-electron tomography. Nucleus 2017, 8, 475–481. [Google Scholar] [CrossRef] [PubMed]
- Ben-Harush, K.; Wiesel, N.; Frenkiel-Krispin, D.; Moeller, D.; Soreq, E.; Aebi, U.; Herrmann, H.; Gruenbaum, Y.; Medalia, O. The supramolecular organization of the C. elegans nuclear lamin filament. J. Mol. Biol. 2009, 386, 1392–1402. [Google Scholar] [CrossRef] [PubMed]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef] [PubMed]
- Schulz, I.; Baumann, O.; Samereier, M.; Zoglmeier, C.; Gräf, R. Dictyostelium Sun1 is a dynamic membrane protein of both nuclear membranes and required for centrosomal association with clustered centromeres. Eur. J. Cell Biol. 2009, 88, 621–638. [Google Scholar] [CrossRef] [PubMed]
- Tikhonenko, I.; Magidson, V.; Gräf, R.; Khodjakov, A.; Koonce, M.P. A kinesin-mediated mechanism that couples centrosomes to nuclei. Cell Mol. Life Sci. 2013, 70, 1285–1296. [Google Scholar] [CrossRef]
- Beck, M.; Medalia, O. Structural and functional insights into nucleocytoplasmic transport. Histol. Histopathol. 2008, 23, 1025–1033. [Google Scholar]
- López-Jiménez, A.T.; Cardenal-Muñoz, E.; Leuba, F.; Gerstenmaier, L.; Barisch, C.; Hagedorn, M.; King, J.S.; Soldati, T. The ESCRT and autophagy machineries cooperate to repair ESX-1-dependent damage at the Mycobacterium-containing vacuole but have opposite impact on containing the infection. PLoS Pathog. 2018, 14, e1007501. [Google Scholar] [CrossRef]
- Blanc, C.; Charette, S.J.; Mattei, S.; Aubry, L.; Smith, E.W.; Cosson, P.; Letourneur, F. Dictyostelium Tom1 participates to an ancestral ESCRT-0 complex. Traffic Cph. Den. 2009, 10, 161–171. [Google Scholar] [CrossRef]
- Mattei, S.; Klein, G.; Satre, M.; Aubry, L. Trafficking and developmental signaling: Alix at the crossroads. Eur. J. Cell Biol. 2006, 85, 925–936. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grafe, M.; Batsios, P.; Meyer, I.; Lisin, D.; Baumann, O.; Goldberg, M.W.; Gräf, R. Supramolecular Structures of the Dictyostelium Lamin NE81. Cells 2019, 8, 162. https://doi.org/10.3390/cells8020162
Grafe M, Batsios P, Meyer I, Lisin D, Baumann O, Goldberg MW, Gräf R. Supramolecular Structures of the Dictyostelium Lamin NE81. Cells. 2019; 8(2):162. https://doi.org/10.3390/cells8020162
Chicago/Turabian StyleGrafe, Marianne, Petros Batsios, Irene Meyer, Daria Lisin, Otto Baumann, Martin W. Goldberg, and Ralph Gräf. 2019. "Supramolecular Structures of the Dictyostelium Lamin NE81" Cells 8, no. 2: 162. https://doi.org/10.3390/cells8020162
APA StyleGrafe, M., Batsios, P., Meyer, I., Lisin, D., Baumann, O., Goldberg, M. W., & Gräf, R. (2019). Supramolecular Structures of the Dictyostelium Lamin NE81. Cells, 8(2), 162. https://doi.org/10.3390/cells8020162





