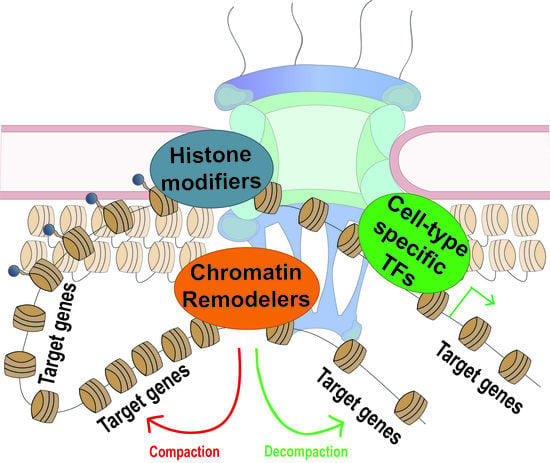
Nuclear Pore Proteins in Regulation of Chromatin State
Abstract
1. Introduction
2. NPCs, Nups, and Chromatin Structure
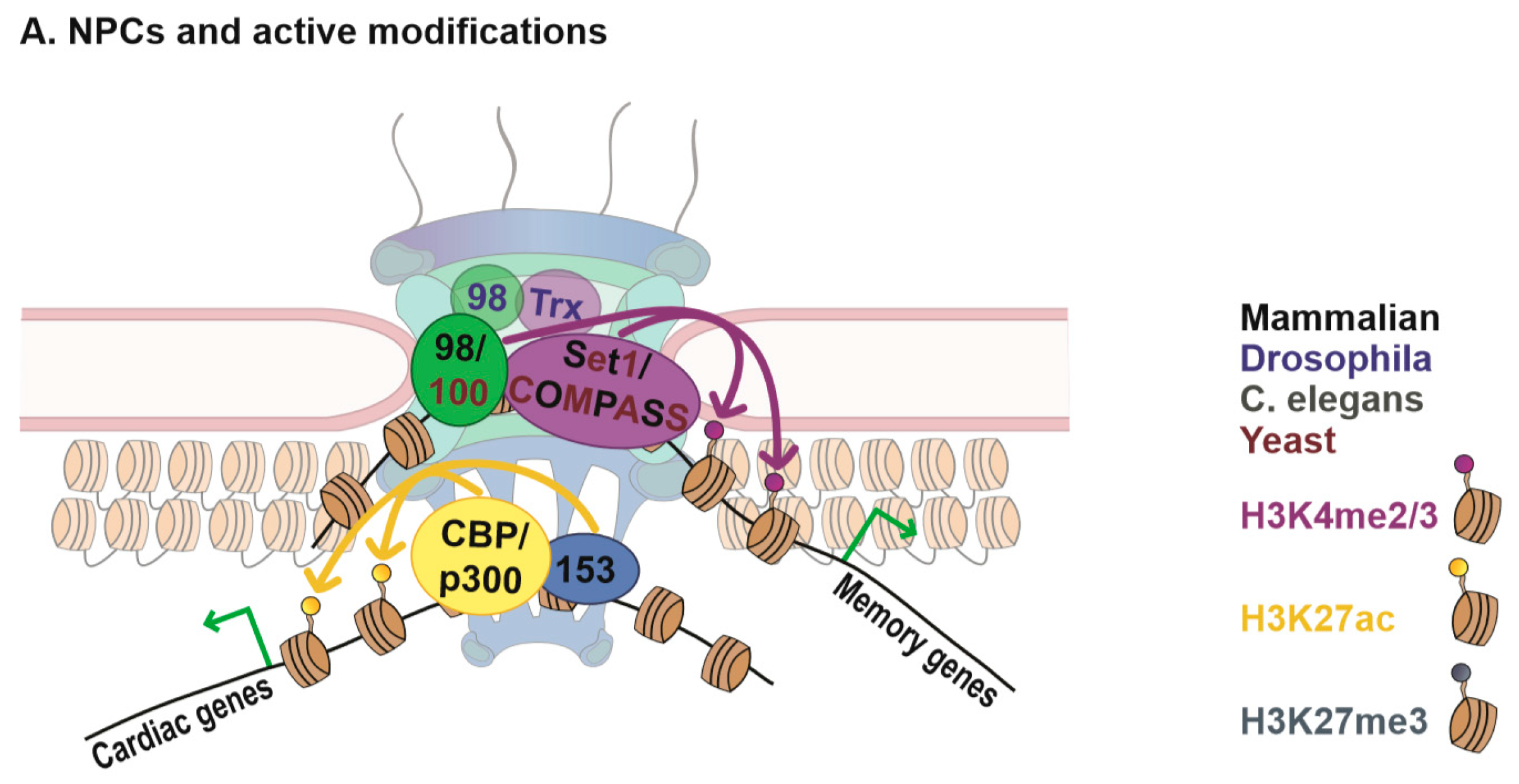
2.1. Nups and Histone Modifications
2.1.1. Nup98 and Methylation of Histone H3 Lysine K4
2.1.2. Nups and Histone Acetylation
2.1.3. High Resolution Imaging of Chromatin at Nuclear Pores
2.1.4. Activation and Repression Dichotomy–Nup155/Nup170p and Chromatin Silencing
2.1.5. Nups and Polycomb Repression
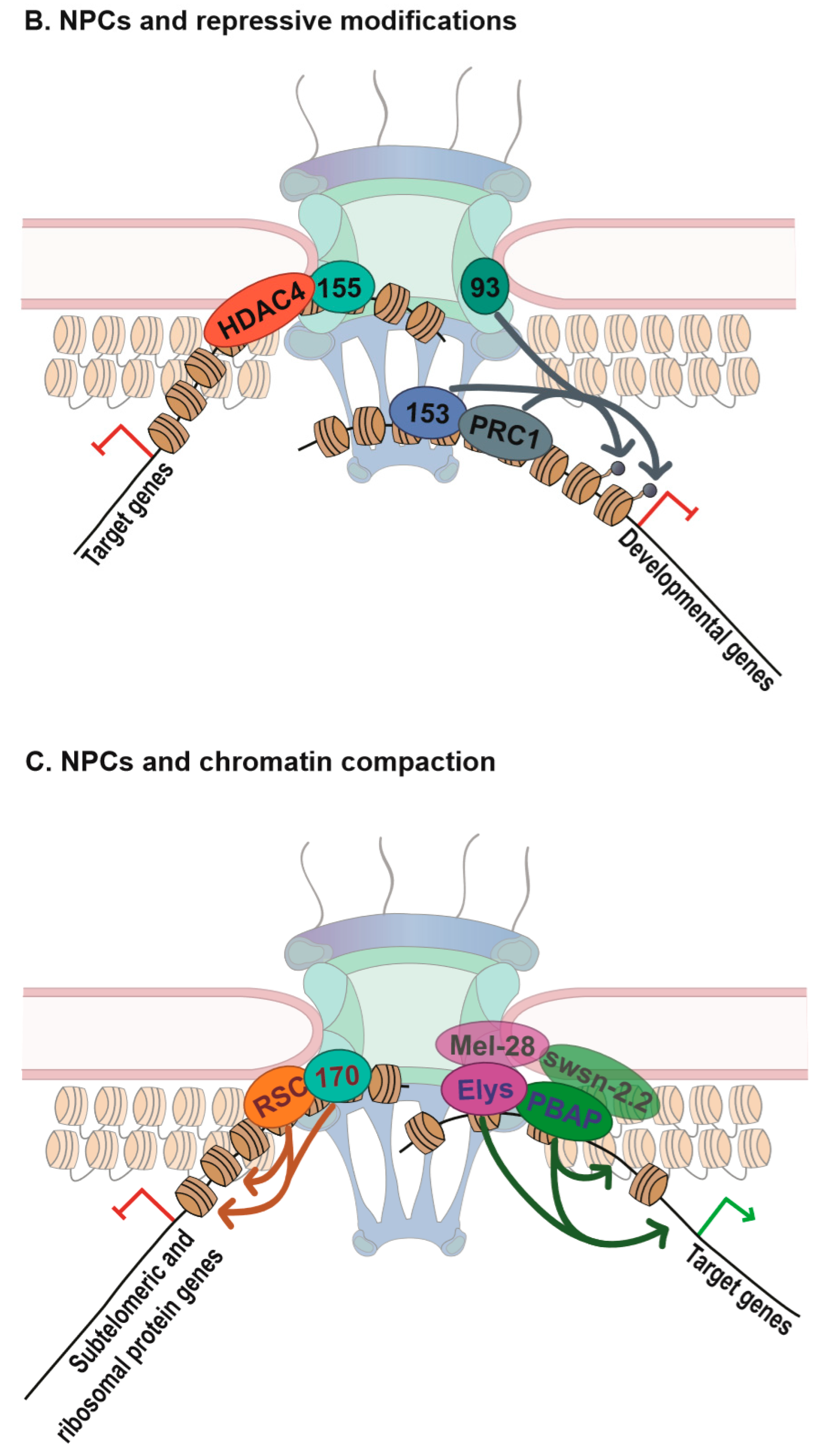
2.2. Nups and Chromatin Remodeling
2.2.1. Nup170p and RSC
2.2.2. Elys and Chromatin Remodeling
2.3. Nups and Large-Scale Chromatin Structure
2.3.1. Nups in Dosage Compensation
2.3.2. Mtor/TPR and Nuclear Organization
2.3.3. Nups and Transposon Silencing
3. NPCs and Transcription
3.1. Nup98 and Hox Genes
3.2. Nup153, Nup93, and Regulation of Cell Identity
3.3. Nups and Transcription Factors
4. Concluding Remarks and Future Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Watson, M.L. Further Observations on the Nuclear Envelope of the Animal Cell. J. Cell Biol. 1959, 6, 147–156. [Google Scholar] [CrossRef]
- Weintraub, H.; Groudine, M. Chromosomal subunits in active genes have an altered conformation. Science 1976, 193, 848–856. [Google Scholar] [CrossRef]
- Hutchison, N.; Weintraub, H. Localization of DNAase I-sensitive sequences to specific regions of interphase nuclei. Cell 1985, 43, 471–482. [Google Scholar] [CrossRef]
- Blobel, G. Gene gating: A hypothesis. Proc. Natl. Acad. Sci. USA 1985, 82, 8527–8529. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, M.A. Nuclear pore complexes as hubs for gene regulation. Nucleus 2018, 9, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Ptak, C.; Wozniak, R.W. Nucleoporins and chromatin metabolism. Curr. Opin. Cell Biol. 2016, 40, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Ibarra, A.; Hetzer, M.W. Nuclear pore proteins and the control of genome functions. Genes Dev. 2015, 29, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Géli, V.; Lisby, M. Recombinational DNA repair is regulated by compartmentalization of DNA lesions at the nuclear pore complex. BioEssays 2015, 37, 1287–1292. [Google Scholar] [CrossRef] [PubMed]
- Pascual-Garcia, P.; Capelson, M. Nuclear pores as versatile platforms for gene regulation. Curr. Opin. Genet. Dev. 2014, 25, 110–117. [Google Scholar] [CrossRef]
- Bukata, L.; Parker, S.L.; Angelo, M.A.D. Nuclear pore complexes in the maintenance of genome integrity. Curr. Opin. Cell Biol. 2013, 25, 378–386. [Google Scholar] [CrossRef]
- Capelson, M.; Liang, Y.; Schulte, R.; Mair, W.; Wagner, U.; Hetzer, M.W. Chromatin-Bound Nuclear Pore Components Regulate Gene Expression in Higher Eukaryotes. Cell 2010, 140, 372–383. [Google Scholar] [CrossRef] [PubMed]
- Kalverda, B.; Pickersgill, H.; Shloma, V.V.; Fornerod, M. Nucleoporins Directly Stimulate Expression of Developmental and Cell-Cycle Genes Inside the Nucleoplasm. Cell 2010, 140, 360–371. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Franks, T.M.; Marchetto, M.C.; Gage, F.H.; Hetzer, M.W. Dynamic Association of NUP98 with the Human Genome. PLoS Genet. 2013, 9, e1003308. [Google Scholar] [CrossRef] [PubMed]
- Light, W.H.; Freaney, J.; Sood, V.; Thompson, A.; D’Urso, A.; Horvath, C.M.; Brickner, J.H. A Conserved Role for Human Nup98 in Altering Chromatin Structure and Promoting Epigenetic Transcriptional Memory. PLoS Biol. 2013, 11, e1001524. [Google Scholar] [CrossRef]
- Griffis, E.R.; Craige, B.; Dimaano, C.; Ullman, K.S.; Powers, M.A. Distinct Functional Domains within Nucleoporins Nup153 and Nup98 Mediate Transcription-dependent Mobility. Mol. Biol. Cell 2004, 15, 1991–2002. [Google Scholar] [CrossRef]
- Rabut, G.; Doye, V.; Ellenberg, J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat. Cell Biol. 2004, 6, 1114–1121. [Google Scholar] [CrossRef]
- Tolsma, T.O.; Hansen, J.C. Post-translational modifications and chromatin dynamics. Essays Biochem. 2019, 63, 89–96. [Google Scholar]
- D’Urso, A.; Brickner, J.H. Mechanisms of epigenetic memory. Trends Genet. 2014, 30, 230–236. [Google Scholar] [CrossRef]
- D’Urso, A.; Brickner, J.H. Epigenetic transcriptional memory. Curr. Genet. 2017, 63, 435–439. [Google Scholar] [CrossRef]
- Bernstein, B.E.; Kamal, M.; Lindblad-Toh, K.; Bekiranov, S.; Bailey, D.K.; Huebert, D.J.; McMahon, S.; Karlsson, E.K.; Kulbokas, E.J.; Gingeras, T.R.; et al. Genomic Maps and Comparative Analysis of Histone Modifications in Human and Mouse. Cell 2005, 120, 169–181. [Google Scholar] [CrossRef]
- Koch, C.M.; Andrews, R.M.; Flicek, P.; Dillon, S.C.; Karaoz, U.; Clelland, G.K.; Wilcox, S.; Beare, D.M.; Fowler, J.C.; Couttet, P.; et al. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res. 2007, 17, 691–707. [Google Scholar] [CrossRef] [PubMed]
- Franks, T.M.; McCloskey, A.; Shokirev, M.N.; Benner, C.; Rathore, A.; Hetzer, M.W. Nup98 recruits the Wdr82-Set1A/COMPASS complex to promoters to regulate H3K4 trimethylation in hematopoietic progenitor cells. Genes Dev. 2017, 31, 2222–2234. [Google Scholar] [CrossRef] [PubMed]
- Shilatifard, A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem. 2012, 81, 65–95. [Google Scholar] [CrossRef] [PubMed]
- Pascual-Garcia, P.; Debo, B.; Aleman, J.R.; Nguyen, N.H.; Won, K.J.; Capelson Correspondence, M. Metazoan Nuclear Pores Provide a Scaffold for Poised Genes and Mediate Induced Enhancer-Promoter Contacts. Mol. Cell 2017, 66, 63–76. [Google Scholar] [CrossRef]
- Pascual-Garcia, P.; Capelson, M. Nuclear pores in genome architecture and enhancer function. Curr. Opin. Cell Biol. 2019, 58, 126–133. [Google Scholar] [CrossRef]
- Vaquerizas, J.M.; Suyama, R.; Kind, J.; Miura, K.; Luscombe, N.M. Nuclear Pore Proteins Nup153 and Megator Define Transcriptionally Active Regions in the Drosophila Genome. PLoS Genet. 2010, 6, 1000846. [Google Scholar] [CrossRef]
- Dancy, B.M.; Cole, P.A. Protein lysine acetylation by p300/CBP. Chem. Rev. 2015, 115, 2419–2452. [Google Scholar] [CrossRef]
- Jin, Q.; Yu, L.-R.; Wang, L.; Zhang, Z.; Kasper, L.H.; Lee, J.-E.; Wang, C.; Brindle, P.K.; Dent, S.Y.R.; Ge, K. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011, 30, 249–262. [Google Scholar] [CrossRef]
- Creyghton, M.P.; Cheng, A.W.; Welstead, G.G.; Kooistra, T.; Carey, B.W.; Steine, E.J.; Hanna, J.; Lodato, M.A.; Frampton, G.M.; Sharp, P.A.; et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 2010, 107, 21931–21936. [Google Scholar] [CrossRef]
- Nanni, S.; Re, A.; Ripoli, C.; Gowran, A.; Nigro, P.; D’Amario, D.; Amodeo, A.; Crea, F.; Grassi, C.; Pontecorvi, A.; et al. The nuclear pore protein Nup153 associates with chromatin and regulates cardiac gene expression in dystrophic mdx hearts. Cardiovasc. Res. 2016, 112, 555–567. [Google Scholar] [CrossRef]
- Kasper, L.H.; Brindle, P.K.; Schnabel, C.A.; Pritchard, C.E.J.; Cleary, M.L.; van Deursen, J.M.A. CREB Binding Protein Interacts with Nucleoporin-Specific FG Repeats That Activate Transcription and Mediate NUP98-HOXA9 Oncogenicity. Mol. Cell. Biol. 1999, 19, 764–776. [Google Scholar] [CrossRef] [PubMed]
- Ibarra, A.; Benner, C.; Tyagi, S.; Cool, J.; Hetzer, M.W. Nucleoporin-mediated regulation of cell identity genes. Genes Dev. 2016, 30, 2253–2258. [Google Scholar] [CrossRef] [PubMed]
- Fišerová, J.; Efenberková, M.; Sieger, T.; Maninová, M.; Uhlířová, J.; Hozák, P. Chromatin organization at the nuclear periphery as revealed by image analysis of structured illumination microscopy data. J. Cell Sci. 2017, 130, 2066–2077. [Google Scholar] [CrossRef] [PubMed]
- Kehat, I.; Accornero, F.; Aronow, B.J.; Molkentin, J.D. Modulation of chromatin position and gene expression by HDAC4 interaction with nucleoporins. J. Cell Biol. 2011, 193, 21–29. [Google Scholar] [CrossRef]
- Seto, E.; Yoshida, M. Erasers of histone acetylation: The histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol. 2014, 6. [Google Scholar] [CrossRef]
- Van de Vosse, D.W.; Wan, Y.; Lapetina, D.L.; Chen, W.-M.; Chiang, J.-H.; Aitchison, J.D.; Wozniak, R.W. A Role for the Nucleoporin Nup170p in Chromatin Structure and Gene Silencing. Cell 2013, 152, 969–983. [Google Scholar] [CrossRef]
- Xu, F.; Zhang, Q.; Zhang, K.; Xie, W.; Grunstein, M. Sir2 deacetylates histone H3 lysine 56 to regulate telomeric heterochromatin structure in yeast. Mol. Cell 2007, 27, 890–900. [Google Scholar] [CrossRef]
- Brickner, J. Genetic and epigenetic control of the spatial organization of the genome. Mol. Biol. Cell 2017, 28, 364–369. [Google Scholar] [CrossRef]
- Lapetina, D.L.; Ptak, C.; Roesner, U.K.; Wozniak, R.W. Yeast silencing factor Sir4 and a subset of nucleoporins form a complex distinct from nuclear pore complexes. J. Cell Biol. 2017, 216, 3145–3159. [Google Scholar] [CrossRef]
- Jacinto, F.V.; Benner, C.; Hetzer, M.W. The nucleoporin Nup153 regulates embryonic stem cell pluripotency through gene silencing. Genes Dev. 2015, 29, 1224–1238. [Google Scholar] [CrossRef]
- Sachani, S.S.; Landschoot, L.S.; Zhang, L.; White, C.R.; MacDonald, W.A.; Golding, M.C.; Mann, M.R.W. Nucleoporin 107, 62 and 153 mediate Kcnq1ot1 imprinted domain regulation in extraembryonic endoderm stem cells. Nat. Commun. 2018, 9, 2795. [Google Scholar] [CrossRef] [PubMed]
- Labade, A.S.; Karmodiya, K.; Sengupta, K. HOXA repression is mediated by nucleoporin Nup93 assisted by its interactors Nup188 and Nup205. Epigenet. Chromatin 2016, 9. [Google Scholar] [CrossRef] [PubMed]
- Pascual-Garcia, P.; Jeong, J.; Capelson, M. Nucleoporin Nup98 Associates with Trx/MLL and NSL Histone-Modifying Complexes and Regulates Hox Gene Expression. Cell Rep. 2014, 9, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, T.M.; Pascual-Garcia, P.; Gozalo, A.; Little, S.C.; Capelson, M. Chromatin targeting of nuclear pore proteins induces chromatin decondensation. J. Cell Biol. 2019, jcb.201807139. [Google Scholar] [CrossRef]
- Ertl, I.; Porta-De-La-Riva, M.; Gómez-Orte, E.; Rubio-Peña, K.; Aristizábal-Corrales, D.; Cornes, E.; Fontrodona, L.; Osteikoetxea, X.; Ayuso, C.; Askjaer, P.; et al. Functional Interplay of Two Paralogs Encoding SWI/SNF Chromatin-Remodeling Accessory Subunits During Caenorhabditis elegans Development. Genetics 2016, 202, 961–975. [Google Scholar] [CrossRef]
- Hargreaves, D.C.; Crabtree, G.R. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011, 21, 396–420. [Google Scholar] [CrossRef]
- Li, B.; Carey, M.; Workman, J.L. The Role of Chromatin during Transcription. Cell 2007, 128, 707–719. [Google Scholar] [CrossRef]
- Gottesfeld, J.M.; Michael, X.; Carey, F. Chromatin and transcription. J. Biol. Chem. 2018, 13775. [Google Scholar] [CrossRef]
- Gómez-Saldivar, G.; Fernandez, A.; Hirano, Y.; Mauro, M.; Lai, A.; Ayuso, C.; Haraguchi, T.; Hiraoka, Y.; Piano, F.; Askjaer, P. Identification of Conserved MEL-28/ELYS Domains with Essential Roles in Nuclear Assembly and Chromosome Segregation. PLoS Genet. 2016, 12. [Google Scholar] [CrossRef]
- Hampoelz, B.; Andres-Pons, A.; Kastritis, P.; Beck, M. Structure and Assembly of the Nuclear Pore Complex. Annu. Rev. Biophys. 2019, 48, 515–536. [Google Scholar] [CrossRef]
- Rasala, B.A.; Ramos, C.; Harel, A.; Forbes, D.J. Capture of AT-rich Chromatin by ELYS Recruits POM121 and NDC1 to Initiate Nuclear Pore Assembly. Mol. Biol. Cell 2008, 19, 3982–3996. [Google Scholar] [CrossRef]
- Inoue, A.; Zhang, Y. Nucleosome assembly is required for nuclear pore complex assembly in mouse zygotes. Nat. Struct. Mol. Biol. 2014, 21, 609–616. [Google Scholar] [CrossRef]
- Zierhut, C.; Jenness, C.; Kimura, H.; Funabiki, H. Nucleosomal regulation of chromatin composition and nuclear assembly revealed by histone depletion. Nat. Struct. Mol. Biol. 2014, 21, 617–625. [Google Scholar] [CrossRef]
- Kimura, N.; Takizawa, M.; Okita, K.; Natori, O.; Igarashi, K.; Ueno, M.; Nakashima, K.; Nobuhisa, I.; Taga, T. Identification of a novel transcription factor, ELYS, expressed predominantly in mouse foetal haematopoietic tissues. Genes Cells 2002, 7, 435–446. [Google Scholar] [CrossRef]
- Peterson, C.L.; Herskowitz, I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell 1992, 68, 573–583. [Google Scholar] [CrossRef]
- Fernandez, A.G.; Mis, E.K.; Lai, A.; Mauro, M.; Quental, A.; Bock, C.; Piano, F. Uncovering buffered pleiotropy: A genome-scale screen for mel-28 genetic interactors in caenorhabditis elegans. G3 Genes Genomes Genet. 2014, 4, 185–196. [Google Scholar] [CrossRef][Green Version]
- Gdula, D.A.; Sandaltzopoulos, R.; Tsukiyama, T.; Ossipow, V.; Wu, C. Inorganic pyrophosphatase is a component of the Drosophila nucleosome remodeling factor complex. Genes Dev. 1998, 12, 3206–3216. [Google Scholar] [CrossRef]
- Mizuguchi, G.; Wu, C. Nucleosome Remodeling Factor NURF and In Vitro Transcription of Chromatin. In Chromatin Protocols; Humana Press: Totowa, NJ, USA, 1999; pp. 333–342. [Google Scholar]
- Fasci, D.; Van Ingen, H.; Scheltema, R.A.; Heck, A.J.R. Histone interaction landscapes visualized by crosslinking mass spectrometry in intact cell nuclei. Mol. Cell. Proteomics 2018, 17, 2018–2033. [Google Scholar] [CrossRef]
- Rochman, M.; Postnikov, Y.; Correll, S.; Malicet, C.; Wincovitch, S.; Karpova, T.S.; McNally, J.G.; Wu, X.; Bubunenko, N.A.; Grigoryev, S.; et al. The Interaction of NSBP1/HMGN5 with Nucleosomes in Euchromatin Counteracts Linker Histone-Mediated Chromatin Compaction and Modulates Transcription. Mol. Cell 2009, 35, 642–656. [Google Scholar] [CrossRef]
- Sonneville, R.; Craig, G.; Labib, K.; Gartner, A.; Blow, J.J. Both Chromosome Decondensation and Condensation Are Dependent on DNA Replication in C. elegans Embryos. Cell Rep. 2015, 12, 405–417. [Google Scholar] [CrossRef]
- Aze, A.; Fragkos, M.; Bocquet, S.; Cau, J.; Méchali, M. RNAs coordinate nuclear envelope assembly and DNA replication through ELYS recruitment to chromatin. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef]
- Liu, Z.; Yan, M.; Liang, Y.; Mo, W.; Fontoura, B.M.; Zhang Correspondence, L.; Liu, Z.; Liu, M.; Zhang, K.; Shao, D.; et al. Nucleoporin Seh1 Interacts with Olig2/Brd7 to Promote Oligodendrocyte Differentiation and Myelination. Neuron 2019, 102, 1–15. [Google Scholar] [CrossRef]
- Lucchesi, J.C. Transcriptional modulation of entire chromosomes: dosage compensation. J. Genet. 2018, 97, 357–364. [Google Scholar] [CrossRef]
- Mendjan, S.; Taipale, M.; Kind, J.; Holz, H.; Gebhardt, P.; Schelder, M.; Vermeulen, M.; Buscaino, A.; Duncan, K.; Mueller, J.; et al. Nuclear Pore Components Are Involved in the Transcriptional Regulation of Dosage Compensation in Drosophila. Mol. Cell 2006, 21, 811–823. [Google Scholar] [CrossRef]
- Sharma, R.; Jost, D.; Kind, J.; Gómez-Saldivar, G.; van Steensel, B.; Askjaer, P.; Vaillant, C.; Meister, P. Differential spatial and structural organization of the X chromosome underlies dosage compensation in C. elegans. Genes Dev. 2014, 28, 2591–2596. [Google Scholar] [CrossRef]
- Grimaud, C.; Becker, P.B. The dosage compensation complex shapes the conformation of the X chromosome in Drosophila. Genes Dev. 2009, 23, 2490–2495. [Google Scholar] [CrossRef]
- Raich, N.; Mahmoudi, S.; Emre, D.; Karess, R.E. Mad1 influences interphase nucleoplasm organization and chromatin regulation in Drosophila. Open Biol. 2018, 8. [Google Scholar] [CrossRef]
- Lee, S.H.; Sterling, H.; Burlingame, A.; McCormick, F. Tpr directly binds to Mad1 and Mad2 and is important for the Mad1-Mad2-mediated mitotic spindle checkpoint. Genes Dev. 2008, 22, 2926–2931. [Google Scholar] [CrossRef]
- Lince-Faria, M.; Maffini, S.; Orr, B.; Ding, Y.; Cláudia Florindo, C.; Sunkel, C.E.; Tavares, A.; Johansen, J.; Johansen, K.M.; Maiato, H. Spatiotemporal control of mitosis by the conserved spindle matrix protein Megator. J. Cell Biol. 2009, 184, 647–657. [Google Scholar] [CrossRef]
- Krull, S.; Dö rries, J.; rn Boysen, B.; Reidenbach, S.; Magnius, L.; Norder, H.; Thyberg, J.; Cordes, V.C. Protein Tpr is required for establishing nuclear pore-associated zones of heterochromatin exclusion. EMBO J. 2010, 29, 1659–1673. [Google Scholar] [CrossRef]
- Capelson, M.; Hetzer, M.W. The role of nuclear pores in gene regulation, development and disease. EMBO Rep. 2009, 10, 697–705. [Google Scholar] [CrossRef]
- Marini, B.; Kertesz-Farkas, A.; Ali, H.; Lucic, B.; Lisek, K.; Manganaro, L.; Pongor, S.; Luzzati, R.; Recchia, A.; Mavilio, F.; et al. Nuclear architecture dictates HIV-1 integration site selection. Nature 2015, 521, 227–231. [Google Scholar] [CrossRef]
- Lelek, M.; Casartelli, N.; Pellin, D.; Rizzi, E.; Souque, P.; Severgnini, M.; Di Serio, C.; Fricke, T.; Diaz-Griffero, F.; Zimmer, C.; et al. Chromatin organization at the nuclear pore favours HIV replication. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef]
- Ilyin, A.A.; Ryazansky, S.S.; Doronin, S.A.; Olenkina, O.M.; Mikhaleva, E.A.; Yakushev, E.Y.; Abramov, Y.A.; Belyakin, S.N.; Ivankin, A.V.; Pindyurin, A.V.; et al. Piwi interacts with chromatin at nuclear pores and promiscuously binds nuclear transcripts in Drosophila ovarian somatic cells. Nucleic Acids Res. 2017, 45, 7666–7680. [Google Scholar] [CrossRef]
- Parikh, R.Y.; Lin, H.; Gangaraju, V.K. A critical role for nucleoporin 358 (Nup358) in transposon silencing and piRNA biogenesis in Drosophila. J. Biol. Chem. 2018, 293, 9140–9147. [Google Scholar] [CrossRef]
- Handler, D.; Meixner, K.; Pizka, M.; Lauss, K.; Schmied, C.; Gruber, F.S.; Brennecke, J. The genetic makeup of the Drosophila piRNA pathway. Mol. Cell 2013, 50, 762–777. [Google Scholar] [CrossRef]
- Muerdter, F.; Guzzardo, P.M.; Gillis, J.; Luo, Y.; Yu, Y.; Chen, C.; Fekete, R.; Hannon, G.J. A genome-wide RNAi screen draws a genetic framework for transposon control and primary piRNA biogenesis in Drosophila. Mol. Cell 2013, 50, 736–748. [Google Scholar] [CrossRef]
- Nagai, S.; Davoodi, N.; Gasser, S.M. Nuclear organization in genome stability: SUMO connections. Cell Res. 2011, 21, 474–485. [Google Scholar] [CrossRef]
- Casolari, J.M.; Brown, C.R.; Komili, S.; West, J.; Hieronymus, H.; Silver, P.A. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 2004, 117, 427–439. [Google Scholar] [CrossRef]
- Taddei, A.; Van Houwe, G.; Hediger, F.; Kalck, V.; Cubizolles, F.; Schober, H.; Gasser, S.M. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature 2006, 441, 774–778. [Google Scholar] [CrossRef]
- Schmid, M.; Arib, G.; Laemmli, C.; Nishikawa, J.; Durussel, T.; Laemmli, U.K. Nup-PI: The nucleopore-promoter interaction of genes in yeast. Mol. Cell 2006, 21, 379–391. [Google Scholar] [CrossRef] [PubMed]
- Jühlen, R.; Fahrenkrog, B. Moonlighting nuclear pore proteins: Tissue-specific nucleoporin function in health and disease. Histochem. Cell Biol. 2018, 150, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Talamas, J.A.; Capelson, M. Nuclear envelope and genome interactions in cell fate. Front. Genet. 2015, 5, 95. [Google Scholar] [CrossRef] [PubMed]
- Gough, S.M.; Slape, C.I.; Aplan, P.D. NUP98 gene fusions and hematopoietic malignancies: Common themes and new biologic insights. Blood 2011, 118, 6247–6257. [Google Scholar] [CrossRef]
- Oka, M.; Mura, S.; Yamada, K.; Sangel, P.; Hirata, S.; Maehara, K.; Kawakami, K.; Tachibana, T.; Ohkawa, Y.; Kimura, H.; et al. Chromatin-prebound Crm1 recruits Nup98-HoxA9 fusion to induce aberrant expression of Hox cluster genes. Elife 2016, 5, 1–21. [Google Scholar] [CrossRef]
- Xu, H.; Valerio, D.G.; Eisold, M.E.; Sinha, A.; Koche, R.P.; Hu, W.; Chen, C.W.; Chu, S.H.; Brien, G.L.; Park, C.Y.; et al. NUP98 Fusion Proteins Interact with the NSL and MLL1 Complexes to Drive Leukemogenesis. Cancer Cell 2016, 30, 863–878. [Google Scholar] [CrossRef]
- Hnisz, D.; Schuijers, J.; Bradner, J.E.; Correspondence, R.A.Y. Convergence of Developmental and Oncogenic Signaling Pathways at Transcriptional Super-Enhancers. Mol. Cell 2015, 58, 362–370. [Google Scholar] [CrossRef]
- Brown, C.R.; Kennedy, C.J.; Delmar, V.A.; Forbes, D.J.; Silver, P.A. Global histone acetylation induces functional genomic reorganization at mammalian nuclear pore complexes. Genes Dev. 2008, 22, 627–639. [Google Scholar] [CrossRef]
- Toda, T.; Hsu, J.Y.; Linker, S.B.; Jacinto, F.V.; Hetzer, M.W.; Gage Correspondence, F.H. Nup153 Interacts with Sox2 to Enable Bimodal Gene Regulation and Maintenance of Neural Progenitor Cells Data Resources GSE99783 Toda et al. Cell Stem Cell 2017, 21, 618–634. [Google Scholar] [CrossRef]
- Engelen, E.; Akinci, U.; Bryne, J.C.; Hou, J.; Gontan, C.; Moen, M.; Szumska, D.; Kockx, C.; Van Ijcken, W.; Dekkers, D.H.W.; et al. Sox2 cooperates with Chd7 to regulate genes that are mutated in human syndromes. Nat. Genet. 2011, 43, 607–611. [Google Scholar] [CrossRef]
- Raices, M.; Bukata, L.; Sakuma, S.; Borlido, J.; Hernandez, L.S.; Hart, D.O.; D’Angelo, M.A. Nuclear Pores Regulate Muscle Development and Maintenance by Assembling a Localized Mef2C Complex. Dev. Cell 2017, 41, 540–554. [Google Scholar] [CrossRef]
- Brickner, D.G.; Randise-Hinchliff, C.; Corbin, M.L.; Liang, J.M.; Kim, S.; Sump, B.; D’Urso, A.; Kim, S.H.; Satomura, A.; Schmit, H.; et al. The Role of Transcription Factors and Nuclear Pore Proteins in Controlling the Spatial Organization of the Yeast Genome. Dev. Cell 2019, 49, 936–947. [Google Scholar] [CrossRef]
- Sood, V.; Brickner, J.H. Nuclear pore interactions with the genome. Curr. Opin. Genet. Dev. 2014, 25, 43–49. [Google Scholar] [CrossRef]
- Schneider, M.; Hellerschmied, D.; Schubert, T.; Amlacher, S.; Vinayachandran, V.; Reja, R.; Pugh, B.F.; Clausen, T.; Köhler, A. The Nuclear Pore-Associated TREX-2 Complex Employs Mediator to Regulate Gene Expression. Cell 2015, 162, 1016–1028. [Google Scholar] [CrossRef]
- Luthra, R.; Kerr, S.C.; Harreman, M.T.; Apponi, L.H.; Fasken, M.B.; Ramineni, S.; Chaurasia, S.; Valentini, S.R.; Corbett, A.H. Actively transcribed GAL genes can be physically linked to the nuclear pore by the SAGA chromatin modifying complex. J. Biol. Chem. 2007, 282, 3042–3049. [Google Scholar] [CrossRef]
- Brown, C.E.; Howe, L.; Sousa, K.; Alley, S.C.; Carrozza, M.J.; Tan, S.; Workman, J.L. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 2001, 292, 2333–2337. [Google Scholar] [CrossRef]
- Raices, M.; D’Angelo, M.A. Nuclear pore complex composition: A new regulator of tissue-specific and developmental functions. Nat. Rev. Mol. Cell Biol. 2012, 13, 687–699. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuhn, T.M.; Capelson, M. Nuclear Pore Proteins in Regulation of Chromatin State. Cells 2019, 8, 1414. https://doi.org/10.3390/cells8111414
Kuhn TM, Capelson M. Nuclear Pore Proteins in Regulation of Chromatin State. Cells. 2019; 8(11):1414. https://doi.org/10.3390/cells8111414
Chicago/Turabian StyleKuhn, Terra M., and Maya Capelson. 2019. "Nuclear Pore Proteins in Regulation of Chromatin State" Cells 8, no. 11: 1414. https://doi.org/10.3390/cells8111414
APA StyleKuhn, T. M., & Capelson, M. (2019). Nuclear Pore Proteins in Regulation of Chromatin State. Cells, 8(11), 1414. https://doi.org/10.3390/cells8111414