An RBPJ-Drosophila Model Reveals Dependence of RBPJ Protein Stability on the Formation of Transcription–Regulator Complexes
Abstract
1. Introduction
2. Materials and Methods
2.1. Generation of Mouse RBPJ Constructs and Establishment of RBPJwt and RBPJLLL Mutant Flies
2.2. Fly Work and Documentation of Adult Phenotypes
2.3. Immunochemistry
2.4. RT-PCR and Quantitative RT-qPCR
2.5. Yeast Protein–Protein Interaction Assays
- NICD mouse UP: 5’ -GAT GAA TTC CGA CGA CGA CAT GGC CAG CTC TTG T-3’;
- NICD mouse LP: 5’ -CGG GTC GAC TTA GCT TGC TGG TGC ACC CAC G -3’;
- MamL UP: 5’ -GAT AGA TCT CCA TGG CAC TGC CGC GGC ACA-3’; and
- MamL LP: 5’ -CTT CTC GAG TTA GGT GGC GAT GGA TCC CGG G-3’.
2.6. Statistical Analysis
3. Results
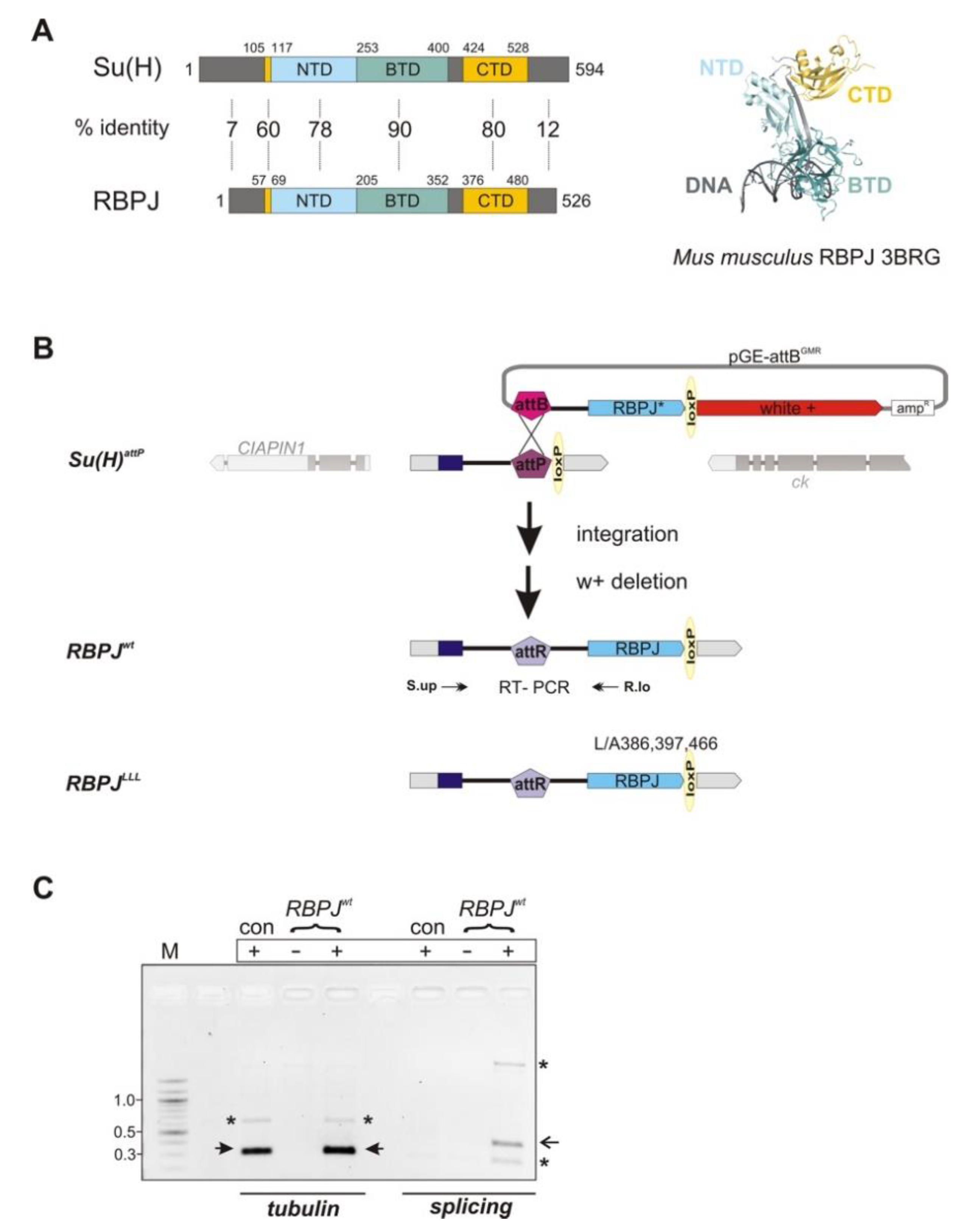
3.1. Replacement of the Su(H) Locus with Murine RBPJ by Genome Engineering in Drosophila
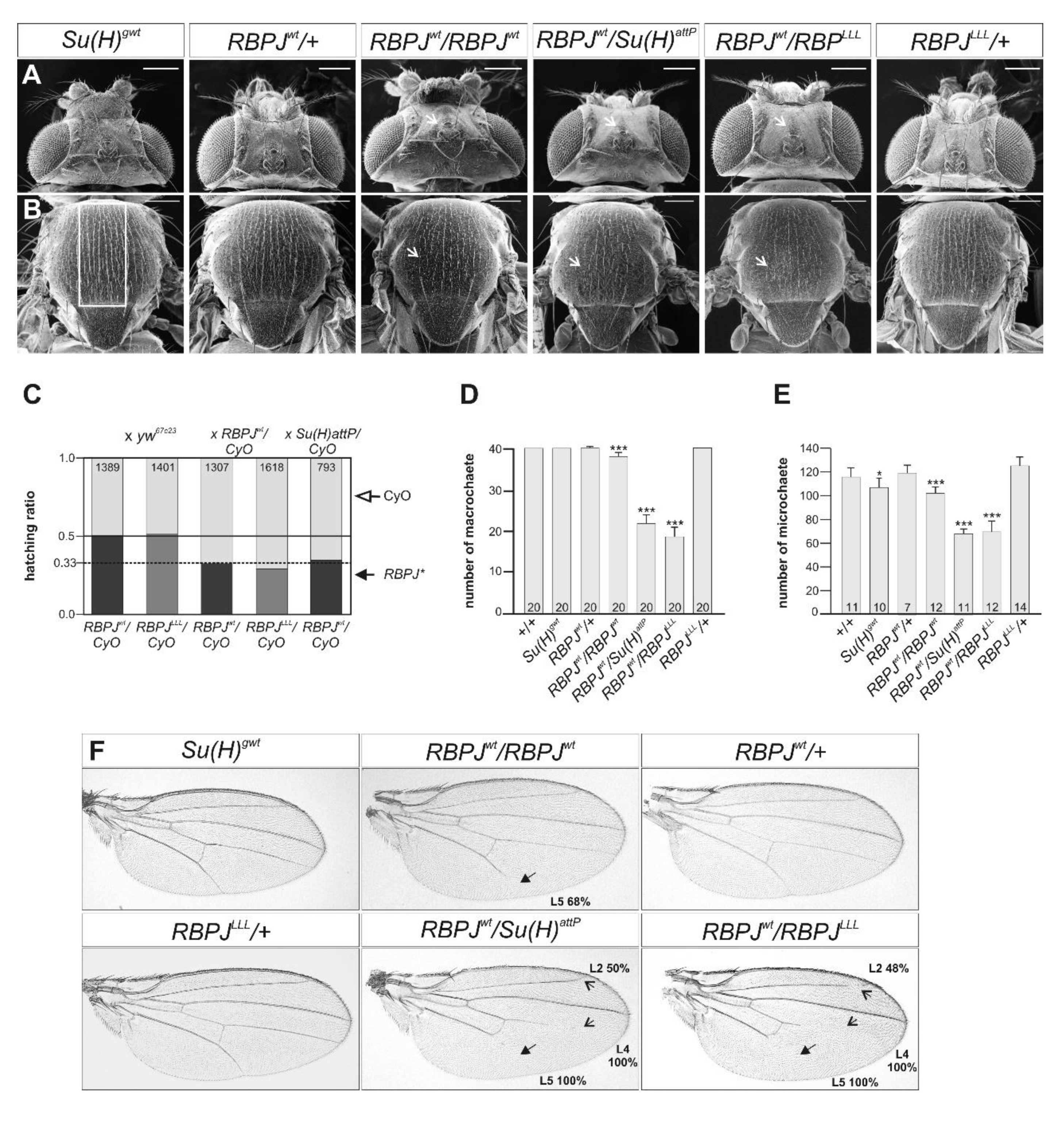
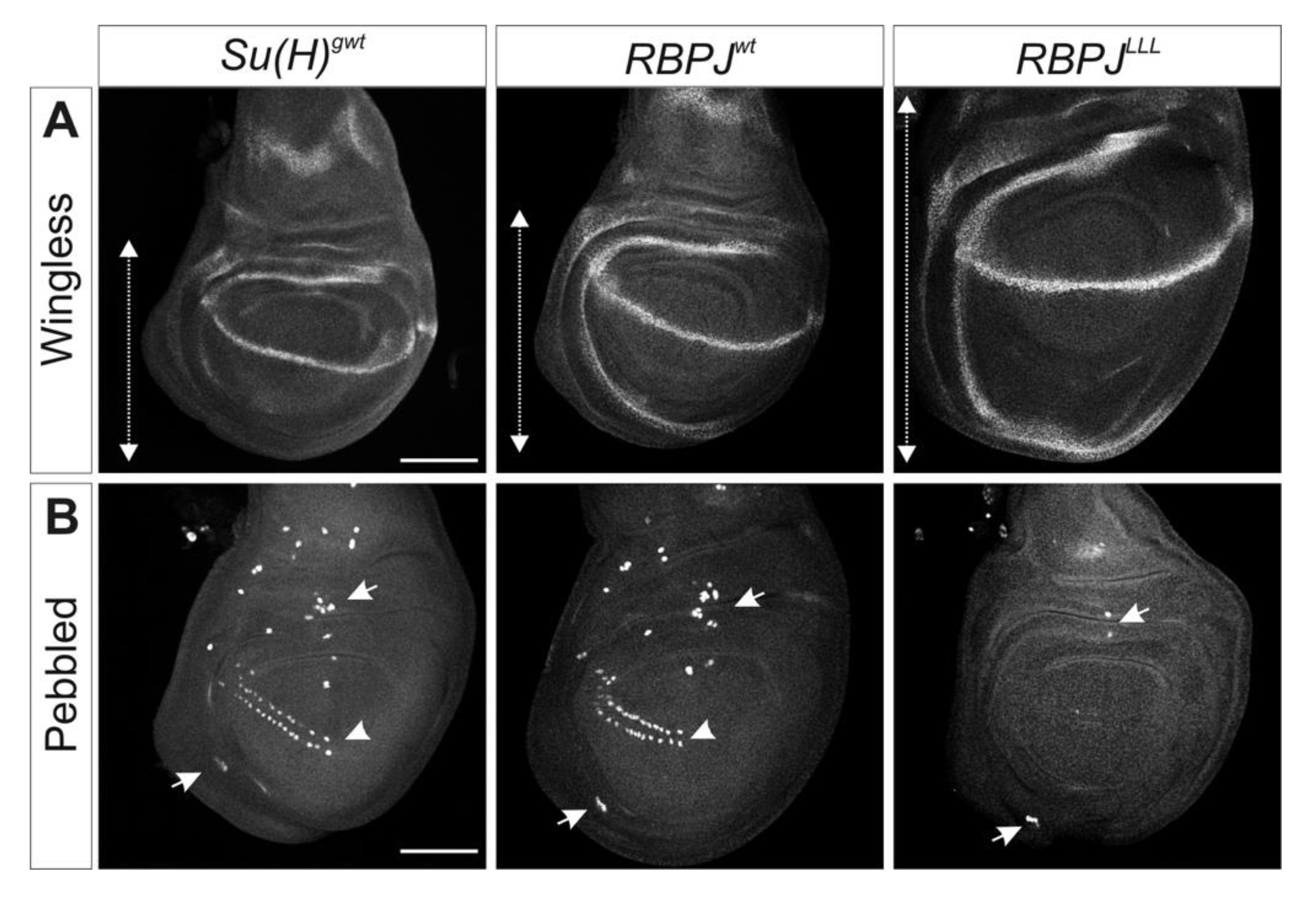
3.2. RBPJwt Flies Are Viable and Show Characteristics of a Gain in Notch Activity
3.3. RBPJLLL Flies Are Lethal Due to a Failure of Repressor Complex Assembly
3.4. The Interaction between RBPJ and Hairless Is Impaired
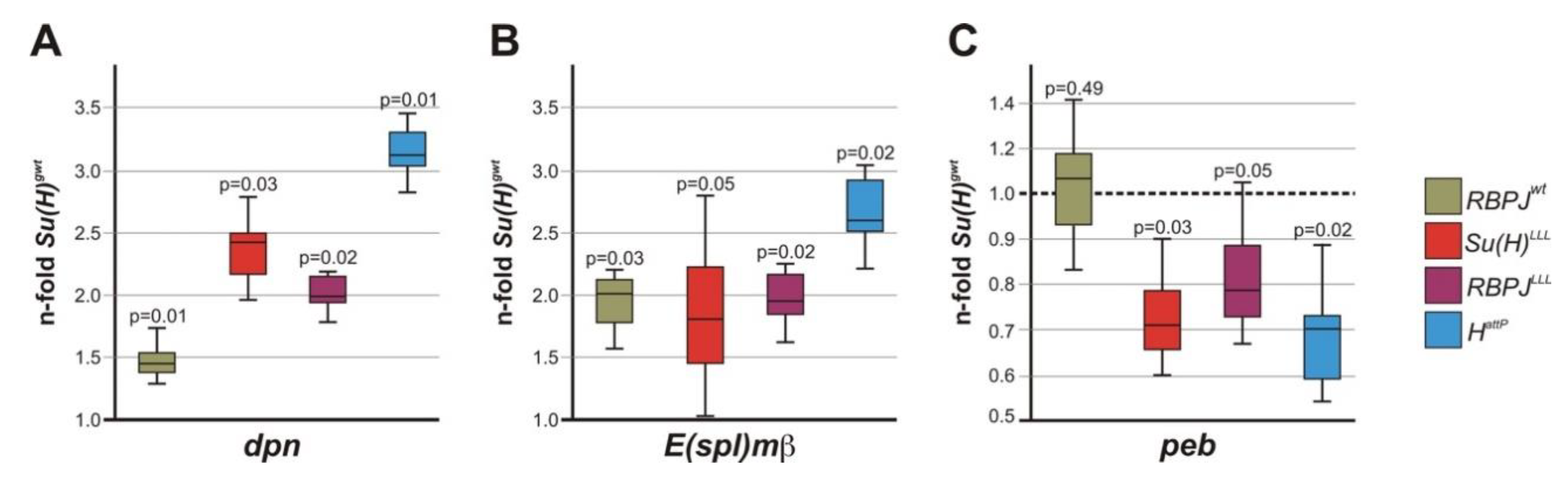
3.5. Notch Activity Is Increased RBPJLLL Homozygous Mutants
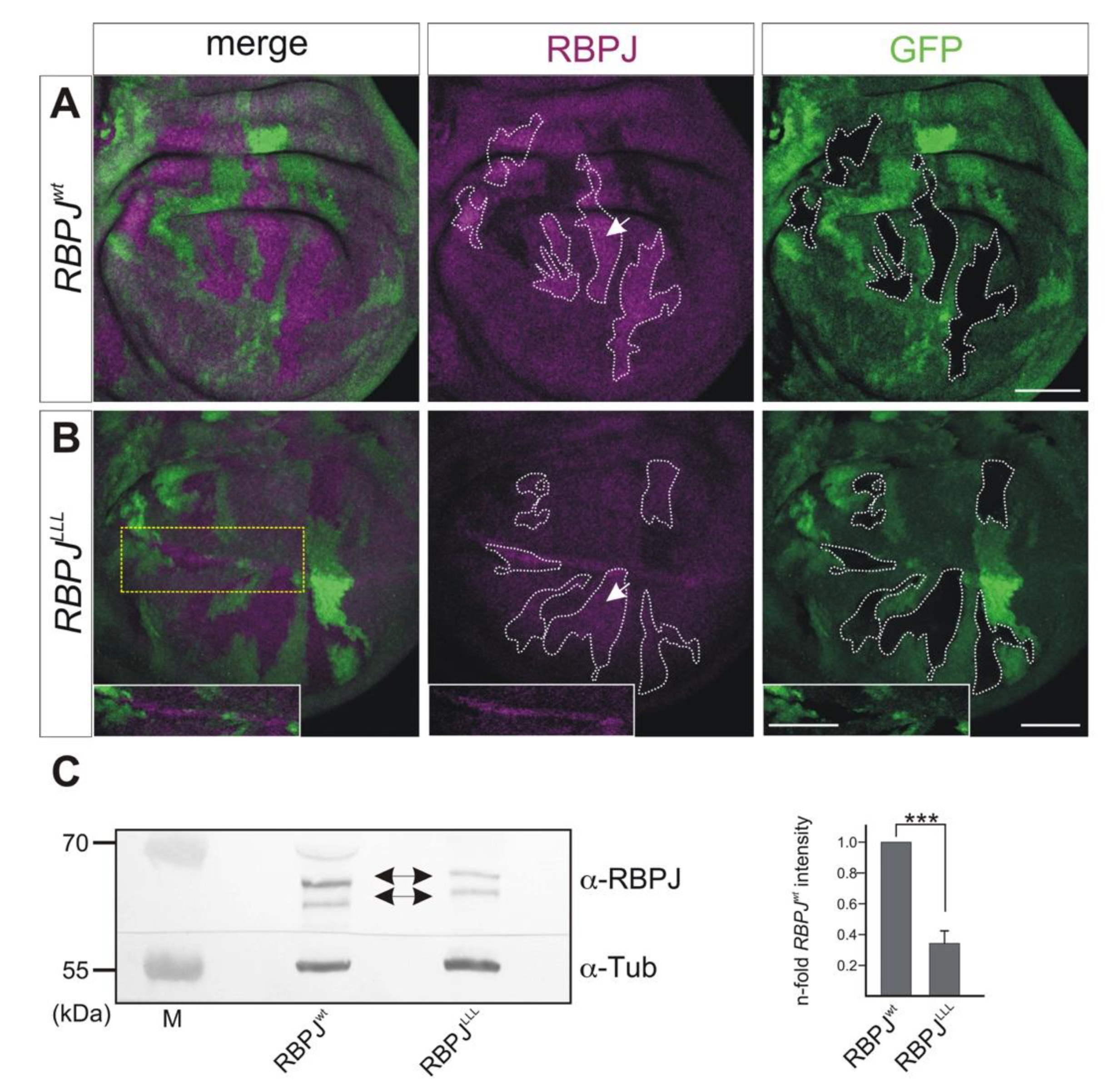
3.6. Stability of RBPJ Protein Depends on Its Recruitment into Repressor or Activator Complexes
4. Discussion
4.1. The RBPJ-H Repressor Complex
4.2. Regulation of Mouse RBPJ Availability in Drosophila
4.3. Transcriptional Regulation of Notch Target Genes by RBPJ in Drosophila
4.4. Perspectives of the RBPJ Fly Model
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Louvi, A.; Artavanis-Tsakonas, S. Notch and disease: A growing field. Semin. Cell Dev. Biol. 2012, 23, 473–480. [Google Scholar]
- Bray, S.J. Notch signalling in context. Nat. Rev. Mol. Biol. 2016, 17, 722–735. [Google Scholar] [CrossRef]
- Kopan, R.; Ilagan, M.X.G. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell 2009, 137, 216–233. [Google Scholar]
- Kovall, R.A.; Blacklow, S.C. Mechanistic insights into Notch receptor signaling from structural and biochemical studies. Curr. Top. Dev. Biol. 2010, 92, 31–71. [Google Scholar]
- Borggrefe, T.; Oswald, F. Setting the stage for Notch: The Drosophila Su(H)-Hairless repressor complex. PLoS Biol. 2016, 14, e1002524. [Google Scholar] [CrossRef]
- Gomez-Lamarca, M.J.; Falo-Sanjuan, J.; Stojnic, R.; Rehman, S.A.; Muresan, L.; Jones, M.L.; Pilidge, Z.; Cerda-Moya, G.; Yuan, Z.; Baloul, S.; et al. Activation of the Notch signaling pathway in vivo elicits changes in CSL nuclear dynamics. Dev. Cell 2018, 44, 611–623. [Google Scholar]
- Oswald, F.; Rodriguez, P.; Giaimo, B.D.; Antonello, Z.A.; Mira, L.; Mittler, G.; Thiel, V.N.; Collins, K.J.; Tabaja, N.; Cizelsky, W.; et al. A phospho-dependent mechanism involving NCoR and KMT2D controls a permissive chromatin state at Notch target genes. Nucleic Acids Res. 2016, 44, 4703–4720. [Google Scholar]
- Moshkin, Y.M.; Kan, T.W.; Goodfellow, H.; Bezstarosti, K.; Maeda, R.K.; Pilyugin, M.; Karch, F.; Bray, S.J.; Demmers, J.A.; Verrijzer, C.P. Histone chaperones ASF1 and NAP1 differentially modulate removal of active histone marks by LID-RPD3 complexes during NOTCH silencing. Mol. Cell 2009, 35, 782–793. [Google Scholar]
- Liefke, R.; Oswald, F.; Alvarado, C.; Ferres-Marco, D.; Mittler, G.; Rodriguez, P.; Dominguez, M.; Borggrefe, T. Histone demethylase KDM5A is an integral part of the core Notch-RBP-J repressor complex. Genes Dev. 2010, 24, 590–601. [Google Scholar] [CrossRef]
- Mulligan, P.; Yang, F.; Di Stefano, L.; Ji, J.Y.; Ouyang, J.; Nishikawa, J.L.; Toiber, D.; Kulkarni, M.; Wang, Q.; Najafi-Shoushtari, S.H.; et al. A SIRT1-LSD1 corepressor complex regulates notch target gene expression and development. Mol. Cell 2011, 42, 689–699. [Google Scholar]
- Yatim, A.; Benne, C.; Sobhian, B.; Laurent-Chabalier, S.; Deas, O.; Judde, J.G.; Lelievre, J.D.; Levy, Y.; Benkirane, M. NOTCH1 nuclear interactome reveals key regulators of its transcriptional activity and oncogenic function. Mol. Cell 2012, 48, 445–458. [Google Scholar]
- Borggrefe, T.; Oswald, F. The Notch signaling pathway: Transcriptional regulation at Notch target genes. Cell. Mol. Life Sci. 2009, 66, 1631–1646. [Google Scholar]
- Oka, C.; Nakano, T.; Wakeham, A.; de la Pompa, J.L.; Mori, C.; Sakai, T.; Okazaki, S.; Kawaichi, M.; Shiota, K.; Mak, T.W.; et al. Disruption of the mouse RBP-J kappa gene results in early embryonic death. Development 1995, 121, 3291–3301. [Google Scholar]
- Kovall, R.A.; Hendrickson, W.A. Crystal structure of the nuclear effector of Notch signaling, CSL, bound to DNA. EMBO J. 2004, 23, 3441–3451. [Google Scholar]
- Nam, Y.; Sliz, P.; Song, L.; Aster, J.B.; Blacklow, S.C. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell 2006, 124, 973–983. [Google Scholar]
- Wilson, J.J.; Kovall, R.A. Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA. Cell 2006, 124, 985–996. [Google Scholar]
- Furukawa, T.; Maruyama, S.; Kawaichi, M.; Honjo, T. The Drosophila homolog of the immunoglobulin recombination signal binding protein regulates peripheral nervous system development. Cell 1992, 69, 1191–1197. [Google Scholar]
- Schweisguth, F.; Posakony, J.W. Suppressor of Hairless, the Drosophila homolog of the mouse Recombination signal-binding protein gene, controls sensory organ cell fates. Cell 1992, 69, 1199–1212. [Google Scholar]
- Maier, D. The evolution of transcriptional repressors in the Notch signaling pathway: A computational analysis. Hereditas 2019, 156, 5. [Google Scholar]
- Maier, D.; Kurth, P.; Schulz, A.; Russell, A.; Yuan, Z.; Gruber, K.; Kovall, R.A.; Preiss, A. Structural and functional analysis of the repressor complex in the Notch signaling pathway of Drosophila melanogaster. Mol. Biol. Cell 2011, 22, 3242–3252. [Google Scholar]
- Yuan, Z.; Praxenthaler, H.; Tabaja, N.; Torella, R.; Preiss, A.; Maier, D.; Kovall, R.A. Structure and function of the Su(H)-Hairless repressor complex, the major antagonist of the Notch signaling in Drosophila melanogaster. PLoS Biol. 2016, 14, e1002509. [Google Scholar] [CrossRef]
- Brou, C.; Logeat, F.; Lecourtois, M.; Vanderkerckhove, J.; Kourilsky, P.; Schweisguth, F.; Israël, A. Inhibition of the DNA-binding activity of Drosophila Suppressor of Hairless and of its human homolog, KBP2/RBP-J kappa, by direct protein-protein interaction with Drosophila Hairless. Genes Dev. 1994, 8, 2491–2503. [Google Scholar] [CrossRef]
- Kaspar, M.; Klein, T. Functional analysis of murine CBF1 during Drosophila development. Dev. Dyn. 2006, 235, 918–927. [Google Scholar] [CrossRef]
- Praxenthaler, H.; Nagel, A.C.; Schulz, A.; Zimmermann, M.; Meier, M.; Schmid, H.; Preiss, A.; Maier, D. Hairless-binding deficient Suppressor of Hairless alleles reveal Su(H) protein levels are dependent on complex formation with Hairless. PLoS Genet. 2017, 13, e1006774. [Google Scholar] [CrossRef]
- Huang, J.; Zhou, W.; Dong, W.; Watson, A.M.; Hong, Y. Directed, efficient, and versatile modifications of the Drosophila genome by genomic engineering. Proc. Natl. Acad. Sci. USA 2009, 106, 8284–8289. [Google Scholar] [CrossRef]
- Praxenthaler, H.; Smylla, T.K.; Nagel, A.C.; Preiss, A.; Maier, D. Generation of new Hairless alleles by genomic engineering at the Hairless locus in Drosophila melanogaster. PLoS ONE 2015, 10, e0140007. [Google Scholar] [CrossRef]
- Bischof, J.; Maeda, R.K.; Hediger, M.; Karch, F.; Basler, K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA 2007, 104, 3312–3317. [Google Scholar] [CrossRef]
- Maggert, K.A.; Gong, W.J.; Golic, K.G. Methods for homologous recombination in Drosophila. Methods Mol. Biol. 2008, 420, 155–174. [Google Scholar]
- Maier, D.; Nagel, A.C.; Preiss, A. Protein Kinase D is dispensable for development and survival of Drosophila melanogaster. G3 (Bethesda) 2019, 9, 2477–2487. [Google Scholar] [CrossRef]
- Kober, L.; Zimmermann, M.; Kurz, M.; Bayer, M.; Nagel, A.C. Loss of putzig in the germline impedes germ cell development by inducing cell death and new niche like microenvironments. Sci. Rep. 2019, 9, 9108. [Google Scholar] [CrossRef]
- Hu, Y.; Sopko, R.; Foos, M.; Kelley, C.; Flockhart, I.; Ammeux, N.; Wang, X.; Perkins, L.; Perrimon, N.; Mohr, S.E. FlyPrimerBank: An online database for Drosophila melanogaster gene expression analysis and knockdown evaluation of RNAi reagents. G3 (Bethesda) 2013, 3, 1607–1616. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST) for a group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef] [PubMed]
- Gyuris, J.; Golemis, E.; Chertkov, H.; Brent, R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 1993, 75, 791–803. [Google Scholar] [CrossRef]
- Golemis, E.A.; Brent, R. Searching for interacting proteins with the two-hybrid system III. In The Yeast Two-Hybrid System; Bartel, P.L., Fields, S., Eds.; Oxford University Press: Oxford, UK, 1997; pp. 43–72. [Google Scholar]
- Sambrook, J.; Russel, D. Molecular Cloning: A Laboratory Manual, 3rd ed.; Chapter 18, Protocol 1; Cold Spring Harbor Laboratory Press: Long Island, NY, USA, 2001. [Google Scholar]
- Guarente, L. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 1983, 101, 181–191. [Google Scholar] [PubMed]
- Nagel, A.C.; Krejci, A.; Tenin, G.; Bravo-Patiño, A.; Bray, S.; Maier, D.; Preiss, A. Hairless-mediated repression of Notch target genes requires the combined activity of Groucho and CtBP corepressors. Mol. Cell. Biol. 2005, 25, 10433–10441. [Google Scholar] [CrossRef]
- Friedmann, D.R.; Wilson, J.J.; Kovall, R.A. RAM-induced allostery facilitates assembly of a Notch pathway active transcription complex. J. Biol. Chem. 2008, 283, 14781–14791. [Google Scholar] [CrossRef]
- Rebay, I.; Fehon, R.G.; Artavanis-Tsakonas, S. Specific truncations of Drosophila Notch define dominant activated and dominant negative forms of the receptor. Cell 1993, 74, 319–329. [Google Scholar] [CrossRef]
- Schweisguth, F.; Posakony, J.W. Antagonistic activities of Suppressor of Hairless and Hairless control alternative cell fates in the Drosophila epidermis. Development 1994, 120, 1433–1441. [Google Scholar]
- Bang, A.G.; Bailey, A.M.; Posakony, J.W. Hairless promotes stable commitment to the sensory organ precursor cell fate by negatively regulating the activity of the Notch signaling pathway. Dev. Biol. 1995, 172, 479–494. [Google Scholar] [CrossRef]
- Lee, G.; Liang, C.; Park, G.; Jang, C.; Jung, J.U.; Chung, J. UVRAG is required for organ rotation by regulating Notch endocytosis in Drosophila. Dev. Biol. 2011, 356, 588–597. [Google Scholar] [CrossRef]
- Lindsley, D.L.; Zimm, G.G. The Genome of Drosophila melanogaster; Academic Press: San Diego, CA, USA, 1992. [Google Scholar]
- De Celis, J.F.; Garcia-Bellido, A. Modifications of the Notch function by Abruptex mutations in Drosophila melanogaster. Genetics 1994, 136, 183–194. [Google Scholar] [PubMed]
- Bang, A.G.; Hartenstein, V.; Posakony, J.W. Hairless is required for the development of adult sensory organ precursor cells in Drosophila. Development 1991, 111, 89–104. [Google Scholar] [PubMed]
- Preiss, A.; Nagel, A.C.; Praxenthaler, H.; Maier, D. Complex genetic interactions of novel Suppressor of Hairless alleles deficient in co-repressor binding. PLoS ONE 2018, 13, e0193956. [Google Scholar] [CrossRef] [PubMed]
- Kurth, P.; Preiss, A.; Kovall, R.A.; Maier, D. Molecular analysis of the Notch repressor-complex in Drosophila: Characterization of potential Hairless binding sites on Suppressor of Hairless. PLoS ONE 2011, 6, e27986. [Google Scholar]
- Zhang, J.; Lautar, S. A yeast three-hybrid method to clone ternary protein complex components. Anal. Biochem. 1996, 242, 68–72. [Google Scholar] [CrossRef]
- Petcherski, A.G.; Kimble, J. LAG-3 is a putative transcriptional activator in the C. elegans Notch pathway. Nature 2000, 405, 364–368. [Google Scholar]
- Neumann, C.J.; Cohen, S.M. A hierarchy of cross-regulation involving Notch, wingless, vestigial and cut organizes the dorsal/ventral axis of the Drosophila wing. Development 1996, 122, 3477–3485. [Google Scholar]
- Klein, T.; Seugnet, L.; Haenlin, M.; Martinez-Arias, A. Two different activities of Suppressor of Hairless during wing development in Drosophila. Development 2000, 127, 3553–3566. [Google Scholar]
- Go, M.J.; Eastman, D.S.; Artavanis-Tsakonas, S. Cell proliferation control by Notch signaling in Drosophila development. Development 1998, 125, 2031–2040. [Google Scholar]
- Djiane, A.; Krejci, A.; Bernard, F.; Fexova, S.; Millen, K.; Bray, S.J. Dissecting the mechanisms of Notch induced hyperplasia. EMBO J. 2013, 32, 60–71. [Google Scholar]
- Yip, M.L.R.; Lamka, M.L.; Lipshitz, H.D. Control of germ-band retraction in Drosophila by the zinc-finger protein HINDSIGHT. Development 1997, 124, 2129–2141. [Google Scholar] [PubMed]
- Pickup, A.T.; Lamka, M.L.; Sun, Q.; Yip, M.L.R.; Lipshitz, H.D. Control of photoreceptor cell morphology and integrity during Drosophila eye development. Development 2002, 129, 2247–2258. [Google Scholar] [PubMed]
- Koelzer, S.; Klein, T. A Notch-independent function of Suppressor of Hairless during the development of the bristle sensory organ precursor cell of Drosophila. Development 2003, 130, 1973–1988. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Babaoğlan, A.B.; Housden, B.E.; Furriols, M.; Bray, S.J. Deadpan contributes to the robustness of the Notch response. PLoS ONE 2013, 8, e75632. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.K.K.; Cerda-Moya, G.; Stojnic, R.; Millen, K.; Fischer, B.; Fexova, S.; Skalska, L.; Gomez-Lamarca, M.; Pillidge, Z.; Russell, S.; et al. Role of co-repressor genomic landscapes in shaping the Notch response. PLoS Genet. 2017, 13, e1007096. [Google Scholar] [CrossRef] [PubMed]
- San Juan, B.P.; Andrade-Zapata, I.; Baonza, A. The bHLH factors Dpn and members of the E(spl) complex mediate the function of Notch signalling regulating cell proliferation during wing disc development. Biol. Open 2012, 1, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Bier, E.; Vaessin, H.; Younger-Shepherd, S.; Jan, L.Y.; Jan, Y.N. Deadpan, an essential pan-neuronal gene in Drosophila, encodes a helix-loop-helix protein similar to the hairy gene product. Genes Dev. 1992, 6, 2137–2151. [Google Scholar] [CrossRef]
- Delidakis, C.; Monastirioti, M.; Magadi, S.S. E(spl): Genetic, developmental, and evolutionary aspects of a group of invertebrate Hes proteins with close ties to Notch signaling. Curr. Top. Dev. Biol. 2014, 110, 217–262. [Google Scholar]
- Xu, T.; Rubin, G.M. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 1993, 117, 1223–1237. [Google Scholar]
- Wolf, D.; Smylla, T.K.; Reichmuth, J.; Hoffmeister, P.; Kober, L.; Zimmermann, M.; Turkiewicz, A.; Borggrefe, T.; Nagel, A.C.; Oswald, F.; et al. Nucleo-cytoplasmic shuttling of Drosophila Hairless/Su(H) heterodimer as a means of regulating Notch dependent transcription. Biochim. Biophys. Acta Mol. Cell Res. 2019, 1866, 1520–1532. [Google Scholar] [CrossRef]
- Furriols, M.; Bray, S. Dissecting the mechanisms of Suppressor of Hairless function. Dev. Biol. 2000, 227, 520–532. [Google Scholar] [CrossRef] [PubMed]
- Gho, M.; Lecourtois, M.; Geraud, G.; Posakony, J.W.; Schweisguth, F. Subcellular localization of Suppressor of Hairless in Drosophila sense organ cells during Notch signalling. Development 1996, 122, 1673–1682. [Google Scholar] [PubMed]
- Graveley, B.R.; Brooks, A.N.; Carlson, J.W.; Duff, M.O.; Landolin, J.M.; Yang, L.; Artieri, C.G.; van Baren, M.J.; Boley, N.; Booth, B.W.; et al. The developmental transcriptome of Drosophila melanogaster. Nature 2011, 471, 473–479. [Google Scholar] [CrossRef] [PubMed]
- Fisher, B.; Weiszmann, R.; Frise, E.; Hammonds, A.; Tomancak, P.; Beaton, A.; Berman, B.; Quan, E.; Shu, S.; Lewis, S.; et al. BDGP Insitu Homepage. 2012. Available online: http://insitu.fruitfly.org/cgi-bin/ex/insitu.pl (accessed on 4 June 2019).
- Koelzer, S.; Klein, T. Regulation of expression of Vg and establishment of the dorsoventral compartment boundary in the wing imaginal disc by Suppressor of Hairless. Dev. Biol. 2006, 289, 77–90. [Google Scholar] [CrossRef]
- Fortini, M.E.; Artavanis-Tsakonas, S. The Suppressor of Hairless protein participates in Notch receptor signaling. Cell 1994, 79, 273–282. [Google Scholar] [CrossRef]
- Maier, D.; Nagel, A.C.; Johannes, B.; Preiss, A. Subcellular localization of Hairless protein shows major focus of activity within the nucleus. Mech. Dev. 1999, 89, 195–199. [Google Scholar] [CrossRef]
- Zhou, S.; Hayward, S.D. Nuclear localization of CBF1 is regulated by interactions with the SMRT corepressor complex. Mol. Cell. Biol 2001, 21, 6222–6232. [Google Scholar] [CrossRef]
- Kim, S.M.; Kim, A.Y.; Ann, E.J.; Mo, J.S.; Yoon, J.J.; Park, H.S. Presenilin-2 regulates the degradation of RBP-Jk protein through p38 mitogen-activated protein kinase. J. Cell Sci. 2012, 125, 1296–1308. [Google Scholar] [CrossRef][Green Version]
- Auer, J.S.; Nagel, A.C.; Schulz, A.; Wahl, V.; Preiss, A. MAPK-dependent phosphorylation modulates the activity of Suppressor of Hairless in Drosophila. Cell. Signal. 2015, 27, 115–124. [Google Scholar] [CrossRef]
- Pérez, L.; Milán, M.; Bray, S.; Cohen, S.M. Ligand-binding and signaling properties of the AxM1 form of Notch. Mech. Dev. 2005, 122, 479–486. [Google Scholar] [CrossRef]
- Borggrefe, T.; Liefke, R. Fine-tuning of the intracellular canonical Notch signaling pathway. Cell Cycle 2012, 11, 264–276. [Google Scholar] [CrossRef] [PubMed]
- Terriente-Felix, A.; Li, J.; Collins, S.; Mulligan, A.; Reekie, I.; Bernard, F.; Krejci, A.; Bray, S. Notch cooperates with Lozenge/Runx to lock haemocytes into a differentiation programme. Development 2013, 140, 926–937. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zang, C.; Taing, L.; Arnett, K.L.; Wong, Y.J.; Pear, W.S.; Blacklow, S.C.; Liu, X.S.; Aster, J.C. NOTCH1-RBPJ complexes drive target gene expression through dynamic interactions with superenhancers. Proc. Natl. Acad. Sci. USA 2014, 111, 705–710. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.W.; Avidor-Reiss, T.; Polyanovsky, A.; Posakony, J.W. Complex interplay of three transcription factors in controlling the tormogen differentiation program of Drosophila mechanoreceptors. Dev. Biol. 2009, 329, 386–399. [Google Scholar] [CrossRef] [PubMed]
- Herranz, H.; Eichenlaub, T.; Cohen, S.M. Cancer in Drosophila: Imaginal discs as a model for epithelial tumor formation. Curr. Top. Dev. Biol. 2016, 116, 181–199. [Google Scholar]
- Moulton, M.J.; Letsou, A. Modeling congenital disease and inborn errors of development in Drosophila melanogaster. Dis. Models Mech. 2016, 9, 253–269. [Google Scholar] [CrossRef]
- Siebel, C.; Lendahl, U. Notch signaling in development, tissue homeostasis, and disease. Physiol. Rev. 2017, 97, 1235–1294. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gahr, B.M.; Brändle, F.; Zimmermann, M.; Nagel, A.C. An RBPJ-Drosophila Model Reveals Dependence of RBPJ Protein Stability on the Formation of Transcription–Regulator Complexes. Cells 2019, 8, 1252. https://doi.org/10.3390/cells8101252
Gahr BM, Brändle F, Zimmermann M, Nagel AC. An RBPJ-Drosophila Model Reveals Dependence of RBPJ Protein Stability on the Formation of Transcription–Regulator Complexes. Cells. 2019; 8(10):1252. https://doi.org/10.3390/cells8101252
Chicago/Turabian StyleGahr, Bernd M., Franziska Brändle, Mirjam Zimmermann, and Anja C. Nagel. 2019. "An RBPJ-Drosophila Model Reveals Dependence of RBPJ Protein Stability on the Formation of Transcription–Regulator Complexes" Cells 8, no. 10: 1252. https://doi.org/10.3390/cells8101252
APA StyleGahr, B. M., Brändle, F., Zimmermann, M., & Nagel, A. C. (2019). An RBPJ-Drosophila Model Reveals Dependence of RBPJ Protein Stability on the Formation of Transcription–Regulator Complexes. Cells, 8(10), 1252. https://doi.org/10.3390/cells8101252




