Oncogenic Role of Tumor Necrosis Factor α-Induced Protein 8 (TNFAIP8)
Abstract
1. Introduction
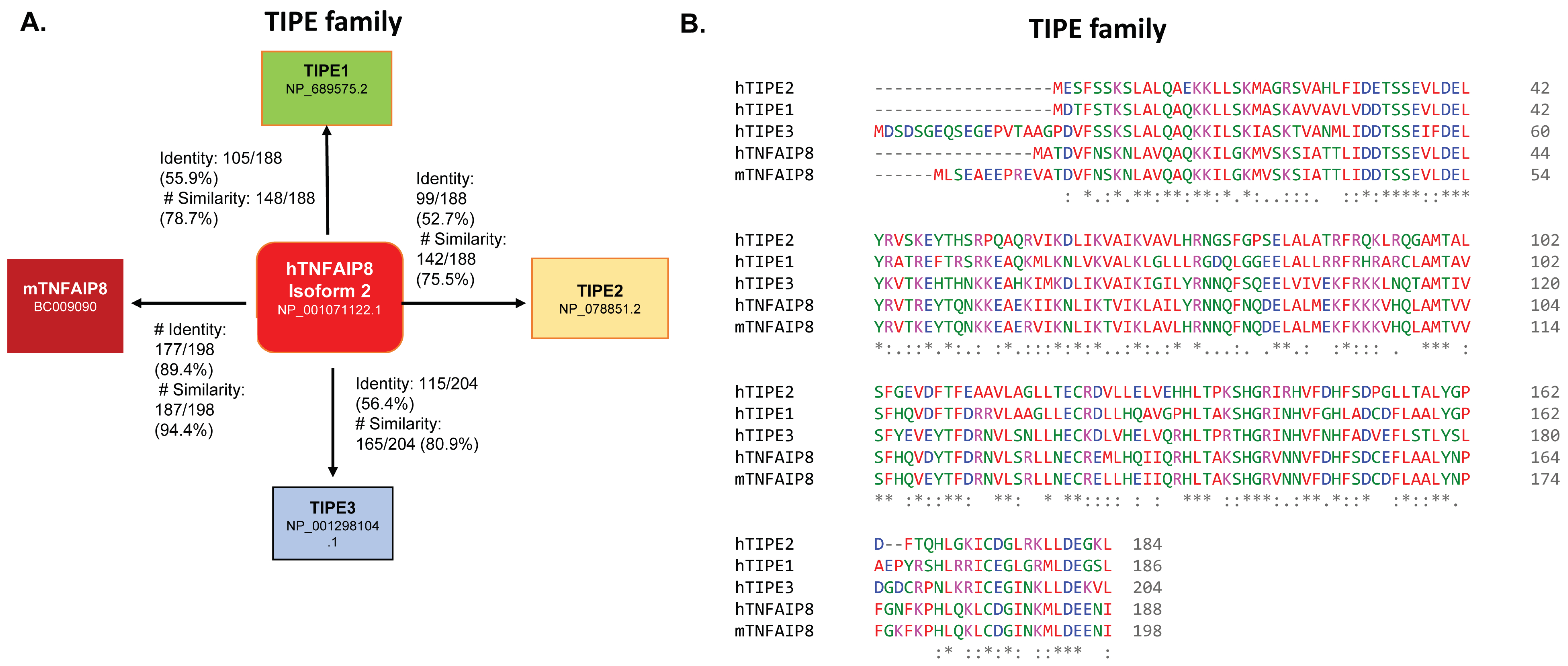
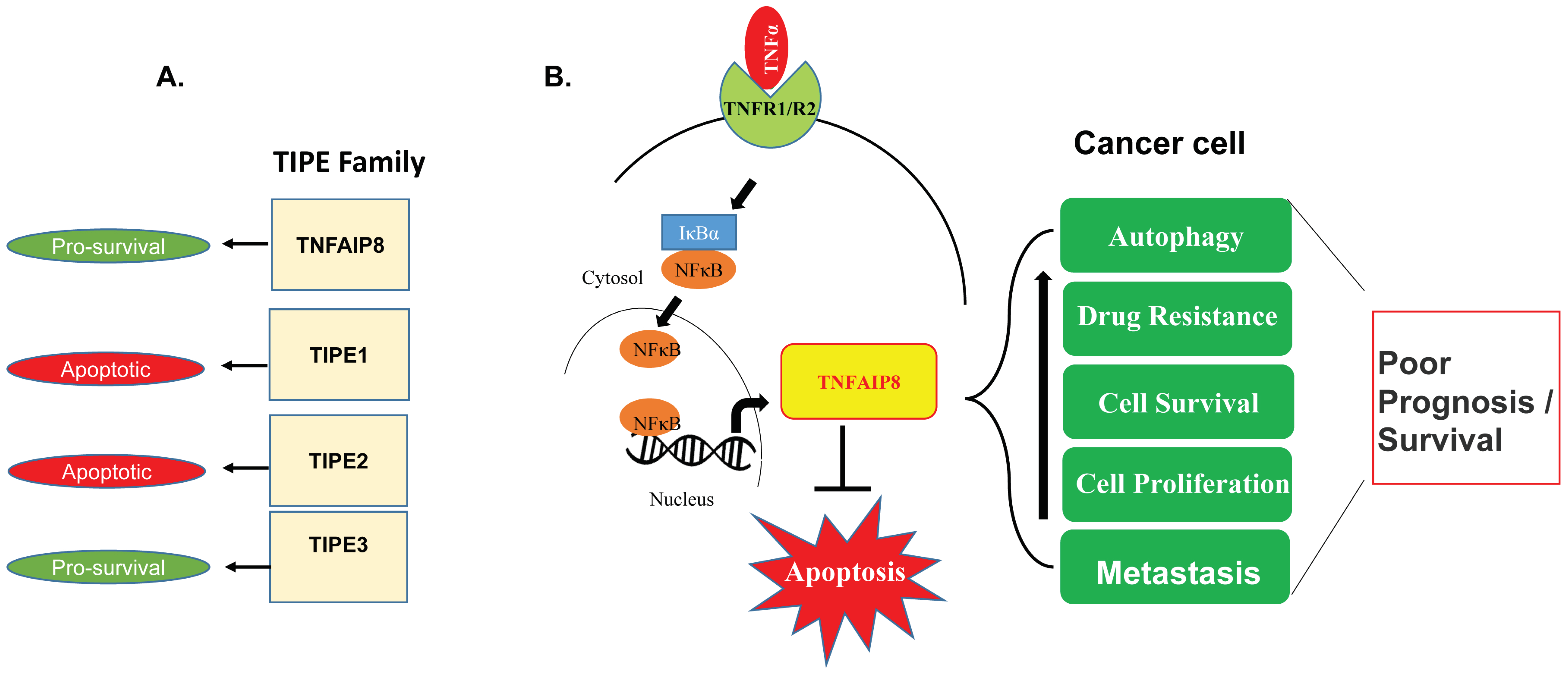
2. TIPE Family Proteins: TIPE1, TIPE2, and TIPE3 and Their Brief Roles in Human Disease
3. Tumor Necrosis Factor α-Inducing Protein 8 (TNFAIP8)
3.1. TNFAIP8 Expression and Regulation
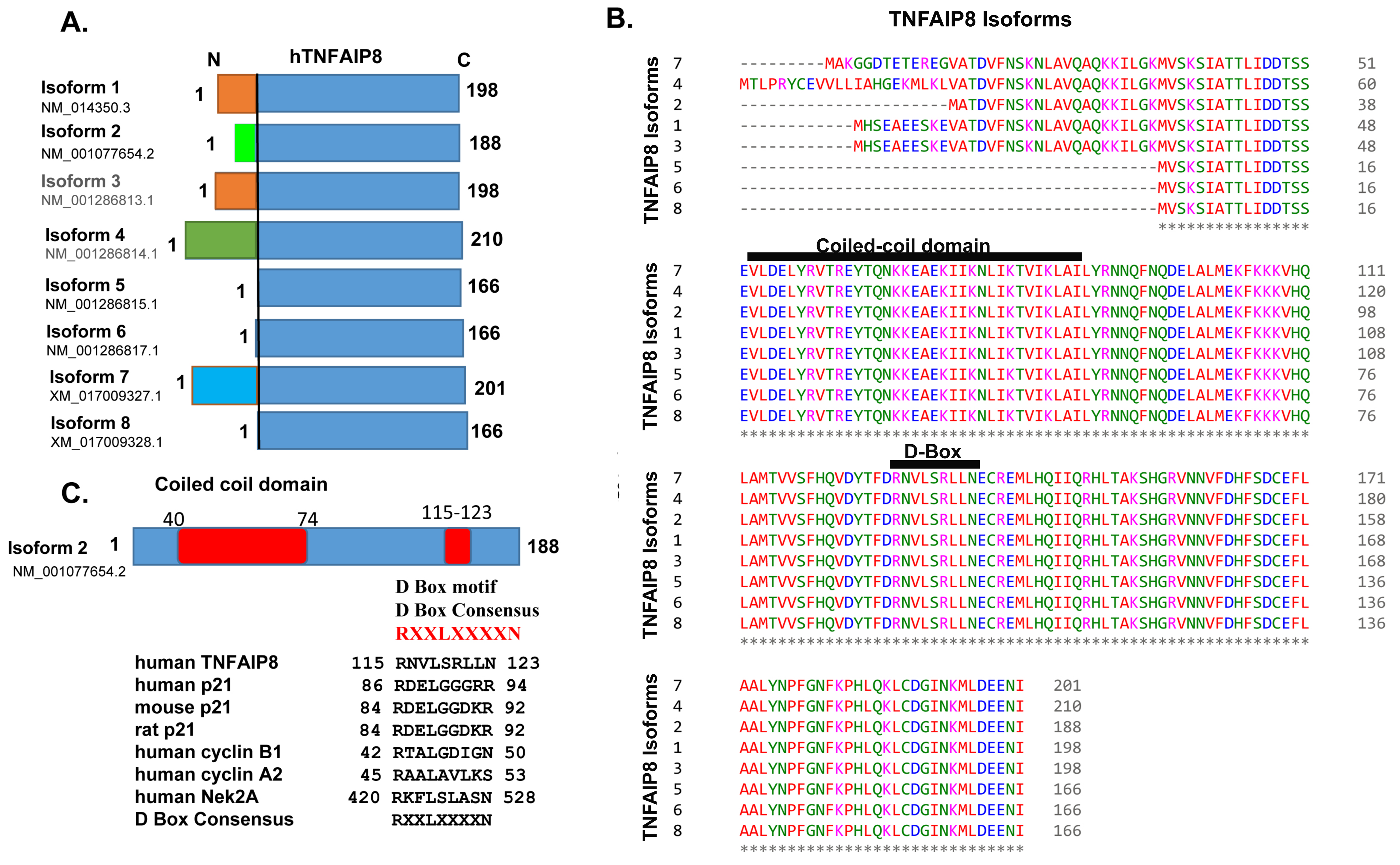
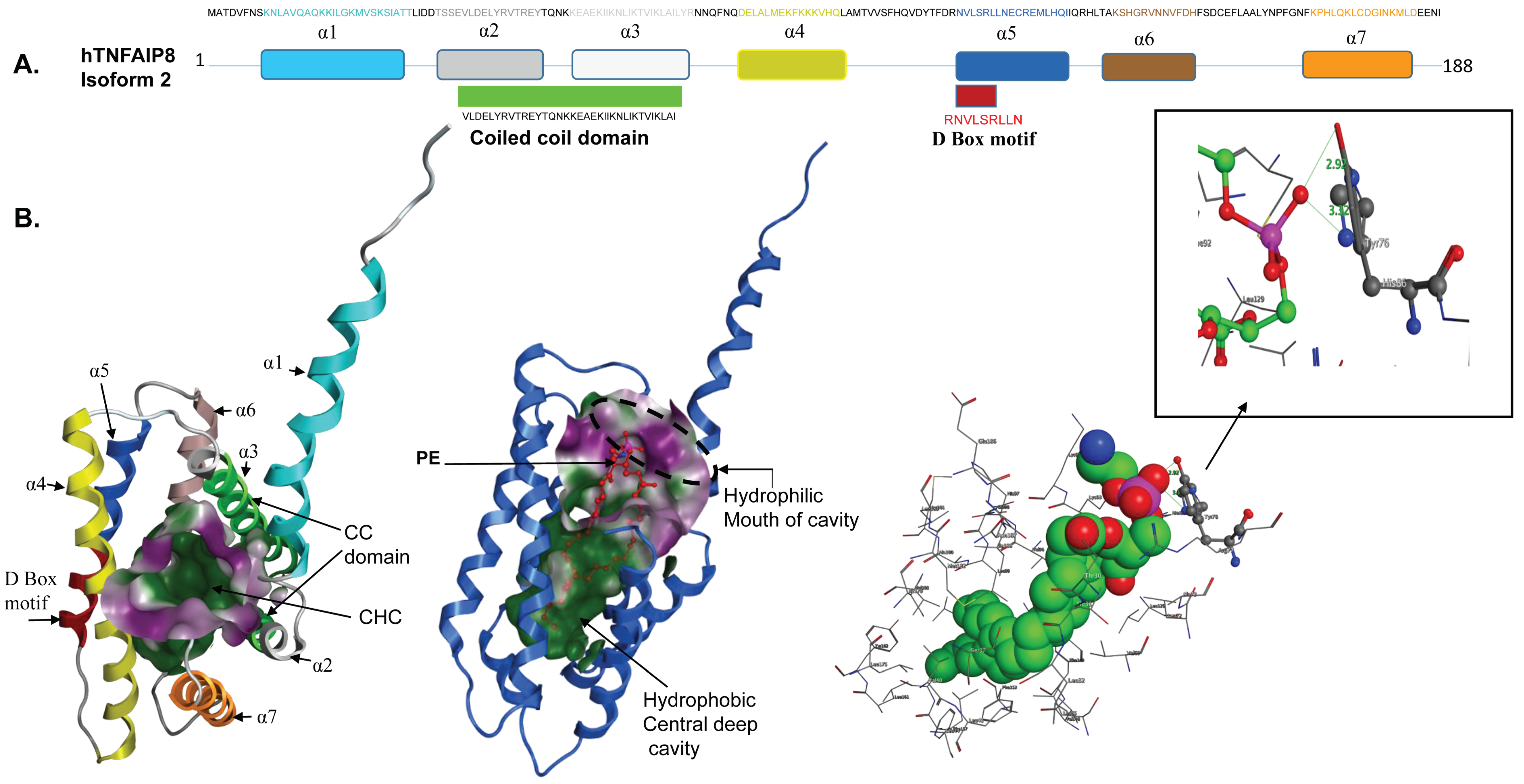
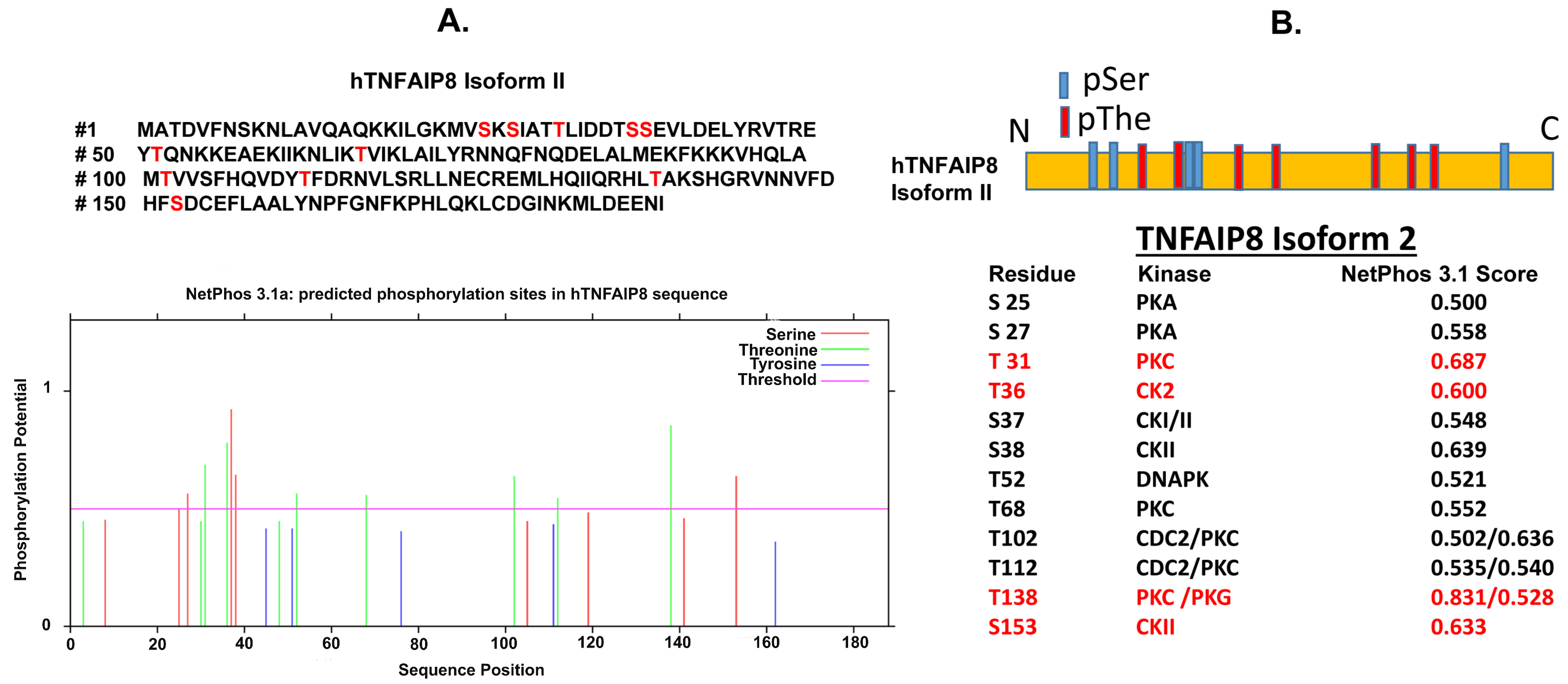
3.2. TNFAIP8 Structure
3.3. TNFAIP8 Interactions and Signaling
3.4. TNFAIP8 Modification
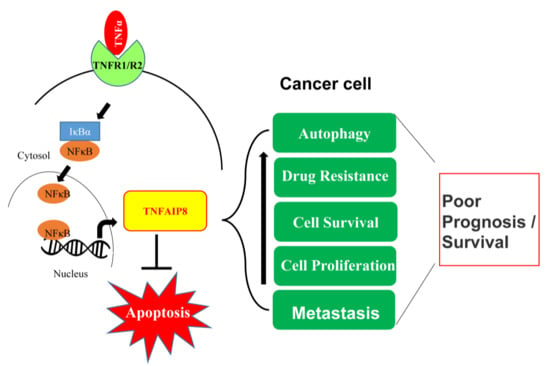
4. Oncogenic Roles of TNFAIP8
5. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Germano, G.; Allavena, P.; Mantovani, A. Cytokines as a key component of cancer-related inflammation. Cytokine 2008, 43, 374–379. [Google Scholar] [CrossRef] [PubMed]
- Colotta, F.; Allavena, P.; Sica, A.; Garlanda, C.; Mantovani, A. Cancer-related inflammation, the seventh hallmark of cancer: Links to genetic instability. Carcinogenesis 2009, 30, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Garlanda, C.; Allavena, P. Molecular pathways and targets in cancer-related inflammation. Ann. Med. 2010, 42, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Swardfager, W.; Lanctot, K.; Rothenburg, L.; Wong, A.; Cappell, J.; Herrmann, N. A meta-analysis of cytokines in Alzheimer’s disease. Biol. Psychiatry 2010, 68, 930–941. [Google Scholar] [CrossRef] [PubMed]
- Dowlati, Y.; Herrmann, N.; Swardfager, W.; Liu, H.; Sham, L.; Reim, E.K.; Lanctot, K.L. A meta-analysis of cytokines in major depression. Biol. Psychiatry 2010, 67, 446–457. [Google Scholar] [CrossRef] [PubMed]
- Brynskov, J.; Foegh, P.; Pedersen, G.; Ellervik, C.; Kirkegaard, T.; Bingham, A.; Saermark, T. Tumour necrosis factor alpha converting enzyme (TACE) activity in the colonic mucosa of patients with inflammatory bowel disease. Gut 2002, 51, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Hauner, H.; Petruschke, T.; Russ, M.; Rohrig, K.; Eckel, J. Effects of tumour necrosis factor alpha (TNF alpha) on glucose transport and lipid metabolism of newly-differentiated human fat cells in cell culture. Diabetologia 1995, 38, 764–771. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lin, Y. Tumor necrosis factor and cancer, buddies or foes? Acta Pharmacol. Sin. 2008, 29, 1275–1288. [Google Scholar] [CrossRef] [PubMed]
- Ham, B.; Fernandez, M.C.; D’Costa, Z.; Brodt, P. The diverse roles of the TNF axis in cancer progression and metastasis. Trends Cancer Res. 2016, 11, 1–27. [Google Scholar]
- Sidiropoulos, P.I.; Boumpas, D.T. Differential drug resistance to anti-tumour necrosis factor agents in rheumatoid arthritis. Ann. Rheum Dis. 2006, 65, 701–703. [Google Scholar] [CrossRef]
- Kumar, D.; Whiteside, T.L.; Kasid, U. Identification of a novel tumor necrosis factor-alpha-inducible gene, SCC-S2, containing the consensus sequence of a death effector domain of fas-associated death domain-like interleukin- 1beta-converting enzyme-inhibitory protein. J. Biol. Chem. 2000, 275, 2973–2978. [Google Scholar] [CrossRef] [PubMed]
- Day, T.F.; Mewani, R.R.; Starr, J.; Li, X.; Chakravarty, D.; Ressom, H.; Zou, X.; Eidelman, O.; Pollard, H.B.; Srivastava, M.; et al. Transcriptome and proteome analyses of TNFAIP8 knockdown cancer cells reveal new insights into molecular determinants of cell survival and tumor progression. Methods Mol. Biol. 2017, 1513, 83–100. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Liang, X.; Gao, L.; Ma, H.; Liu, X.; Pan, Y.; Yan, W.; Shan, H.; Wang, Z.; Chen, Y.H.; et al. TIPE1 induces apoptosis by negatively regulating Rac1 activation in hepatocellular carcinoma cells. Oncogene 2015, 34, 2566–2574. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, Y.; Hu, C.; Ni, X.; Huang, X. Tumor necrosis factor alpha-induced protein 8-like 1 promotes apoptosis by regulating B-cell leukemia/lymphoma-2 family proteins in RAW264.7 cells. Oncol. Lett. 2016, 12, 3506–3512. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Wang, F.H.; Whiteside, T.L.; Kasid, U. Identification of seven differentially displayed transcripts in human primary and matched metastatic head and neck squamous cell carcinoma cell lines: Implications in metastasis and/or radiation response. Oral Oncol. 1997, 33, 197–203. [Google Scholar] [CrossRef]
- Horrevoets, A.J.; Fontijn, R.D.; van Zonneveld, A.J.; de Vries, C.J.; ten Cate, J.W.; Pannekoek, H. Vascular endothelial genes that are responsive to tumor necrosis factor-alpha in vitro are expressed in atherosclerotic lesions, including inhibitor of apoptosis protein-1, stannin, and two novel genes. Blood 1999, 93, 3418–3431. [Google Scholar] [PubMed]
- Niture, S.; Ramalinga, M.; Kedir, H.; Patacsil, D.; Niture, S.S.; Li, J.; Mani, H.; Suy, S.; Collins, S.; Kumar, D. TNFAIP8 promotes prostate cancer cell survival by inducing autophagy. Oncotarget 2018, 9, 26884–26899. [Google Scholar] [CrossRef]
- Lowe, J.M.; Nguyen, T.A.; Grimm, S.A.; Gabor, K.A.; Peddada, S.D.; Li, L.; Anderson, C.W.; Resnick, M.A.; Menendez, D.; Fessler, M.B. The novel p53 target TNFAIP8 variant 2 is increased in cancer and offsets p53-dependent tumor suppression. Cell Death Differ. 2017, 24, 181–191. [Google Scholar] [CrossRef]
- Bradley, J.R. TNF-mediated inflammatory disease. J. Pathol. 2008, 214, 149–160. [Google Scholar] [CrossRef]
- Theiss, A.L.; Simmons, J.G.; Jobin, C.; Lund, P.K. Tumor necrosis factor (TNF) alpha increases collagen accumulation and proliferation in intestinal myofibroblasts via TNF receptor 2. J. Biol. Chem. 2005, 280, 36099–36109. [Google Scholar] [CrossRef]
- Sun, H.; Gong, S.; Carmody, R.J.; Hilliard, A.; Li, L.; Sun, J.; Kong, L.; Xu, L.; Hilliard, B.; Hu, S.; et al. TIPE2, a negative regulator of innate and adaptive immunity that maintains immune homeostasis. Cell 2008, 133, 415–426. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Kallakury, B.V.; Ross, J.S.; Mewani, R.R.; Sheehan, C.E.; Sakabe, I.; Luta, G.; Kumar, D.; Yadavalli, S.; Starr, J.; et al. The significance of TNFAIP8 in prostate cancer response to radiation and docetaxel and disease recurrence. Int. J. Cancer 2013, 133, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, C.; Lage, C.R.; Yoder, J.A.; Postlethwait, J.H.; Kim, C.H. Evolutionary divergence of the vertebrate TNFAIP8 gene family: Applying the spotted gar orthology bridge to understand ohnolog loss in teleosts. PLoS ONE 2017, 12, e0179517. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Zhang, G.; Hao, C.; Wang, Y.; Lou, Y.; Zhang, W.; Wang, J.; Liu, S. The expression of TIPE1 in murine tissues and human cell lines. Mol. Immunol. 2011, 48, 1548–1555. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Ha, J.Y.; Kim, J.S.; Kang, Y.H.; Bok, E.; Kim, Y.S.; Son, J.H. Tnfaip8 l1/Oxi-beta binds to FBXW5, increasing autophagy through activation of TSC2 in a Parkinson’s disease model. J. Neurochem. 2014, 129, 527–538. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Ma, Y.; Cheng, J.; Li, X.; Zheng, H.; Jiang, L.; Zhou, R. TIPE1 function as a prognosis predictor and negative regulator of lung cancer. Oncotarget 2017, 8, 78496–78506. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Song, L.; Fan, Y.; Li, X.; Li, Y.; Chen, J.; Zhu, F.; Guo, C.; Shi, Y.; Zhang, L. Down-regulation of TIPE2 mRNA expression in peripheral blood mononuclear cells from patients with systemic lupus erythematosus. Clin. Immunol. 2009, 133, 422–427. [Google Scholar] [CrossRef]
- Ma, Y.; Liu, X.; Wei, Z.; Wang, X.; Wang, Z.; Zhong, W.; Li, Y.; Zhu, F.; Guo, C.; Zhang, L.; et al. The expression and significance of TIPE2 in peripheral blood mononuclear cells from asthmatic children. Scand. J. Immunol. 2013, 78, 523–528. [Google Scholar] [CrossRef]
- Xi, W.; Hu, Y.; Liu, Y.; Zhang, J.; Wang, L.; Lou, Y.; Qu, Z.; Cui, J.; Zhang, G.; Liang, X.; et al. Roles of TIPE2 in hepatitis B virus-induced hepatic inflammation in humans and mice. Mol. Immunol. 2011, 48, 1203–1208. [Google Scholar] [CrossRef]
- Qin, B.; Wei, T.; Wang, L.; Ma, N.; Tang, Q.; Liang, Y.; Yang, Z.; Zhou, L.; Zhong, R. Decreased expression of TIPE2 contributes to the hyperreactivity of monocyte to Toll-like receptor ligands in primary biliary cirrhosis. J. Gastroenterol. Hepatol. 2016, 31, 1177–1183. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhu, T.; Liu, W.; Qu, X.; Chen, Y.; Ren, P.; Wang, Z.; Wei, X.; Zhang, Y.; Yi, F. TIPE2 acts as a negative regulator linking NOD2 and inflammatory responses in myocardial ischemia/reperfusion injury. J. Mol. Med. 2015, 93, 1033–1043. [Google Scholar] [CrossRef] [PubMed]
- Gus-Brautbar, Y.; Johnson, D.; Zhang, L.; Sun, H.; Wang, P.; Zhang, S.; Zhang, L.; Chen, Y.H. The anti-inflammatory TIPE2 is an inhibitor of the oncogenic Ras. Mol. Cell 2012, 45, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Zhao, M.; Dong, T.; Zhou, C.; Peng, Y.; Zhou, X.; Fan, B.; Ma, W.; Han, M.; Liu, S. Tumor necrosis factor-alpha-induced protein-8 like-2 (TIPE2) upregulates p27 to decrease gastic cancer cell proliferation. J. Cell Biochem. 2015, 116, 1121–1129. [Google Scholar] [CrossRef]
- Liu, Q.Q.; Zhang, F.F.; Wang, F.; Qiu, J.H.; Luo, C.H.; Zhu, G.Y.; Liu, Y.F. TIPE2 Inhibits lung cancer growth attributing to promotion of apoptosis by regulating some apoptotic molecules expression. PLoS ONE 2015, 10, e0126176. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Qi, H.; Hou, S.; Jin, X. TIPE2 mRNA overexpression correlates with TNM staging in renal cell carcinoma tissues. Oncol. Lett. 2013, 6, 571–575. [Google Scholar] [CrossRef] [PubMed]
- Li, X.M.; Su, J.R.; Yan, S.P.; Cheng, Z.L.; Yang, T.T.; Zhu, Q. A novel inflammatory regulator TIPE2 inhibits TLR4-mediated development of colon cancer via caspase-8. Cancer Biomark 2014, 14, 233–240. [Google Scholar] [CrossRef]
- Zhu, Y.; Tao, M.; Wu, J.; Meng, Y.; Xu, C.; Tian, Y.; Zhou, X.; Xiang, J.; Zhang, H.; Xie, Y. Adenovirus-directed expression of TIPE2 suppresses gastric cancer growth via induction of apoptosis and inhibition of AKT and ERK1/2 signaling. Cancer Gene Ther. 2016, 23, 98–106. [Google Scholar] [CrossRef]
- Fayngerts, S.A.; Wu, J.; Oxley, C.L.; Liu, X.; Vourekas, A.; Cathopoulis, T.; Wang, Z.; Cui, J.; Liu, S.; Sun, H.; et al. TIPE3 is the transfer protein of lipid second messengers that promote cancer. Cancer Cell 2014, 26, 465–478. [Google Scholar] [CrossRef]
- Cui, J.; Hao, C.; Zhang, W.; Shao, J.; Zhang, N.; Zhang, G.; Liu, S. Identical expression profiling of human and murine TIPE3 protein reveals links to its functions. J. Histochem. Cytochem. 2015, 63, 206–216. [Google Scholar] [CrossRef]
- Lian, K.; Ma, C.; Hao, C.; Li, Y.; Zhang, N.; Chen, Y.H.; Liu, S. TIPE3 protein promotes breast cancer metastasis through activating AKT and NF-κB signaling pathways. Oncotarget 2017, 8, 48889–48904. [Google Scholar] [CrossRef] [PubMed]
- You, Z.; Ouyang, H.; Lopatin, D.; Polver, P.J.; Wang, C.Y. Nuclear factor-κB-inducible death effector domain-containing protein suppresses tumor necrosis factor-mediated apoptosis by inhibiting caspase-8 activity. J. Biol. Chem. 2001, 276, 26398–26404. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.J.; Liu, X.; Gafken, P.R.; Kioussi, C.; Leid, M. A chicken ovalbumin upstream promoter transcription factor I (COUP-TFI) complex represses expression of the gene encoding tumor necrosis factor alpha-induced protein 8 (TNFAIP8). J. Biol. Chem. 2009, 284, 6156–6168. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Yu, P.; Duan, X.; Liu, C.; Xu, S.; Chen, Y.; Tan, Y.; Qiang, Y.; Shen, J.; Tao, Z. Genome-wide analysis of androgen receptor binding sites in prostate cancer cells. Exp. Ther. Med. 2015, 9, 2319–2324. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yu, Q.; Cho, A.H.; Rondeau, G.; Welsh, J.; Adamson, E.; Mercola, D.; McClelland, M. Survey of differentially methylated promoters in prostate cancer cell lines. Neoplasia 2005, 7, 748–760. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Yu, Y.H.; Zhang, Z.H.; Yang, Y. Significant hypomethylation of TNFAIP8 and increased expression in the placenta and peripheral blood cells from early-onset preeclamptic patients. Int. J. Clin. Exp. Med. 2016, 9, 10384. [Google Scholar]
- Salihu, H.M.; Das, R.; Morton, L.; Huang, H.; Paothong, A.; Wilson, R.E.; Aliyu, M.H.; Salemi, J.L.; Marty, P.J. Racial differences in DNA-methylation of CpG sites within preterm-promoting genes and gene variants. Matern Child Health J. 2016, 20, 1680–1687. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zheng, Q.; Deng, Y.; Cheng, C.S.; Kallenbach, N.R.; Lu, M. A seven-helix coiled coil. Proc. Natl. Acad Sci. USA 2006, 103, 15457–15462. [Google Scholar] [CrossRef]
- Burkhard, P.; Stetefeld, J.; Strelkov, S.V. Coiled coils: A highly versatile protein folding motif. Trends Cell Biol. 2001, 11, 82–88. [Google Scholar] [CrossRef]
- Gustems, M.; Woellmer, A.; Rothbauer, U.; Eck, S.H.; Wieland, T.; Lutter, D.; Hammerschmidt, W. c-Jun/c-Fos heterodimers regulate cellular genes via a newly identified class of methylated DNA sequence motifs. Nucleic Acids Res. 2014, 42, 3059–3072. [Google Scholar] [CrossRef]
- Amador, V.; Ge, S.; Santamaria, P.G.; Guardavaccaro, D.; Pagano, M. APC/C(Cdc20) controls the ubiquitin-mediated degradation of p21 in prometaphase. Mol. Cell 2007, 27, 462–473. [Google Scholar] [CrossRef]
- Yang, C.H.; Kuo, W.T.; Chuang, Y.T.; Chen, C.Y.; Lin, C.C. Cyclin B1 destruction box-mediated protein instability: The enhanced sensitivity of fluorescent-protein-based reporter gene system. Biomed. Res. Int. 2013, 2013, 732307. [Google Scholar] [CrossRef]
- Geley, S.; Kramer, E.; Gieffers, C.; Gannon, J.; Peters, J.M.; Hunt, T. Anaphase-promoting complex/cyclosome-dependent proteolysis of human cyclin A starts at the beginning of mitosis and is not subject to the spindle assembly checkpoint. J. Cell Biol. 2001, 153, 137–148. [Google Scholar] [CrossRef]
- Hames, R.S.; Wattam, S.L.; Yamano, H.; Bacchieri, R.; Fry, A.M. APC/C-mediated destruction of the centrosomal kinase Nek2A occurs in early mitosis and depends upon a cyclin A-type D-box. EMBO J. 2001, 20, 7117–7127. [Google Scholar] [CrossRef]
- Kuo, P.L.; Huang, Y.L.; Hsieh, C.C.; Lee, J.C.; Lin, B.W.; Hung, L.Y. STK31 is a cell-cycle regulated protein that contributes to the tumorigenicity of epithelial cancer cells. PLoS ONE 2014, 9, e93303. [Google Scholar] [CrossRef]
- Harper, J.W.; Burton, J.L.; Solomon, M.J. The anaphase-promoting complex: It’s not just for mitosis any more. Genes Dev. 2002, 16, 2179–2206. [Google Scholar] [CrossRef]
- Dong, Q.; Fu, L.; Zhao, Y.; Xie, C.; Li, Q.; Wang, E. TNFAIP8 interacts with LATS1 and promotes aggressiveness through regulation of Hippo pathway in hepatocellular carcinoma. Oncotarget 2017, 8, 15689–15703. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, J.; Fan, C.; Li, H.; Sun, H.; Gong, S.; Chen, Y.H.; Shi, Y. Crystal structure of TIPE2 provides insights into immune homeostasis. Nat. Struct. Mol. Biol. 2009, 16, 89–90. [Google Scholar] [CrossRef]
- Kim, J.S.; Park, J.; Kim, M.S.; Ha, J.Y.; Jang, Y.W.; Shin, D.H.; Son, J.H. The Tnfaip8-PE complex is a novel upstream effector in the anti-autophagic action of insulin. Sci. Rep. 2017, 7, 6248. [Google Scholar] [CrossRef]
- Laliberte, B.; Wilson, A.M.; Nafisi, H.; Mao, H.; Zhou, Y.Y.; Daigle, M.; Albert, P.R. TNFAIP8: A new effector for Galpha(i) coupling to reduce cell death and induce cell transformation. J. Cell Physiol. 2010, 225, 865–874. [Google Scholar] [CrossRef]
- Chittaranjan, S.; Xu, J.; Kuzyk, M.; Dullat, H.K.; Wilton, J.; DeVorkin, L.; Lebovitz, C.; Morin, G.B.; Marra, M.A.; Gorski, S.M. The Drosophila TIPE family member Sigmar interacts with the Ste20-like kinase Misshapen and modulates JNK signaling, cytoskeletal remodeling and autophagy. Biol. Open 2015, 4, 672–684. [Google Scholar] [CrossRef]
- Monastyrska, I.; Rieter, E.; Klionsky, D.J.; Reggiori, F. Multiple roles of the cytoskeleton in autophagy. Biol. Rev. Camb. Philos. Soc. 2009, 84, 431–448. [Google Scholar] [CrossRef]
- Kristensen, A.R.; Gsponer, J.; Foster, L.J. A high-throughput approach for measuring temporal changes in the interactome. Nat. Methods 2012, 9, 907–909. [Google Scholar] [CrossRef]
- Rolland, T.; Tasan, M.; Charloteaux, B.; Pevzner, S.J.; Zhong, Q.; Sahni, N.; Yi, S.; Lemmens, I.; Fontanillo, C.; Mosca, R.; et al. A proteome-scale map of the human interactome network. Cell 2014, 159, 1212–1226. [Google Scholar] [CrossRef]
- Ewing, R.M.; Chu, P.; Elisma, F.; Li, H.; Taylor, P.; Climie, S.; McBroom-Cerajewski, L.; Robinson, M.D.; O’Connor, L.; Li, M.; et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol. Syst. Biol. 2007, 3, 89. [Google Scholar] [CrossRef]
- Padmavathi, G.; Banik, K.; Monisha, J.; Bordoloi, D.; Bano, S.; Arfuso, F.; Sethi, G.; Lu, F.; Kunnumakkara, A.B. Novel tumor necrosis factor-alpha induced protein eight (TNFAIP8/TIPE) family: Functions and downstream targets involved in cancer progression. Cancer Lett. 2018, 432, 260–271. [Google Scholar] [CrossRef]
- Kumar, D.; Gokhale, P.; Broustas, C.; Chakravarty, D.; Ahmad, I.; Kasid, U. Expression of SCC-S2, an antiapoptotic molecule, correlates with enhanced proliferation and tumorigenicity of MDA-MB 435 cells. Oncogene 2004, 23, 612–616. [Google Scholar] [CrossRef]
- Zhang, C.; Chakravarty, D.; Sakabe, I.; Mewani, R.R.; Boudreau, H.E.; Kumar, D.; Ahmad, I.; Kasid, U.N. Role of SCC-S2 in experimental metastasis and modulation of VEGFR-2, MMP-1, and MMP-9 expression. Mol. Ther. 2006, 13, 947–955. [Google Scholar] [CrossRef]
- Xiao, M.; Xu, Q.; Lou, C.; Qin, Y.; Ning, X.; Liu, T.; Zhao, X.; Jia, S.; Huang, Y. Overexpression of TNFAIP8 is associated with tumor aggressiveness and poor prognosis in patients with invasive ductal breast carcinoma. Hum. Pathol. 2017, 62, 40–49. [Google Scholar] [CrossRef]
- Romanuik, T.L.; Ueda, T.; Le, N.; Haile, S.; Yong, T.M.; Thomson, T.; Vessella, R.L.; Sadar, M.D. Novel biomarkers for prostate cancer including noncoding transcripts. Am. J. Pathol. 2009, 175, 2264–2276. [Google Scholar] [CrossRef]
- Wang, L.; Song, Y.; Men, X. Variance of TNFAIP8 expression between tumor tissues and tumor-infiltrating CD4+ and CD8+ T cells in non-small cell lung cancer. Tumour Biol. 2014, 35, 2319–2325. [Google Scholar] [CrossRef]
- Han, Y.; Tang, Z.; Zhao, Y.; Li, Q.; Wang, E. TNFAIP8 regulates Hippo pathway through interacting with LATS1 to promote cell proliferation and invasion in lung cancer. Mol. Carcinog. 2018, 57, 159–166. [Google Scholar] [CrossRef]
- Monteith, J.A.; Mellert, H.; Sammons, M.A.; Kuswanto, L.A.; Sykes, S.M.; Resnick-Silverman, L.; Manfredi, J.J.; Berger, S.L.; McMahon, S.B. A rare DNA contact mutation in cancer confers p53 gain-of-function and tumor cell survival via TNFAIP8 induction. Mol. Oncol. 2016, 10, 1207–1220. [Google Scholar] [CrossRef]
- Porturas, T.P.; Sun, H.; Buchlis, G.; Lou, Y.; Liang, X.; Cathopoulis, T.; Fayngerts, S.; Johnson, D.S.; Wang, Z.; Chen, Y.H. Crucial roles of TNFAIP8 protein in regulating apoptosis and Listeria infection. J. Immunol. 2015, 194, 5743–5750. [Google Scholar] [CrossRef]
- Gao, H.Y.; Huo, F.C.; Wang, H.Y.; Pei, D.S. MicroRNA-9 inhibits the gastric cancer cell proliferation by targeting TNFAIP8. Cell Prolif. 2017, 50, e12331. [Google Scholar] [CrossRef]
- Yang, M.; Zhao, Q.; Wang, X.; Liu, T.; Yao, G.; Lou, C.; Zhang, Y. TNFAIP8 overexpression is associated with lymph node metastasis and poor prognosis in intestinal-type gastric adenocarcinoma. Histopathology 2014, 65, 517–526. [Google Scholar] [CrossRef]
- Xing, B.; Ren, C. Tumor-suppressive miR-99a inhibits cell proliferation via targeting of TNFAIP8 in osteosarcoma cells. Am. J. Transl. Res. 2016, 8, 1082–1090. [Google Scholar]
- Sun, Z.; Liu, X.; Song, J.H.; Cheng, Y.; Liu, Y.; Jia, Y.; Meltzer, S.J.; Wang, Z. TNFAIP8 overexpression: A potential predictor of lymphatic metastatic recurrence in pN0 esophageal squamous cell carcinoma after Ivor Lewis esophagectomy. Tumour Biol. 2016, 37, 10923–10934. [Google Scholar] [CrossRef]
- Hadisaputri, Y.E.; Miyazaki, T.; Suzuki, S.; Yokobori, T.; Kobayashi, T.; Tanaka, N.; Inose, T.; Sohda, M.; Kuwano, H. TNFAIP8 overexpression: Clinical relevance to esophageal squamous cell carcinoma. Ann. Surg. Oncol. 2012, 19, 589–596. [Google Scholar] [CrossRef]
- Liu, T.; Xia, B.; Lu, Y.; Xu, Y.; Lou, G. TNFAIP8 overexpression is associated with platinum resistance in epithelial ovarian cancers with optimal cytoreduction. Hum. Pathol. 2014, 45, 1251–1257. [Google Scholar] [CrossRef]
- Liu, T.; Gao, H.; Chen, X.; Lou, G.; Gu, L.; Yang, M.; Xia, B.; Yin, H. TNFAIP8 as a predictor of metastasis and a novel prognostic biomarker in patients with epithelial ovarian cancer. Br. J. Cancer 2013, 109, 1685–1692. [Google Scholar] [CrossRef]
- Liu, T.; Gao, H.; Yang, M.; Zhao, T.; Liu, Y.; Lou, G. Correlation of TNFAIP8 overexpression with the proliferation, metastasis, and disease-free survival in endometrial cancer. Tumour Biol. 2014, 35, 5805–5814. [Google Scholar] [CrossRef]
- Shi, T.Y.; Cheng, X.; Yu, K.D.; Sun, M.H.; Shao, Z.M.; Wang, M.Y.; Zhu, M.L.; He, J.; Li, Q.X.; Chen, X.J.; et al. Functional variants in TNFAIP8 associated with cervical cancer susceptibility and clinical outcomes. Carcinogenesis 2013, 34, 770–778. [Google Scholar] [CrossRef]
- Liu, K.; Qin, C.K.; Wang, Z.Y.; Liu, S.X.; Cui, X.P.; Zhang, D.Y. Expression of tumor necrosis factor-alpha-induced protein 8 in pancreas tissues and its correlation with epithelial growth factor receptor levels. Asian-Pac. J. Cancer Prev. 2012, 13, 847–850. [Google Scholar] [CrossRef]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niture, S.; Dong, X.; Arthur, E.; Chimeh, U.; Niture, S.S.; Zheng, W.; Kumar, D. Oncogenic Role of Tumor Necrosis Factor α-Induced Protein 8 (TNFAIP8). Cells 2019, 8, 9. https://doi.org/10.3390/cells8010009
Niture S, Dong X, Arthur E, Chimeh U, Niture SS, Zheng W, Kumar D. Oncogenic Role of Tumor Necrosis Factor α-Induced Protein 8 (TNFAIP8). Cells. 2019; 8(1):9. https://doi.org/10.3390/cells8010009
Chicago/Turabian StyleNiture, Suryakant, Xialan Dong, Elena Arthur, Uchechukwu Chimeh, Samiksha S. Niture, Weifan Zheng, and Deepak Kumar. 2019. "Oncogenic Role of Tumor Necrosis Factor α-Induced Protein 8 (TNFAIP8)" Cells 8, no. 1: 9. https://doi.org/10.3390/cells8010009
APA StyleNiture, S., Dong, X., Arthur, E., Chimeh, U., Niture, S. S., Zheng, W., & Kumar, D. (2019). Oncogenic Role of Tumor Necrosis Factor α-Induced Protein 8 (TNFAIP8). Cells, 8(1), 9. https://doi.org/10.3390/cells8010009