SpermQ–A Simple Analysis Software to Comprehensively Study Flagellar Beating and Sperm Steering
Abstract
1. Introduction
2. Materials and Methods
2.1. Mouse Sperm
2.2. Human Sperm
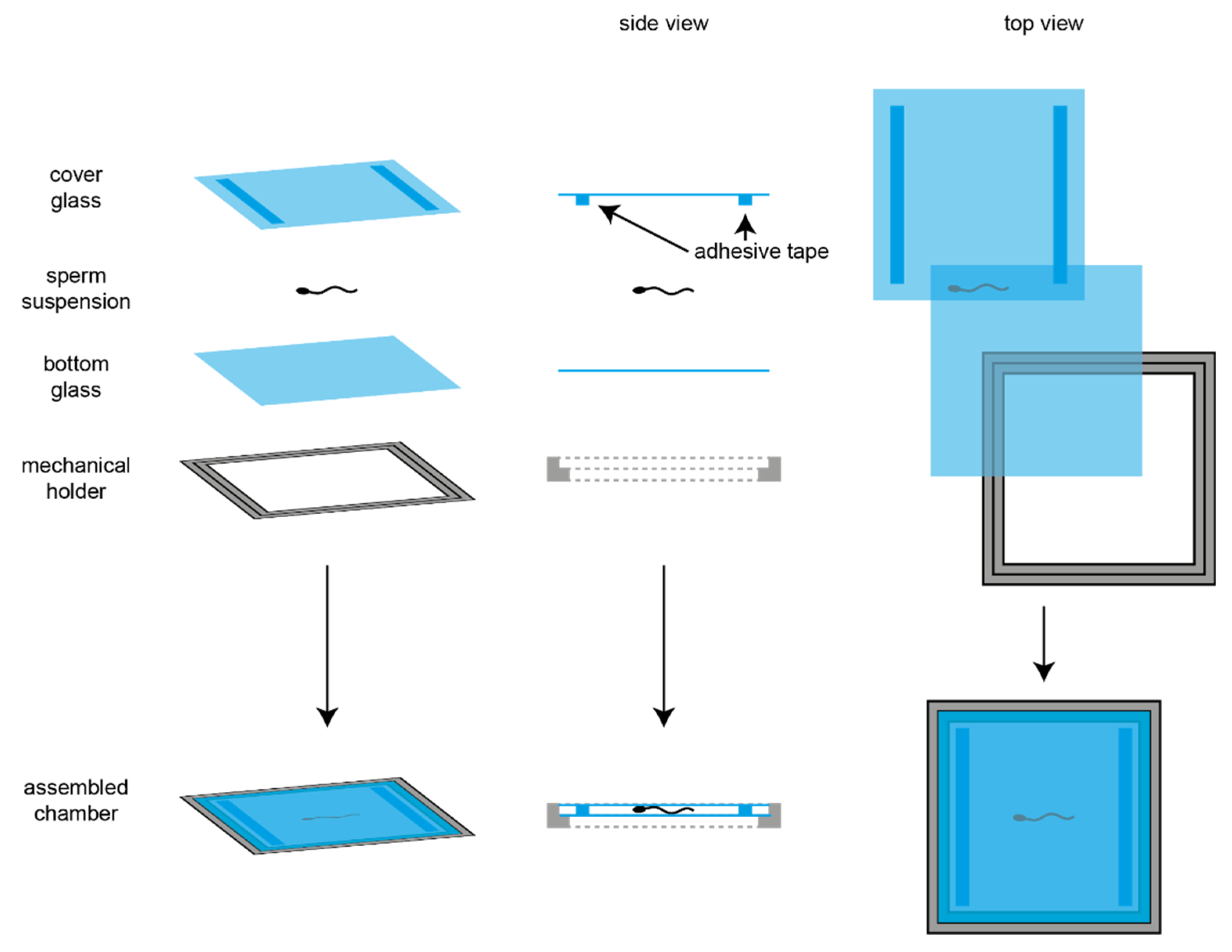
2.3. Imaging
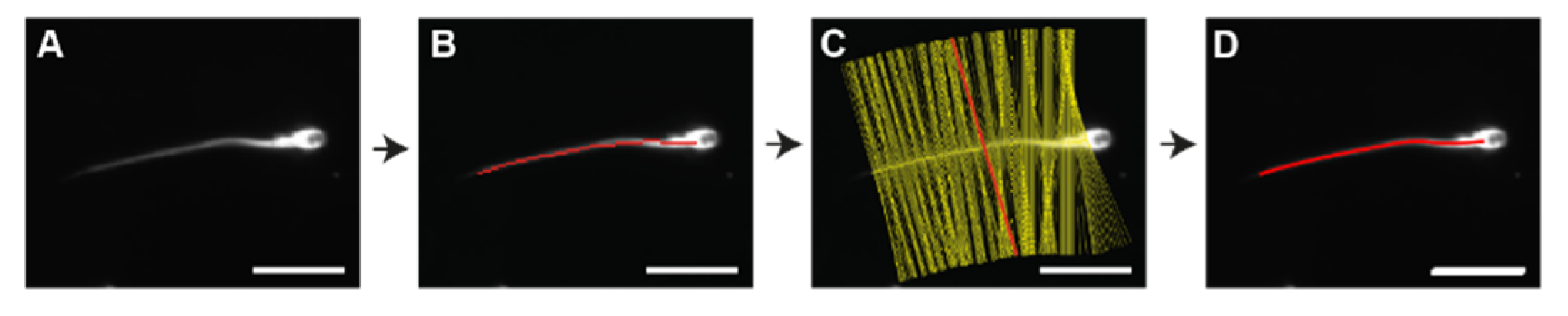
2.4. Image Analysis
2.5. Analysis Workflow Underpinning SpermQ
2.6. Software
2.7. Software Availability
3. Results
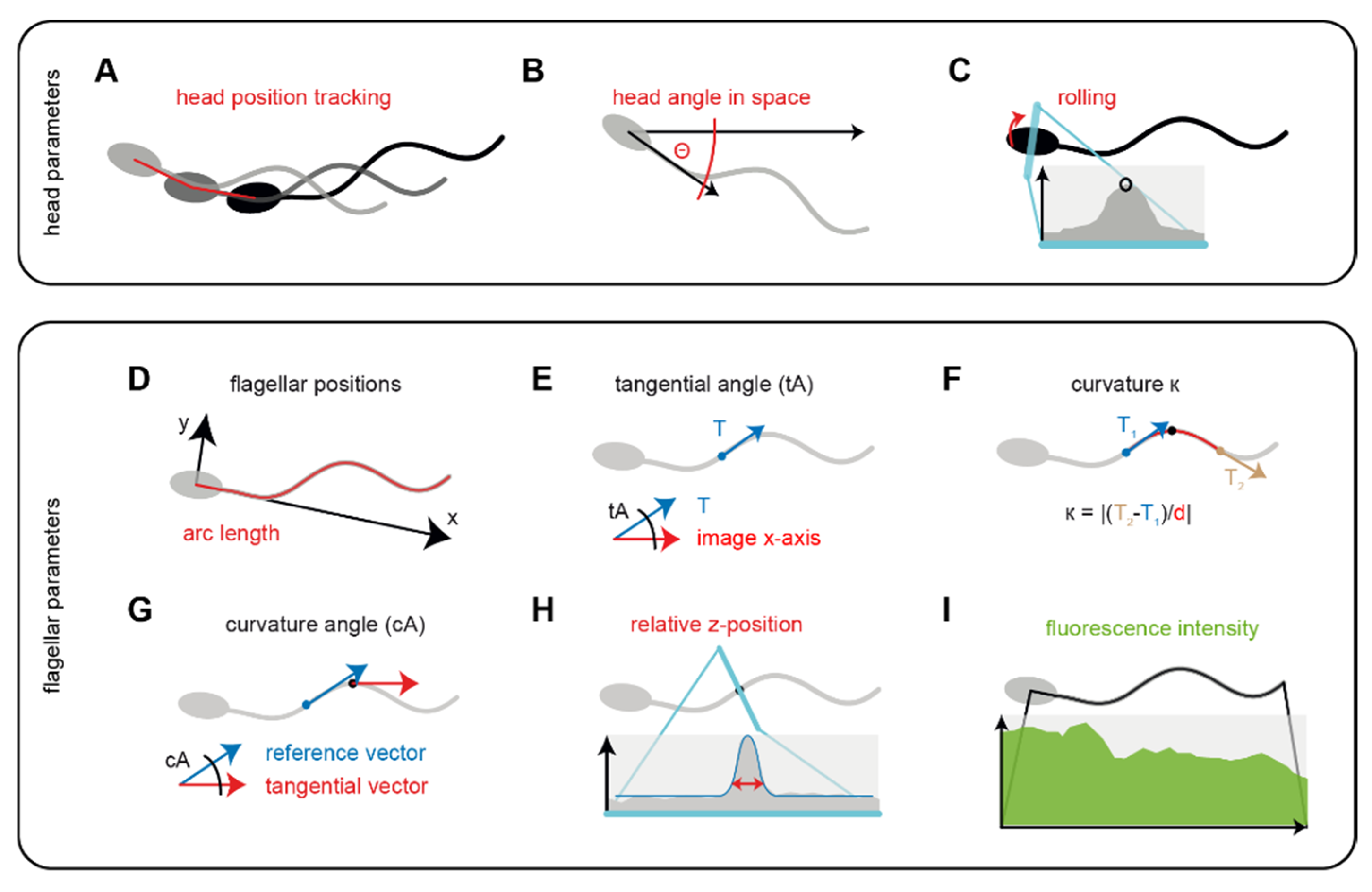
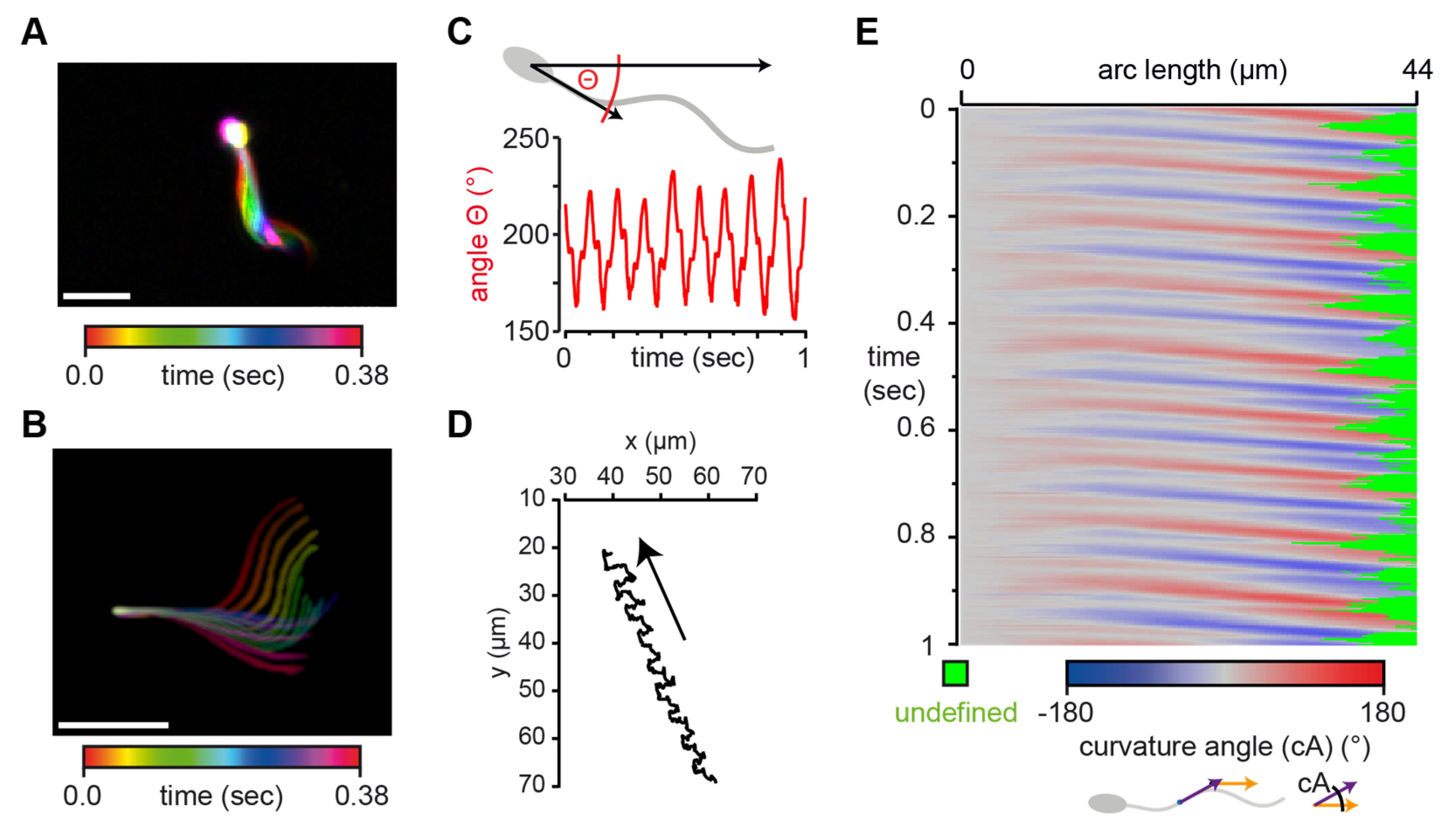
3.1. Analysis and Workflow Performed by the Software SpermQ
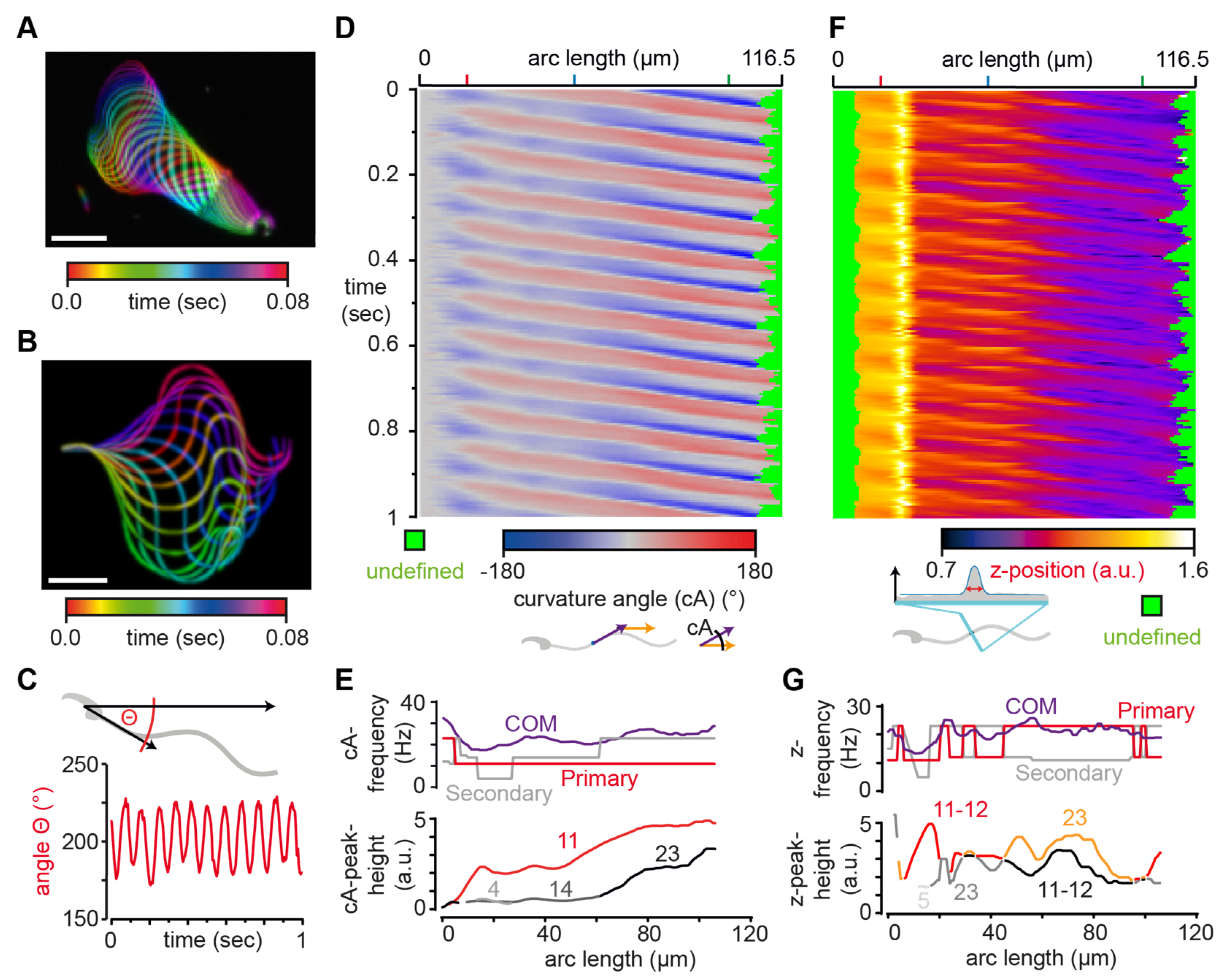
3.2. SpermQ Analysis of the Beat of a Head-Tethered Mouse Sperm
3.3. SpermQ Analysis of the Beat of a Freely-Swimming Human Sperm
3.4. SpermQ Is a Readily-Applicable Software
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wachten, D.; Jikeli, J.F.; Kaupp, U.B. Sperm Sensory Signaling. Cold Spring Harb. Perspect. Biol. 2017, 9, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, L.; Benjamin, M.F.; Gompper, G.; Kaupp, U.B. The computational sperm cell. Trends Cell Biol. 2014, 24, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, B.M.; Riedel-Kruse, I.H.; Howard, J.; Jülicher, F. High-precision tracking of sperm swimming fine structure provides strong test of resistive force theory. J. Exp. Biol. 2010, 213, 1226–1234. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, S.F. Asymmetric waveforms in echinoderm sperm flagella. J. Exp. Biol. 1977, 71, 157–170. [Google Scholar] [PubMed]
- Jikeli, J.F.; Alvarez, L.; Friedrich, B.M.; Wilson, L.G.; Pascal, R.; Colin, R.; Pichlo, M.; Rennhack, A.; Brenker, C.; Kaupp, U.B. Sperm navigation along helical paths in 3D chemoattractant landscapes. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Rikmenspoel, R. The tail movement of bull spermatozoa. Observations and model calculations. Biophys. J. 1965, 5, 365–392. [Google Scholar] [CrossRef]
- Rikmenspoel, R.; van Harpen, G.; Eijkhout, P. Cinematographic observations of the movements of bull sperm cells. Phys. Med. Biol. 1960, 5, 167–181. [Google Scholar] [CrossRef]
- Böhmer, M.; Van, Q.; Weyand, I.; Hagen, V.; Beyermann, M.; Matsumoto, M.; Hoshi, M.; Hildebrand, E.; Kaupp, U.B. Ca2+ spikes in the flagellum control chemotactic behavior of sperm. EMBO J. 2005, 24, 2741–2752. [Google Scholar] [CrossRef]
- Bukatin, A.; Kukhtevich, I.; Stoop, N.; Dunkel, J.; Kantsler, V. Bimodal rheotactic behavior reflects flagellar beat asymmetry in human sperm cells. Proc. Natl. Acad. Sci. USA 2015, 112, 15904–15909. [Google Scholar] [CrossRef]
- Saggiorato, G.; Alvarez, L.; Jikeli, J.F.; Kaupp, U.B.; Gompper, G.; Elgeti, J. Human sperm steer with second harmonics of the flagellar beat. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef]
- Jansen, V.; Alvarez, L.; Balbach, M.; Strünker, T.; Hegemann, P.; Kaupp, U.B.; Wachten, D. Controlling fertilization and cAMP signaling in sperm by optogenetics. eLife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Krähling, A.M.; Alvarez, L.; Debowski, K.; Van, Q.; Gunkel, M.; Irsen, S.; Al-Amoudi, A.; Strünker, T.; Kremmer, E.; Krause, E.; et al. CRIS-a novel cAMP-binding protein controlling spermiogenesis and the development of flagellar bending. PLoS Genet. 2013, 9. [Google Scholar] [CrossRef]
- Chen, S.R.; Chen, M.; Deng, S.L.; Hao, X.X.; Wang, X.X.; Liu, Y.X. Sodium-hydrogen exchanger NHA1 and NHA2 control sperm motility and male fertility. Cell Death. Dis. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Hirano, Y.; Shibahara, H.; Shimada, K.; Yamanaka, S.; Suzuki, T.; Takamizawa, S.; Motoyama, M.; Suzuki, M. Accuracy of sperm velocity assessment using the Sperm Quality Analyzer V. Reprod. Med. Biol. 2003, 2, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Holt, W.; Watson, P.; Curry, M.; Holt, C. Reproducibility of computer-aided semen analysis: Comparison of five different systems used in a practical workshop. Fertil. Steril. 1994, 62, 1277–1282. [Google Scholar] [CrossRef]
- Wang, D.; King, S.M.; Quill, T.A.; Doolittle, L.K.; Garbers, D.L. A new sperm-specific Na+/H+ exchanger required for sperm motility and fertility. Nat. Cell Biol. 2003, 5, 1117–1122. [Google Scholar] [CrossRef] [PubMed]
- Balbach, M.; Beckert, V.; Hansen, J.N.; Wachten, D. Shedding light on the role of cAMP in mammalian sperm physiology. Mol. Cell. Endocrinol. 2018, 468, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Jansen, V.; Jikeli, J.F.; Hamzeh, H.; Alvarez, L.; Dombrowski, M.; Balbach, M.; Strünker, T.; Seifert, R.; Kaupp, U.B.; et al. A novel biosensor to study cAMP dynamics in cilia and flagella. eLife 2016, 5. [Google Scholar] [CrossRef]
- Strünker, T.; Goodwin, N.; Brenker, C.; Kashikar, N.D.; Weyand, I.; Seifert, R.; Kaupp, U.B. The CatSper channel mediates progesterone-induced Ca2+ influx in human sperm. Nature 2011, 471, 382–386. [Google Scholar] [CrossRef] [PubMed]
- Arganda-Carreras, I.; Fernandez-Gonzalez, R.; Munoz-Barrutia, A.; Ortiz-De-Solorzano, C. 3D reconstruction of histological sections: Application to mammary gland tissue. Microsc. Res. Tech. 2010, 73, 1019–1029. [Google Scholar] [CrossRef]
- SpermQ_Evaluator. Available online: https://github.com/IIIImaging/SpermQ_Evaluator (accessed on 22 December 2018).
- Satir, P. Studies on cilia. 3. Further studies on the cilium tip and a “sliding filament” model of ciliary motility. J. Cell Biol. 1968, 39, 77–94. [Google Scholar] [CrossRef] [PubMed]
- Summers, K.E.; Gibbons, I.R. Adenosine triphosphate-induced sliding of tubules in trypsin-treated flagella of sea-urchin sperm. Proc. Natl. Acad. Sci. USA 1971, 68, 3092–3096. [Google Scholar] [CrossRef] [PubMed]
- Brokaw, C.J. Direct measurements of sliding between outer doublet microtubules in swimming sperm flagella. Science 1989, 243, 1593–1596. [Google Scholar] [CrossRef] [PubMed]
- Gaffney, E.A.; Gadelha, H.; Smith, D.J.; Blake, J.R.; Kirkman-Brown, J.C. Mammalian sperm motility: Observation and theory. Annu. Rev. Fluid Mech. 2011, 43, 501–528. [Google Scholar] [CrossRef]
- Bayly, P.V.; Wilson, K.S. Analysis of unstable modes distinguishes mathematical models of flagellar motion. J. R. Soc. Interface 2015, 12, 20150124. [Google Scholar] [CrossRef] [PubMed]
- Blum, J.J.; Hines, M. Biophysics of flagellar motility. Q. Rev. Biophys. 1979, 12, 103–180. [Google Scholar] [CrossRef] [PubMed]
- Brokaw, C.J. Flagellar movement: A sliding filament model. Science 1972, 178, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Camalet, S.; Duke, T.; Jülicher, F.; Prost, J. Auditory sensitivity provided by self-tuned critical oscillations of hair cells. Proc. Natl. Acad. Sci. USA 2000, 97, 3183–3188. [Google Scholar] [CrossRef]
- Lindemann, C.B. A model of flagellar and ciliary functioning which uses the forces transverse to the axoneme as the regulator of dynein activation. Cell Motil. Cytoskelet. 1994, 29, 141–154. [Google Scholar] [CrossRef]
- Oriola, D.; Gadelha, H.; Casademunt, J. Nonlinear amplitude dynamics in flagellar beating. R. Soc. Open Sci. 2017, 4. [Google Scholar] [CrossRef]
- Sartori, P.; Geyer, V.F.; Howard, J.; Jülicher, F. Curvature regulation of the ciliary beat through axonemal twist. Phys. Rev. E 2016, 94. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Klindt, G.S.; Riedel-Kruse, I.H.; Jülicher, F.; Friedrich, B.M. Active phase and amplitude fluctuations of flagellar beating. Phys. Rev. Lett. 2014, 113. [Google Scholar] [CrossRef] [PubMed]
- Riedel-Kruse, I.H.; Hilfinger, A.; Howard, J.; Jülicher, F. How molecular motors shape the flagellar beat. HFSP J. 2007, 1, 192–208. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.J.; Gaffney, E.A.; Gadelha, H.; Kapur, N.; Kirkman-Brown, J.C. Bend propagation in the flagella of migrating human sperm, and its modulation by viscosity. Cell Motil. Cytoskelet. 2009, 66, 220–236. [Google Scholar] [CrossRef] [PubMed]
- Amann, R.P.; Waberski, D. Computer-assisted sperm analysis (CASA): Capabilities and potential developments. Theriogenology 2014, 81, 5–17.e3. [Google Scholar] [CrossRef] [PubMed]
- Davis, R.O.; Katz, D.F. Computer-aided sperm analysis (CASA): Image digitization and processing. Biomater. Artif. Cells Artif. Organs 1989, 17, 93–116. [Google Scholar] [CrossRef]
- Moruzzi, J.F.; Wyrobek, A.J.; Mayall, B.H.; Gledhill, B.L. Quantification and classification of human sperm morphology by computer-assisted image analysis. Fertil. Steril. 1988, 50, 142–152. [Google Scholar] [CrossRef]
- Boryshpolets, S.; Kowalski, R.K.; Dietrich, G.J.; Dzyuba, B.; Ciereszko, A. Different computer-assisted sperm analysis (CASA) systems highly influence sperm motility parameters. Theriogenology 2013, 80, 758–765. [Google Scholar] [CrossRef]
| Parameter | Mouse Sperm | Human Sperm | |
|---|---|---|---|
| A | Thresholding Method | Li | Triangle |
| B | Gauss Sigma | 3.0 | 2.0 |
| C | Repeat gauss fit after binarization | false | true |
| D | Blur only inside ROI selection | true | false |
| E | Upscaling of point list (fold) | 3 | 3 |
| F | Add head center-of-mass as first point | false | false |
| G | Unify start points | false | false |
| H | Filter points by gauss fits | false | false |
| I | Maximum vector length (points) | 14 | 20 |
| J | Normal radius for gauss fit (µm)—defines the distance of each flagellar point to the start and end of the corresponding normal line = half-length of each normal line | 5.0 | 6.0 |
| K | Exclude head from correction or deletion | true | false |
| L | Smooth normal for XY gauss fit | true | true |
| M | Save Roi-sets or vectors and normal | Set true to generate Figure 1C | |
| N | Accepted xy distance of points for fit-width-smoothing (µm) | 9.6 | 9.6 |
| O | # (+/−)-consecutive points for xy- and fit-width-smoothing | 15 | 15 |
| P | Distance of point to first point to form the reference vector (µm) | 10.0 | 6.4 |
| Q | Curvature: reference point distance | 10.0 | 10.0 |
| R | FFT: Grouped consecutive time-steps | 200 | 500 |
| S | FFT: Do not analyze (frequency results of flagellar parameters for the) initial … µm from head | 0.0 | 0.0 |
| T | Head rotation matrix radius | 10 | 10 |
| Parameter | Indication |
|---|---|
| Arc length | Position on the flagellum. |
| Head position | Trajectory of the swimming path. |
| Head angle in space (Θ) | Main beat frequency, rolling frequency (freely-swimming sperm), orientation in space, curvature of the trajectory. |
| Maximum intensity in head (rolling) | Rolling frequency (freely-swimming sperm), main beat frequency. |
| x coordinate | Oscillations on the longitudinal axis through the head. |
| y coordinate | Beat amplitude, asymmetry of the whole beat waveform |
| Tangential angle | custom analysis approaches. |
| Curvature/curvature angle | Localization of the origin and propagation of waves on the flagellum, wave kinetics, beat frequency, local beat amplitude, local beat asymmetry. |
| z position | Qualitative value to describe the kinetics of the flagellar beat in the direction vertical to the focal plane. |
| Intensity | Intensity profile of the cell along the whole flagellum, instructive when the cell is labelled with a fluorescence indicator; intensity also depends on the position of the flagellum in z. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hansen, J.N.; Rassmann, S.; Jikeli, J.F.; Wachten, D. SpermQ–A Simple Analysis Software to Comprehensively Study Flagellar Beating and Sperm Steering. Cells 2019, 8, 10. https://doi.org/10.3390/cells8010010
Hansen JN, Rassmann S, Jikeli JF, Wachten D. SpermQ–A Simple Analysis Software to Comprehensively Study Flagellar Beating and Sperm Steering. Cells. 2019; 8(1):10. https://doi.org/10.3390/cells8010010
Chicago/Turabian StyleHansen, Jan N., Sebastian Rassmann, Jan F. Jikeli, and Dagmar Wachten. 2019. "SpermQ–A Simple Analysis Software to Comprehensively Study Flagellar Beating and Sperm Steering" Cells 8, no. 1: 10. https://doi.org/10.3390/cells8010010
APA StyleHansen, J. N., Rassmann, S., Jikeli, J. F., & Wachten, D. (2019). SpermQ–A Simple Analysis Software to Comprehensively Study Flagellar Beating and Sperm Steering. Cells, 8(1), 10. https://doi.org/10.3390/cells8010010





