HIF-1α and HIF-2α Differently Regulate the Radiation Sensitivity of NSCLC Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Hypoxia Exposure
2.2. CRISPR/Cas9-Mediated Knockout of Human HIF1α and HIF2α Genes
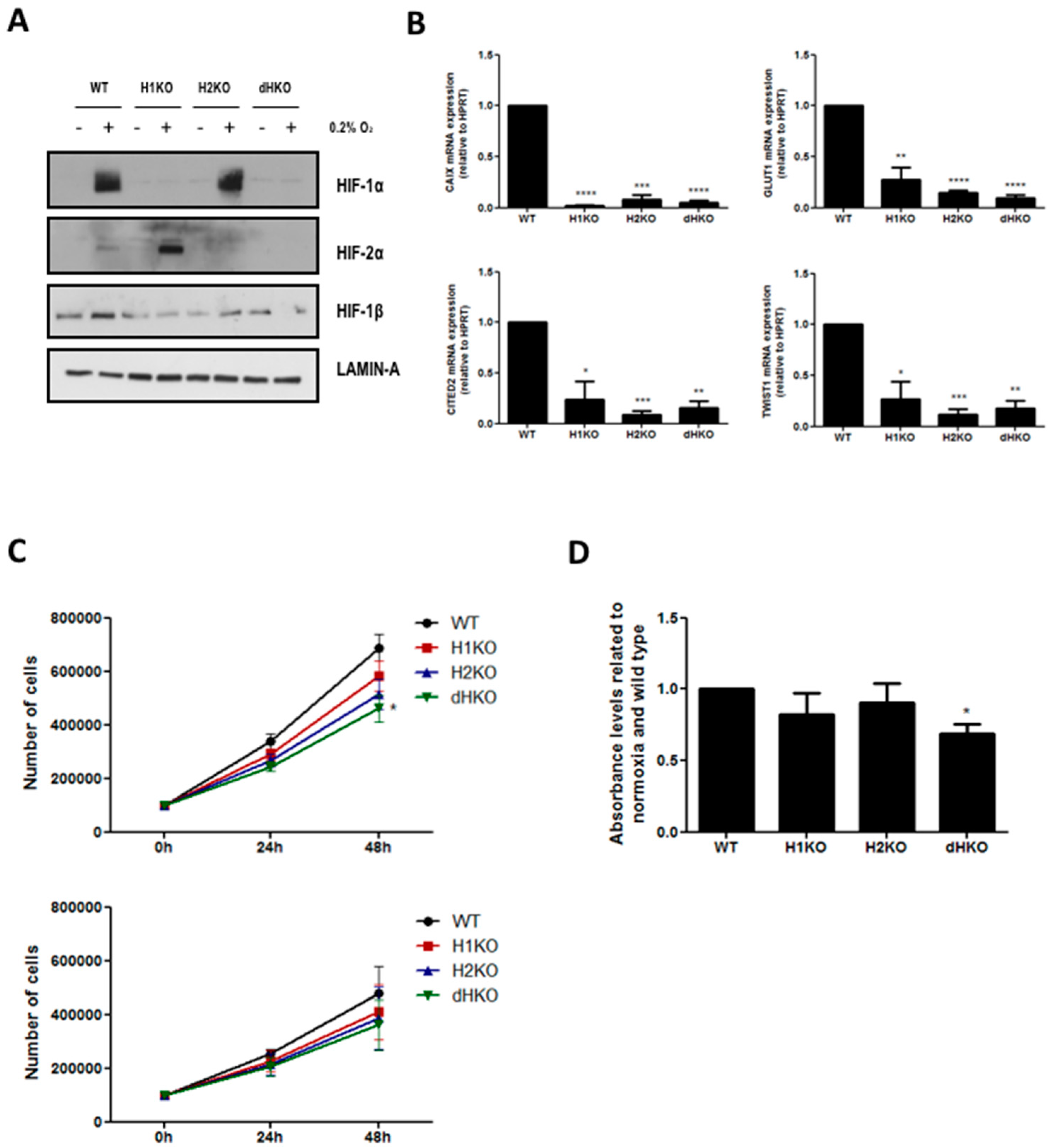
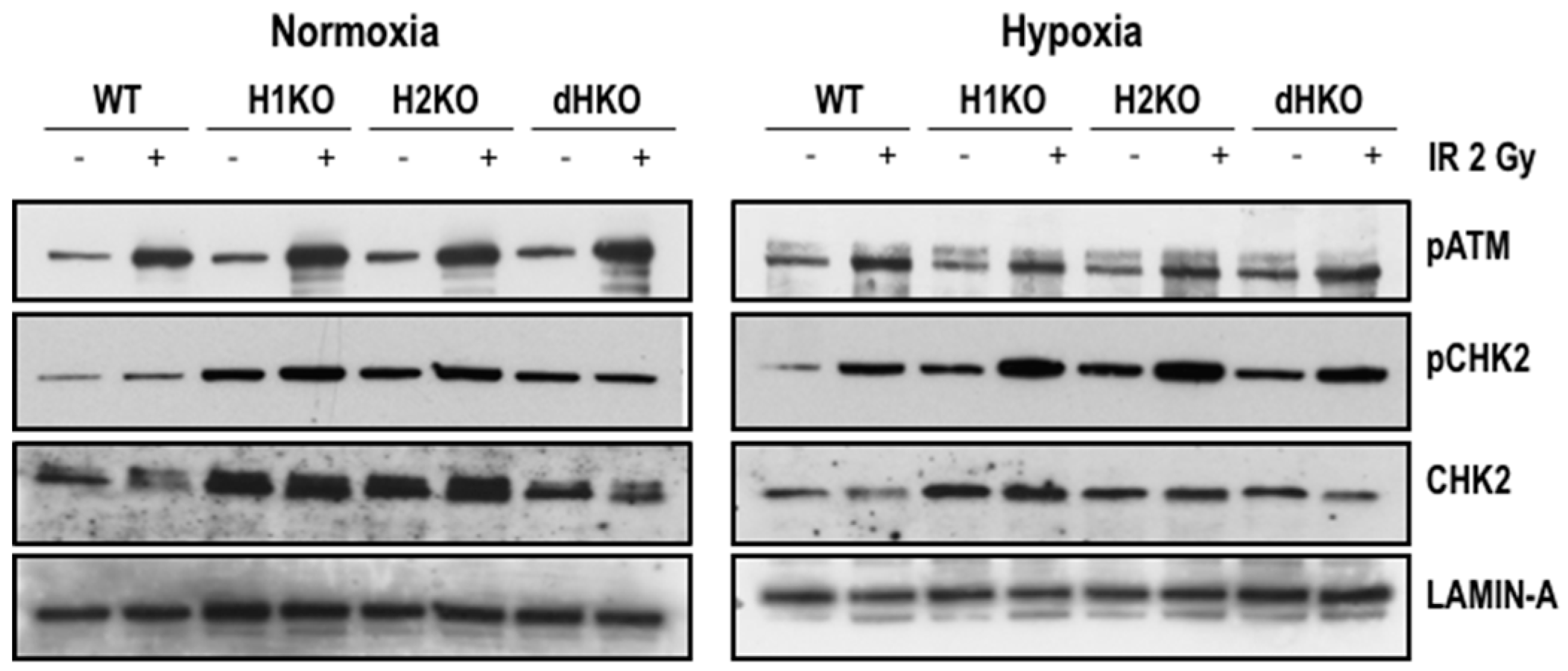
2.3. Western Blotting and Real Time Quantitative PCR
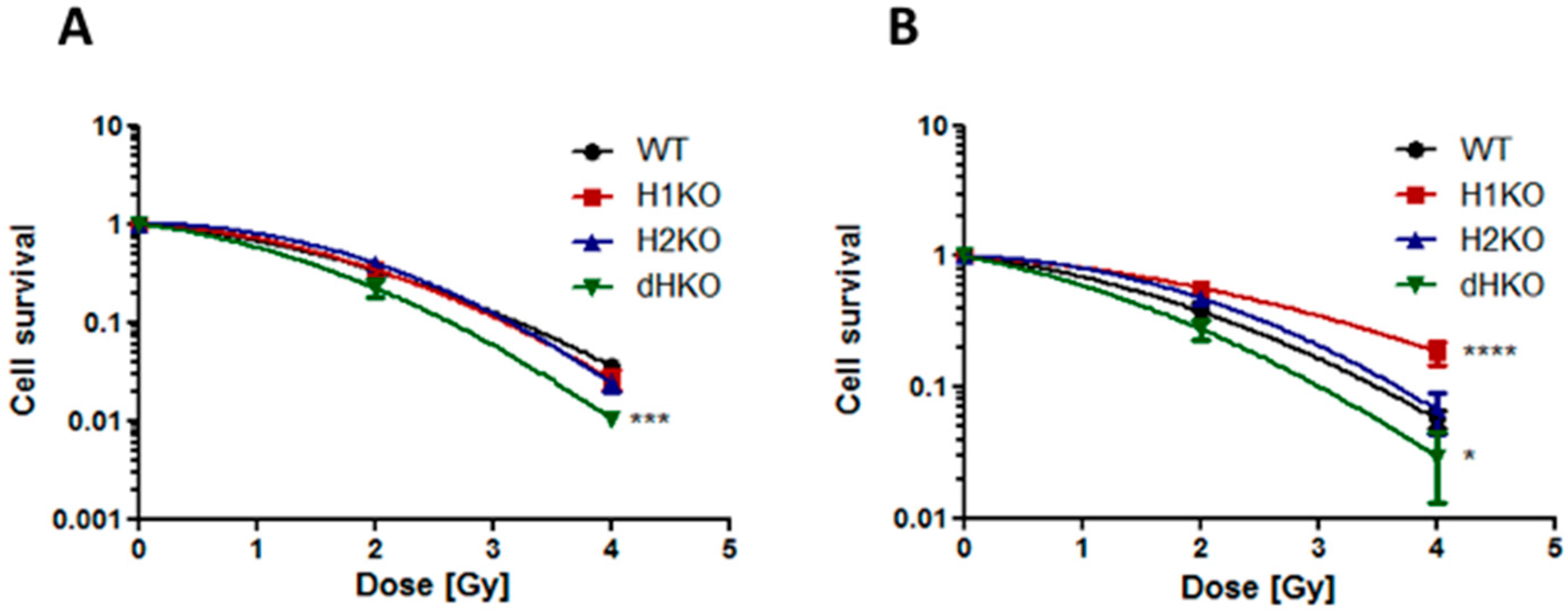
2.4. Proliferation and Clonogenic Survival Assay
2.5. Cell Viability—Hypoxia Tolerance
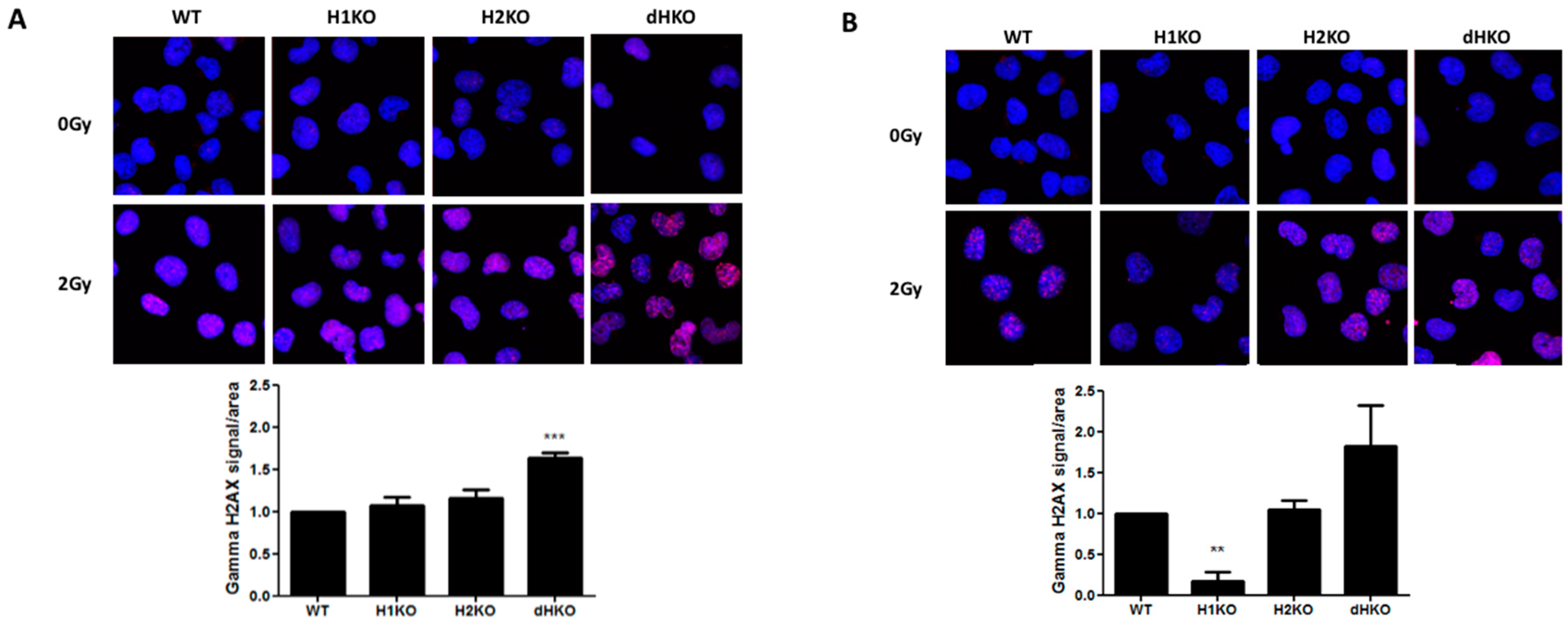
2.6. γ-H2AX Immunocytochemistry
2.7. Image Analysis
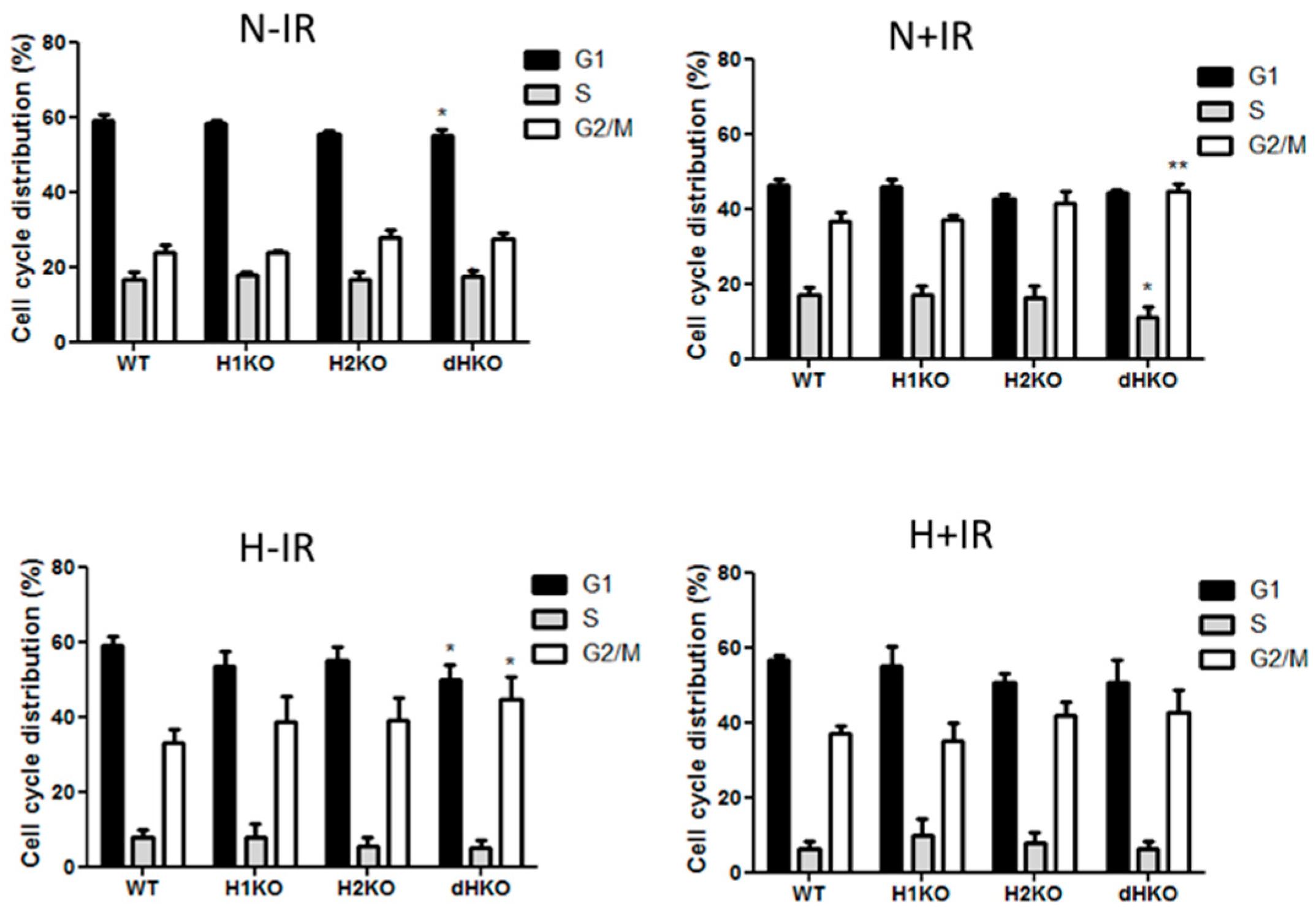
2.8. Cell Cycle Analysis
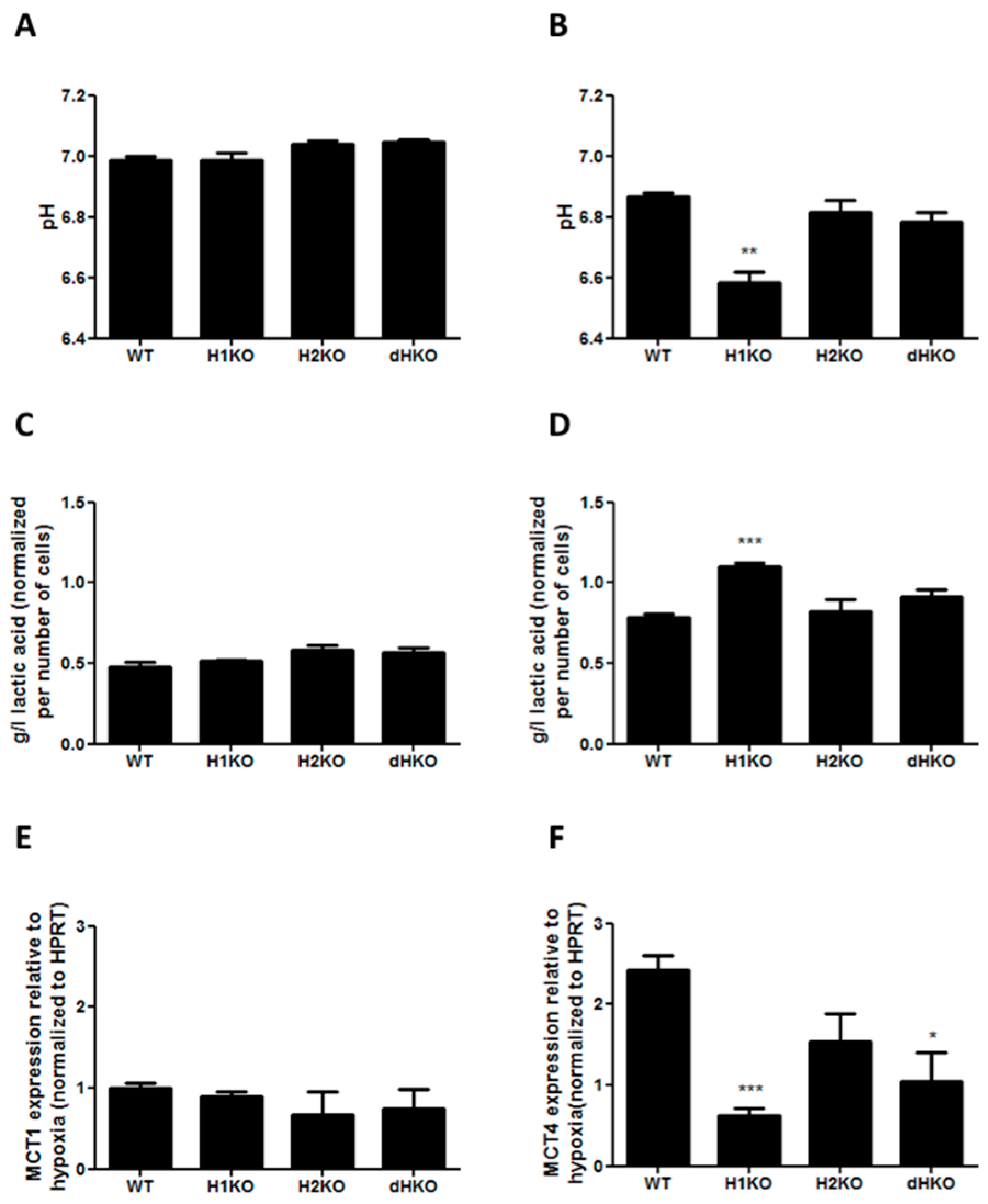
2.9. pH and Extracellular L-Lactic Acid Measurements
2.10. Metabolic Profiling
2.11. Statistics
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Harris, A.L. Hypoxia—A key regulatory factor in tumour growth. Nat. Rev. Cancer 2002, 2, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Bristow, R.G.; Hill, R.P. Hypoxia and metabolism. Hypoxia, DNA repair and genetic instability. Nat. Rev. Cancer 2008, 8, 180–192. [Google Scholar] [CrossRef] [PubMed]
- Rofstad, E.K.; Sundfør, K.; Lyng, H.; Tropé, C.G. Hypoxia-induced treatment failure in advanced squamous cell carcinoma of the uterine cervix is primarily due to hypoxia-induced radiation resistance rather than hypoxia-induced metastasis. Br. J. Cancer 2000, 83, 354–359. [Google Scholar] [CrossRef] [PubMed]
- Horsman, M.R.; Overgaard, J. The impact of hypoxia and its modification of the outcome of radiotherapy. J. Radiat. Res. 2016, 57 (Suppl. 1), i90–i98. [Google Scholar] [CrossRef] [PubMed]
- Bruick, R.K.; McKnight, S.L. A conserved family of prolyl-4-hydroxylases that modify HIF. Science 2001, 294, 1337–1340. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.; Bae, S.H.; Jeong, J.W.; Kim, S.H.; Kim, K.W. Hypoxia-inducible factor (HIF-1)alpha: Its protein stability and biological functions. Exp. Mol. Med. 2004, 36, 1–12. [Google Scholar] [CrossRef]
- Wenger, R.H.; Stiehl, D.P.; Camenisch, G. Integration of oxygen signaling at the consensus HRE. Sci. STKE 2005, 2005, re12. [Google Scholar] [CrossRef]
- Hu, C.J.; Iyer, S.; Sataur, A.; Covello, K.L.; Chodosh, L.A.; Simon, M.C. Differential regulation of the transcriptional activities of hypoxia-inducible factor 1 alpha (HIF-1alpha) and HIF-2alpha in stem cells. Mol. Cell. Biol. 2006, 26, 3514–3526. [Google Scholar] [CrossRef]
- Rosenberger, C.; Mandriota, S.; Jürgensen, J.S.; Wiesener, M.S.; Hörstrup, J.H.; Frei, U.; Ratcliffe, P.; Maxwell, P.H.; Bachmann, S.; Eckardt, K.U. Expression of hypoxia-inducible factor-1alpha and -2alpha in hypoxic and ischemic rat kidneys. J. Am. Soc. Nephrol. 2002, 13, 1721–1732. [Google Scholar] [CrossRef]
- Schodel, J.; Oikonomopoulos, S.; Ragoussis, J.; Pugh, C.W.; Ratcliffe, P.J.; Mole, D.R. High-resolution genome-wide mapping of HIF-binding sites by ChIP-seq. Blood 2011, 117, e207–e217. [Google Scholar] [CrossRef]
- Gort, E.H.; Groot, A.J.; van der Wall, E.; van Diest, P.J.; Vooijs, M.A. Hypoxic regulation of metastasis via hypoxia-inducible factors. Curr. Mol. Med. 2008, 8, 60–67. [Google Scholar] [PubMed]
- Ryan, H.E.; Lo, J.; Johnson, R.S. HIF-1 alpha is required for solid tumor formation and embryonic vascularization. EMBO J. 1998, 17, 3005–3015. [Google Scholar] [CrossRef] [PubMed]
- Williams, K.J.; Telfer, B.A.; Xenaki, D.; Sheridan, M.R.; Desbaillets, I.; Peters, H.J.; Honess, D.; Harris, A.L.; Dachs, G.U.; van der Kogel, A.; et al. Enhanced response to radiotherapy in tumours deficient in the function of hypoxia-inducible factor-1. Radiother. Oncol. 2005, 75, 89–98. [Google Scholar] [CrossRef]
- Luo, Z.; Bai, M.; Xiao, X.; Zhang, W.; Liu, X.; Yang, X.; Li, S.; Huan, Y.; Wu, Z.; Zhang, X.; et al. Silencing of HIF-1alpha enhances the radiation sensitivity of human glioma growth in vitro and in vivo. Neuropharmacology 2015, 89, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Williams, K.J.; Telfer, B.A.; Airley, R.E.; Peters, H.P.; Sheridan, M.R.; van der Kogel, A.J.; Harris, A.L.; Stratford, I.J. A protective role for HIF-1 in response to redox manipulation and glucose deprivation: Implications for tumorigenesis. Oncogene 2002, 21, 282–290. [Google Scholar] [CrossRef] [PubMed]
- Damaghi, M.; Wojtkowiak, J.W.; Gillies, R.J. pH sensing and regulation in cancer. Front. Physiol. 2013, 4, 370. [Google Scholar] [CrossRef] [PubMed]
- Van Kuijk, S.J.; Gieling, R.G.; Niemans, R.; Lieuwes, N.G.; Biemans, R.; Telfer, B.A.; Haenen, G.R.; Yaromina, A.; Lambin, P.; Dubois, L.J.; et al. The sulfamate small molecule CAIX inhibitor S4 modulates doxorubicin efficacy. PLoS ONE 2016, 11, e0161040. [Google Scholar] [CrossRef]
- Groussard, C.; Morel, I.; Chevanne, M.; Monnier, M.; Cillard, J.; Delamarche, A. Free radical scavenging and antioxidant effects of lactate ion: An in vitro study. J. Appl. Physiol. 2000, 89, 169–175. [Google Scholar] [CrossRef]
- Moreno Roig, E.; Yaromina, A.; Houben, R.; Groot, A.J.; Dubois, L.; Vooijs, M. Prognostic role of hypoxia-inducible factor-2α tumor cell expression in cancer patients: A meta-analysis. Front. Oncol. 2018, 8. [Google Scholar] [CrossRef]
- Sun, J.C.; He, F.; Yi, W.; Wan, M.H.; Li, R.; Wei, X.; Wu, R.; Niu, D.L. High expression of HIF-2alpha and its anti-radiotherapy effect in lung cancer stem cells. Genet. Mol. Res. 2015, 14, 18110–18120. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [PubMed]
- Brustugun, O.T. Hypoxia as a cause of treatment failure in non-small cell carcinoma of the lung. Semin. Radiat. Oncol. 2015, 25, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.H.; Qian, C.; Yuan, K. Correlations of hypoxia-inducible factor-1alpha/hypoxia-inducible factor-2alpha expression with angiogenesis factors expression and prognosis in non-small cell lung cancer. Chin. Med. J. 2011, 124, 11–18. [Google Scholar] [PubMed]
- Isono, T.; Chano, T.; Yoshida, T.; Kageyama, S.; Kawauchi, A.; Suzaki, M.; Yuasa, T. Hydroxyl-HIF2-alpha is potential therapeutic target for renal cell carcinomas. Am. J. Cancer Res. 2016, 6, 2263–2276. [Google Scholar] [PubMed]
- Anelli, V.; Gault, C.R.; Cheng, A.B.; Obeid, L.M. Sphingosine kinase 1 is up-regulated during hypoxia in U87MG glioma cells. Role of hypoxia-inducible factors 1 and 2. J. Biol. Chem. 2008, 283, 3365–3375. [Google Scholar] [CrossRef] [PubMed]
- Carroll, V.A.; Ashcroft, M. Role of hypoxia-inducible factor (HIF)-1alpha versus HIF-2alpha in the regulation of HIF target genes in response to hypoxia, insulin-like growth factor-I, or loss of von Hippel-Lindau function: Implications for targeting the HIF pathway. Cancer Res. 2006, 66, 6264–6270. [Google Scholar] [CrossRef] [PubMed]
- Schulz, K.; Milke, L.; Rübsamen, D.; Menrad, H.; Schmid, T.; Brüne, B. HIF-1alpha protein is upregulated in HIF-2alpha depleted cells via enhanced translation. FEBS Lett. 2012, 586, 1652–1657. [Google Scholar] [CrossRef]
- Holmquist-Mengelbier, L.; Fredlund, E.; Löfstedt, T.; Noguera, R.; Navarro, S.; Nilsson, H.; Pietras, A.; Vallon-Christersson, J.; Borg, A.; Gradin, K.; et al. Recruitment of HIF-1alpha and HIF-2alpha to common target genes is differentially regulated in neuroblastoma: HIF-2alpha promotes an aggressive phenotype. Cancer Cell 2006, 10, 413–423. [Google Scholar] [CrossRef]
- Tang, N.; Wang, L.; Esko, J.; Giordano, F.J.; Huang, Y.; Gerber, H.P.; Ferrara, N.; Johnson, R.S. Loss of HIF-1alpha in endothelial cells disrupts a hypoxia-driven VEGF autocrine loop necessary for tumorigenesis. Cancer Cell 2004, 6, 485–495. [Google Scholar] [CrossRef]
- Adams, J.M.; Difazio, L.T.; Rolandelli, R.H.; Luján, J.J.; Haskó, G.; Csóka, B.; Selmeczy, Z.; Németh, Z.H. HIF-1: A key mediator in hypoxia. Acta Physiol. Hung. 2009, 96, 19–28. [Google Scholar] [CrossRef]
- Goda, N.; Ryan, H.E.; Khadivi, B.; McNulty, W.; Rickert, R.C.; Johnson, R.S. Hypoxia-inducible factor 1alpha is essential for cell cycle arrest during hypoxia. Mol. Cell. Biol. 2003, 23, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Piret, J.P.; Lecocq, C.; Toffoli, S.; Ninane, N.; Raes, M.; Michiels, C. Hypoxia and CoCl2 protect HepG2 cells against serum deprivation- and t-BHP-induced apoptosis: A possible anti-apoptotic role for HIF-1. Exp. Cell Res. 2004, 295, 340–349. [Google Scholar] [CrossRef] [PubMed]
- Carmeliet, P.; Dor, Y.; Herbert, J.M.; Fukumura, D.; Brusselmans, K.; Dewerchin, M.; Neeman, M.; Bono, F.; Abramovitch, R.; Maxwell, P.; et al. Role of HIF-1alpha in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature 1998, 394, 485–490. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L.; Roth, P.H.; Fang, H.M.; Wang, G.L. Transcriptional regulation of genes encoding glycolytic enzymes by hypoxia-inducible factor 1. J. Biol. Chem. 1994, 269, 23757–23763. [Google Scholar] [PubMed]
- Chandel, N.S.; Maltepe, E.; Goldwasser, E.; Mathieu, C.E.; Simon, M.C.; Schumacker, P.T. Mitochondrial reactive oxygen species trigger hypoxia-induced transcription. Proc. Natl. Acad. Sci. USA 1998, 95, 11715–11720. [Google Scholar] [CrossRef]
- Stegen, S.; van Gastel, N.; Eelen, G.; Ghesquière, B.; D’Anna, F.; Thienpont, B.; Goveia, J.; Torrekens, S.; Van Looveren, R.; Luyten, F.P.; et al. HIF-1alpha promotes glutamine-mediated redox homeostasis and glycogen-dependent bioenergetics to support postimplantation bone cell survival. Cell Metab. 2016, 23, 265–279. [Google Scholar] [CrossRef] [PubMed]
- Moeller, B.J.; Dreher, M.R.; Rabbani, Z.N.; Schroeder, T.; Cao, Y.; Li, C.Y.; Dewhirst, M.W. Pleiotropic effects of HIF-1 blockade on tumor radiosensitivity. Cancer Cell 2005, 8, 99–110. [Google Scholar] [CrossRef]
- Koukourakis, M.I.; Giatromanolaki, A.; Sivridis, E.; Simopoulos, C.; Turley, H.; Talks, K.; Gatter, K.C.; Harris, A.L. Hypoxia-inducible factor (HIF1A and HIF2A), angiogenesis, and chemoradiotherapy outcome of squamous cell head-and-neck cancer. Int. J. Radiat. Oncol. Biol. Phys. 2002, 53, 1192–1202. [Google Scholar] [CrossRef]
- Bhatt, R.S.; Landis, D.M.; Zimmer, M.; Torregrossa, J.; Chen, S.; Sukhatme, V.P.; Iliopoulos, O.; Balk, S.; Bubley, G.J. Hypoxia-inducible factor-2alpha: Effect on radiation sensitivity and differential regulation by an mTOR inhibitor. BJU Int. 2008, 102, 358–363. [Google Scholar] [CrossRef]
- Bertout, J.A.; Majmundar, A.J.; Gordan, J.D.; Lam, J.C.; Ditsworth, D.; Keith, B.; Brown, E.J.; Nathanson, K.L.; Simon, M.C. HIF2alpha inhibition promotes p53 pathway activity, tumor cell death, and radiation responses. Proc. Natl. Acad. Sci. USA 2009, 106, 14391–14396. [Google Scholar] [CrossRef]
- Ivan, M.; Kondo, K.; Yang, H.; Kim, W.; Valiando, J.; Ohh, M.; Salic, A.; Asara, J.M.; Lane, W.S.; Kaelin, W.G., Jr. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: Implications for O2 sensing. Science 2001, 292, 464–468. [Google Scholar] [CrossRef]
- Lee, S.H.; Koo, K.H.; Park, J.W.; Kim, H.J.; Ye, S.K.; Park, J.B.; Park, B.K.; Kim, Y.N. HIF-1 is induced via EGFR activation and mediates resistance to anoikis-like cell death under lipid rafts/caveolae-disrupting stress. Carcinogenesis 2009, 30, 1997–2004. [Google Scholar] [CrossRef] [PubMed]
- Gort, E.H.; Groot, A.J.; Derks van de Ven, T.L.; van der Groep, P.; Verlaan, I.; van Laar, T.; van Diest, P.J.; van der Wall, E.; Shvarts, A. Hypoxia-inducible factor-1alpha expression requires PI 3-kinase activity and correlates with Akt1 phosphorylation in invasive breast carcinomas. Oncogene 2006, 25, 6123–6127. [Google Scholar] [CrossRef] [PubMed]
- Selak, M.A.; Armour, S.M.; MacKenzie, E.D.; Boulahbel, H.; Watson, D.G.; Mansfield, K.D.; Pan, Y.; Simon, M.C.; Thompson, C.B.; Gottlieb, E. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell 2005, 7, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Koivunen, P.; Hirsilä, M.; Remes, A.M.; Hassinen, I.E.; Kivirikko, K.I.; Myllyharju, J. Inhibition of hypoxia-inducible factor (HIF) hydroxylases by citric acid cycle intermediates: Possible links between cell metabolism and stabilization of HIF. J. Biol. Chem. 2007, 282, 4524–4532. [Google Scholar] [CrossRef] [PubMed]
- Moeller, B.J.; Cao, Y.; Li, C.Y.; Dewhirst, M.W. Radiation activates HIF-1 to regulate vascular radiosensitivity in tumors: Role of reoxygenation, free radicals, and stress granules. Cancer Cell 2004, 5, 429–441. [Google Scholar] [CrossRef]
- Kung, A.L.; Wang, S.; Klco, J.M.; Kaelin, W.G.; Livingston, D.M. Suppression of tumor growth through disruption of hypoxia-inducible transcription. Nat. Med. 2000, 6, 1335–1340. [Google Scholar] [CrossRef]
- Helbig, L.; Koi, L.; Brüchner, K.; Gurtner, K.; Hess-Stumpp, H.; Unterschemmann, K.; Baumann, M.; Zips, D.; Yaromina, A. BAY 87-2243, a novel inhibitor of hypoxia-induced gene activation, improves local tumor control after fractionated irradiation in a schedule-dependent manner in head and neck human xenografts. Radiat. Oncol. 2014, 9, 207. [Google Scholar] [CrossRef]
- Burroughs, S.K.; Kaluz, S.; Wang, D.; Wang, K.; Van Meir, E.G.; Wang, B. Hypoxia inducible factor pathway inhibitors as anticancer therapeutics. Future Med. Chem. 2013, 5, 553–572, (1756-8927 (Electronic)). [Google Scholar] [CrossRef]
- Chen, W.; Hill, H.; Christie, A.; Kim, M.S.; Holloman, E.; Pavia-Jimenez, A.; Homayoun, F.; Ma, Y.; Patel, N.; Yell, P.; et al. Targeting renal cell carcinoma with a HIF-2 antagonist. Nature 2016, 539, 112–117, (1476-4687 (Electronic)). [Google Scholar] [CrossRef]
- Cam, H.; Easton, J.B.; High, A.; Houghton, P.J. mTORC1 signaling under hypoxic conditions is controlled by ATM-dependent phosphorylation of HIF-1alpha. Mol. Cell 2010, 40, 509–520. [Google Scholar] [CrossRef]
- Kastan, M.B.; Lim, D.S. The many substrates and functions of ATM. Nat. Rev. Mol. Cell Biol. 2000, 1, 179–186. [Google Scholar] [CrossRef]
- Reinhardt, H.C.; Aslanian, A.S.; Lees, J.A.; Yaffe, M.B. p53-deficient cells rely on ATM- and ATR-mediated checkpoint signaling through the p38MAPK/MK2 pathway for survival after DNA damage. Cancer Cell 2007, 11, 175–189. [Google Scholar] [CrossRef]
- Walenta, S.; Mueller-Klieser, W.F. Lactate: Mirror and motor of tumor malignancy. Semin. Radiat. Oncol. 2004, 14, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Tarnawski, R.; Sokol, M.; Pieniazek, P.; Maciejewski, B.; Walecki, J.; Miszczyk, L.; Krupska, T. 1H-MRS in vivo predicts the early treatment outcome of postoperative radiotherapy for malignant gliomas. Int. J. Radiat. Oncol. Biol. Phys. 2002, 52, 1271–1276. [Google Scholar] [CrossRef]
- Hirschhaeuser, F.; Sattler, U.G.; Mueller-Klieser, W. Lactate: A metabolic key player in cancer. Cancer Res. 2011, 71, 6921–6925. [Google Scholar] [CrossRef] [PubMed]
- Leung, E.; Cairns, R.A.; Chaudary, N.; Vellanki, R.N.; Kalliomaki, T.; Moriyama, E.H.; Mujcic, H.; Wilson, B.C.; Wouters, B.G.; Hill, R.; et al. Metabolic targeting of HIF-dependent glycolysis reduces lactate, increases oxygen consumption and enhances response to high-dose single-fraction radiotherapy in hypoxic solid tumors. BMC Cancer 2017, 17, 418. [Google Scholar] [CrossRef] [PubMed]
- Draoui, N.; Schicke, O.; Seront, E.; Bouzin, C.; Sonveaux, P.; Riant, O.; Feron, O. Antitumor activity of 7-aminocarboxycoumarin derivatives, a new class of potent inhibitors of lactate influx but not efflux. Mol. Cancer Ther. 2014, 13, 1410–1418. [Google Scholar] [CrossRef]
- Xie, J.; Wu, H.; Dai, C.; Pan, Q.; Ding, Z.; Hu, D.; Ji, B.; Luo, Y.; Hu, X. Beyond Warburg effect—Dual metabolic nature of cancer cells. Sci. Rep. 2014, 4, 4927. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moreno Roig, E.; Groot, A.J.; Yaromina, A.; Hendrickx, T.C.; Barbeau, L.M.O.; Giuranno, L.; Dams, G.; Ient, J.; Olivo Pimentel, V.; van Gisbergen, M.W.; et al. HIF-1α and HIF-2α Differently Regulate the Radiation Sensitivity of NSCLC Cells. Cells 2019, 8, 45. https://doi.org/10.3390/cells8010045
Moreno Roig E, Groot AJ, Yaromina A, Hendrickx TC, Barbeau LMO, Giuranno L, Dams G, Ient J, Olivo Pimentel V, van Gisbergen MW, et al. HIF-1α and HIF-2α Differently Regulate the Radiation Sensitivity of NSCLC Cells. Cells. 2019; 8(1):45. https://doi.org/10.3390/cells8010045
Chicago/Turabian StyleMoreno Roig, Eloy, Arjan J. Groot, Ala Yaromina, Tessa C. Hendrickx, Lydie M. O. Barbeau, Lorena Giuranno, Glenn Dams, Jonathan Ient, Veronica Olivo Pimentel, Marike W. van Gisbergen, and et al. 2019. "HIF-1α and HIF-2α Differently Regulate the Radiation Sensitivity of NSCLC Cells" Cells 8, no. 1: 45. https://doi.org/10.3390/cells8010045
APA StyleMoreno Roig, E., Groot, A. J., Yaromina, A., Hendrickx, T. C., Barbeau, L. M. O., Giuranno, L., Dams, G., Ient, J., Olivo Pimentel, V., van Gisbergen, M. W., Dubois, L. J., & Vooijs, M. A. (2019). HIF-1α and HIF-2α Differently Regulate the Radiation Sensitivity of NSCLC Cells. Cells, 8(1), 45. https://doi.org/10.3390/cells8010045





