Abstract
The ubiquitin-proteasome system (UPS) and autophagy are the two major intracellular protein quality control (PQC) pathways that are responsible for cellular proteostasis (homeostasis of the proteome) by ensuring the timely degradation of misfolded, damaged, and unwanted proteins. Ubiquitination serves as the degradation signal in both these systems, but substrates are precisely targeted to one or the other pathway. Determining how and when cells target specific proteins to these two alternative PQC pathways and control the crosstalk between them are topics of considerable interest. The ubiquitin (Ub) recognition code based on the type of Ub-linked chains on substrate proteins was believed to play a pivotal role in this process, but an increasing body of evidence indicates that the PQC pathway choice is also made based on other criteria. These include the oligomeric state of the Ub-binding protein shuttles, their conformation, protein modifications, and the presence of motifs that interact with ATG8/LC3/GABARAP (autophagy-related protein 8/microtubule-associated protein 1A/1B-light chain 3/GABA type A receptor-associated protein) protein family members. In this review, we summarize the current knowledge regarding the Ub recognition code that is bound by Ub-binding proteasomal and autophagic receptors. We also discuss how cells can modify substrate fate by modulating the structure, conformation, and physical properties of these receptors to affect their shuttling between both degradation pathways.
1. Introduction—Protein Quality Control Mechanisms
At the cellular level, cells need to recycle old, abnormal, or dysfunctional proteins and maintain a “healthy” balance between newly-synthesized proteins and existing ones in order to maintain the efficient functioning of cellular pathways and systems. Approximately one-third of newly-synthesized proteins in human cells require removal due to their improper folding, despite the presence of chaperones that prevent protein aggregation and assist in protein folding [1]. Because homeostasis of the cellular proteome (proteostasis) ensures successful cell development, health aging, stress tolerance, and adaptation to changing environmental conditions or even resistance to infections by pathogens, any imbalance in protein homeostasis (triggered, for example, by accumulation of misfolded proteins) is widely accepted to cause diseases [2]. Because of the fundamental importance of proteostasis, cells have evolved a sophisticated housecleaning protein quality control (PQC) system, which is based primarily on two major degradation pathways known as the ubiquitin (Ub)-proteasome system (UPS) and selective macroautophagy (hereafter, autophagy) [2,3].
In the UPS, substrates for degradation are firstly ubiquitinated via an enzymatic cascade involving E1 Ub-activating enzyme(s), E2 Ub-conjugating enzyme(s), and E3 Ub ligases [4]. Subsequently, most of these ubiquitinated (generally poly-ubiquitinated) proteins are recognized and turned over by the proteasome, which is a large multi-subunit complex of proteases and regulatory proteins (for more details about the UPS and proteasome structure, see References [2,5,6,7]). The UPS is responsible for the degradation of most cellular soluble proteins [2]. Proteins targeted to the UPS are individual misfolded or damaged proteins (housecleaning degradation), as well as unneeded or redundant proteins that function in regulatory processes (regulatory degradation). Therefore, there is no doubt that the UPS plays a pivotal role in PQC. However, if ubiquitinated substrates are resistant to proteasomal degradation, or when the UPS is overwhelmed or impaired, these proteins may become aggregation-prone and may form aberrant aggregates that become inaccessible to proteasomal proteases [8,9] because of the intrinsic size limitation of proteins subject to degradation by the UPS. In such cases, to maintain proteostasis, these protein aggregates will be immediately redirected to autophagy, or temporarily stored in aggresomes-perinuclear compartments with high autophagic activity located at the microtubule organizing center (MTOC)- to reduce their toxicity [10,11]. Based on recent studies in mammalian cells, such initial accumulation of misfolded proteins in aggresomes per se is tolerable. However, the absence of replenishment of amino acids and Ub molecules caused by the longer sequestration of many ubiquitinated proteins could become highly dangerous for cell viability. Therefore, as a consequence, an alternative pathway, autophagy, is induced [12,13]. Thus, regardless of whether a protein is degraded instantly or is stored in the aggresome initially, the fate of poly-ubiquitinated proteins destined for degradation is pre-ordained in the sense that, when proteasomal clearance is inhibited, these proteins will be degraded at some point via autophagy, which is a major pathway for degradation of large substrates, such as aggregates and organelles [2].
Autophagy is a catabolic pathway conserved from yeast to mammals and plants. It involves the conserved action of more than 20 specific AuTophaGy related (ATG) proteins that comprise the core machinery, which mediates the formation of a double-membrane structure called the autophagosome. This structure is decorated by ATG8/LC3/GABARAP (autophagy-related protein 8/microtubule-associated protein 1A/1B-light chain 3/GABA type A receptor-associated protein) protein family members. The ATG8/LC3/GABARAP proteins are Ub-like proteins that share structural features with Ub, such as two amino-terminal α helices and a Ub-like core [14,15,16,17,18,19,20]. During autophagosome formation, these proteins undergo a unique Ub-like conjugation to phosphatidylethanolamine on the isolation membrane (phagophore, pre-autophagosomal structure), which expands around the substrate(s) and encloses it in a vesicle (autophagosome) for further delivery to, and clearance within, the vacuole (in yeast and plants) or lysosome (in mammals). ATG8/LC3/GABARAPs are proteins associated with mature autophagosome and, therefore, serve as bona fide markers for this unique vesicle (for more details about the autophagic process and the machinery, see References [21,22,23]).
Autophagy in eukaryotes was initially considered to be a non-selective, bulk system, but specific receptors precisely recognize and deliver cellular substrates (proteins or organelles) exclusively to autophagosomes and then the vacuole/lysosomes [24]. Aggregates of misfolded proteins are among the various substrates that can be selectively targeted for autophagic clearance [25,26,27,28,29,30,31,32,33]. In this process, called aggrephagy, particular autophagic cargo receptors recognize ubiquitinated aggregates and link them to the expanding isolation membrane through their interaction with ATG8/LC3/GABARAP protein family members for subsequent degradation [25].
Based on several differences in mechanisms of action, the UPS and autophagy were initially viewed as two mutually-independent degradative systems in PQC, with distinct receptors and adaptors. However, in view of the fact that Ub and the ubiquitination process (which refers to the conjugation of the Ub to lysine residues, or rarely cysteine, of the target protein destined for degradation) [34,35,36,37] are used by both the UPS and the autophagy pathways, there is increasing evidence that they should not be studied separately because of the dynamic interplay between these two protein degradation systems [38,39,40,41,42,43].
In this review, we will summarize the current knowledge regarding the Ub code (the varying patterns of ubiquitination of a target protein) in the UPS and autophagy contexts. We will also discuss how cells make decisions and modify the fates of substrates based on the structure and properties of the Ub-binding protein shuttles that are used for substrate degradation.
2. Ub and Ubiquitination at the Crossroads of the UPS and Autophagy
Both the UPS and autophagy pathways use ubiquitination as a labeling system for substrate recognition. Ub is a highly-stable, 76 amino acid protein, which is almost identical in all eukaryotes [44]. Such high evolutionary conservation points to the importance of Ub and its recognition code for proteostasis and suggests enormous pressure to preserve the structure of this protein since many of its surfaces are recognized by Ub-binding domains (UBDs) in proteins [44]. Through the coordinated activity of the E1, E2, and E3 enzymes mentioned earlier, Ub is covalently attached via its C-terminal glycine to usually one or more lysine (and rarely cysteine) residues of the target protein. Attachment of just one Ub is named mono-ubiquitination and is a common mechanism of protein post-translational modification that regulates protein function and trafficking (Figure 1). Multiple Ubs may also become linked to several independent lysine residues in one protein causing multi-mono-ubiquitination. Alternatively, through the addition of other Ubs to a Ub previously conjugated to a substrate at a lysine residue, the target may be poly-ubiquitinated (Figure 1).
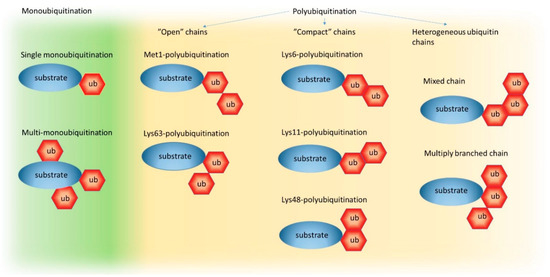
Figure 1.
The ubiquitin code. A schematic representation of possible ubiquitin (Ub) chain formations on a target protein (substrate) from single mono-ubiquitination to mixed and multiply -branched Ub chains. Within polyubiquitin chains, which can exist on one of the seven intrinsic lysine (Lys) residues within the Ub sequence, or methionine (Met) at position 1, Ub can form eight different homogeneous or heterogeneous linkage types. However, only five linkage types have been structurally characterized (Met1-chains, Lys6-chains, Lys11-chains, Lys48-chains, and Lys63-chains) and classified as “open” or “compact” based on their three-dimensional topologies.
Based on the number of Ub molecules attached to each other, short (starting with only two Ub moieties) or long (more than 10 Ubs) chains can be formed. Additionally, since poly-ubiquitination can occur via bond formation on Met1 or any Lys residue within Ub (e.g., Lys6, Lys11, Lys27, Lys29, Lys33, Lys48, Lys63), which, by the way, are well-distributed on the surface and oriented in distinct directions, different types of homogeneous chains (e.g., Lys11-linked, Lys48-linked, or Lys63-linked Ub chains), or chains with mixed topology, can be generated (Figure 1) (for more information about the ubiquitination process and Ub chains, see References [44,45,46]). Such flexibility of the ubiquitination process (from mono- and multi-mono- to poly-ubiquitination) and structural diversity of the Ub chains are commonly accepted as the foundation of the “Ub recognition code” that potentially encodes information about protein fates in the cell. Nuclear magnetic resonance (NMR) and crystal structure analyses of various Ub chains revealed that different chain types adopt distinct conformations with either a so-called “compact” one, where neighboring Ub molecules in close proximity interact with each other and form a higher-order assembly or “open” structures, where linked Ub moieties have greater conformational freedom because they are too far from each other to form tight interactions [44]. Contrary to Met1-linked and Lys63-linked Ub chains that adopt “open” conformations, the Lys6-linked, Lys11-linked, and Lys48-linked Ub chains display “compact” conformations (Figure 1) [44]. Therefore, it is commonly accepted that Ub-binding proteins might recognize and preferentially bind to proteins with particular ubiquitination status, or with specific Ub chain types by, for example, sensing the distance between Ub molecules or the relative orientation of Ub surfaces within a chain [44].
Thus, it is not surprising that many Ub recognition receptors with diverse UBDs have been described. At least 21 different classes of Ub-binding modules have been characterized so far [47]. Among them, the most interesting for the UPS and autophagy degradation systems are: (1) the Ub-associated (UBA) domain present in proteasomal (Rad23, Dsk2, Ddi1) and autophagic receptors (sequestosome 1 (SQSTM1), also known as p62, and neighbor of BRCA1 gene 1 (NBR1) in mammals or Joka2/NBR1 in plants), (2) the coupling of Ub to ER degradation (CUE) domain present in Cue5 protein in yeast and toll-interacting protein (TOLLIP) in mammals, (3) the Ub-interacting motif (UIM) present in the proteasomal protein S5a/Rpn10/Pus1 (in higher eukaryotes, S. cerevisiae, and S. pombe, respectively), (4) zinc fingers such as the zinc-finger Ub-binding domain (ZnF UBP) found in histone deacetylase 6 (HDAC6) or the Ub-binding zinc finger (UBZ) present in the autophagic receptor, nuclear dot protein 52 kDa (NDP52), also known as calcium-binding and coiled-coil domain-containing protein 2 (CALCOCO2) (in mammals), (5) UBD in A20-binding inhibitor of NF-κB (ABIN) proteins and NF-κB essential modulator (NEMO) (UBAN) domain located in another mammalian autophagic receptor, optineurin (OPTN), (6) and a pleckstrin-like receptor for Ub (PRU) domain, present in the Ub receptor at the proteasome, Rpn13.
We present below a brief description of the Ub-binding modules present in several shuttling receptors involved in the UPS and in autophagy.
3. Ub-Binding Proteins Functioning in the UPS
Among the Ub-binding receptors required for the proteasomal clearance of target proteins are those that are part of proteasome itself and others that only temporarily associate with it during substrate delivery. The intrinsic proteasomal (proteasome-associated) Ub receptors, Rpn10 and Rpn13, can only uptake poly-ubiquitinated substrates that are located in a close proximity to proteasomes [48]. In contrast, shuttling Ub-binding receptors, which are not integral subunits of the proteasome, can recognize ubiquitinated cargos located far from the proteasome and bring them for degradation by the proteasomal clearing machinery [49,50,51,52,53,54,55,56,57,58,59]. Interestingly, known classical proteasomal shuttle proteins [60] contain an N-terminal, Ub-like domain (UbL) and a C-terminal Ub-associated domain/s (UBA), which enable them to deliver poly-ubiquitinated proteins to the proteasome for degradation.
The UbL domain is approximately 80 amino acids long and adopts a typical Ub fold, even though the sequence homology with Ub is limited [61]. The UBA domain is even smaller (approximately 40 amino acids) and adopts a specific fold composed of a bundle of three tightly packed α helices separated by two small flexible regions [62,63,64] in which the first and third helices create the major contact site for Ub binding. Yeast Rad23, Dsk2, Ddi1 proteins, and their homologues belong to the UbL-UBA protein family and are known as proteasomal shuttles [60]. Their domain architectures, together with those of known Ub-binding autophagic shuttling receptors [24,65,66,67], are presented in Figure 2.
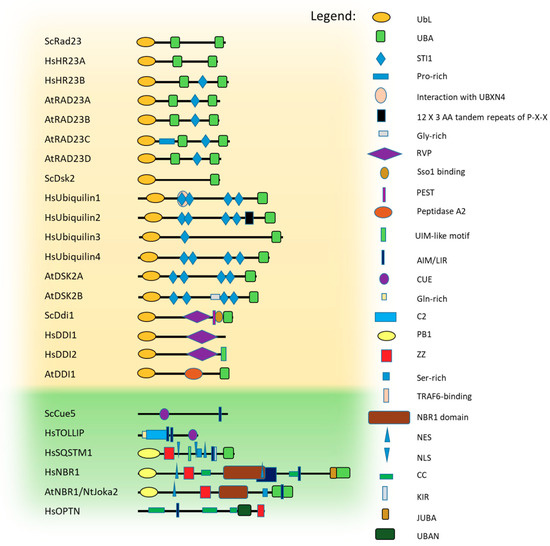
Figure 2.
Ub-binding protein shuttles from yeast, mammals, and plants. Schematic representation of the domain/motif/region organization of the known Ub-binding shuttle receptors used by the proteasomal [59] and autophagic [24,65,66,67] protein quality control (PQC) pathways. The graphical depiction of Ub-binding modules is based on their identification using UniProt and extended by data in the published literature [68,69]. The domain/motif/regions are shown for the proteasomal (S. cerevisiae Rad23 (P32628), H. sapiens HR23A (P54725), H. sapiens HR23B (P54727), A. thaliana RAD23A (Q84L32), A. thaliana RAD23B (Q84L33), A. thaliana RAD23C (Q84L31), A. thaliana RAD23D (Q84L30), S. cerevisiae Dsk2 (P48510), H. sapiens ubiquilin1 (Q9UMX0), H. sapiens ubiquilin2 (Q9UHD9), H. sapiens ubiquilin3 (Q9H347), H. sapiens ubiquilin4 (Q9NRR5), A. thaliana DSK2A (Q9SII9), A. thaliana DSK2B (Q9SII8), S. cerevisiae Ddi1 (P40087), H. sapiens DDI1 (Q8WTU0), H. sapiens DDI2 (Q5TDH0), A. thaliana DDI1 (Q1EBV4)), and autophagic shuttles (S. cerevisiae Cue5 (Q08412), H. sapiens TOLLIP (Q9H0E2), H. sapiens SQSTM1 (Q13501), H. sapiens NBR1 (Q14596), A. thaliana NBR1 (Q9SB64), N. tabacum Joka2 (F8RP79), and H. sapiens OPTN (Q96CV9)). All proteins and domains are drawn to scale. UbL, Ubiquitin-like. UBA, Ubiquitin-associated. STI1, stress-inducible 1. UBXN4, Ubiquitin-regulatory X domain-containing protein 4. RVP, retroviral protease fold domain. PEST, protein rich in Pro, Glu, Ser and Thr. UIM, Ub-interacting motif. AIM/LIR, ATG8-interacting motif/LC3-interacting region. CUE, coupling of Ub-conjugation to ER degradation. C2, Ca+2-dependent membrane-targeting module. PB1, Phox and Bem1. ZZ, Zinc finger. TRAF6, TNF receptor-associated factor 6. NBR1, Neighbor of BRCA1. NES, Nuclear Export Signal. NLS, Nuclear Localization Signal. CC, coiled-coil. KIR, KEAP1 (Kelch-like ECH-associated protein 1)-interacting region. JUBA, Juxta-UBA. UBAN, UBD Ub-binding in A20-binding inhibitor of NF-κB (ABIN) proteins and NF-κB essential modulator (NEMO).
3.1. Rad23
Rad23 was the first UbL-containing protein discovered in yeast [70] and has been the most extensively studied member of the UbL-UBA shuttle family. It is a medium-sized, multi-domain protein containing an N-terminal UbL and two UBAs (central UBA1 and C-terminal UBA2) (Figure 2). Due to various binding modules, Rad23 is considered a scaffold protein since it is involved in a large number of cellular processes including nucleotide excision repair (NER) and, of course, proteasomal degradation of poly-ubiquitinated proteins [71]. The N-terminal UbL domain of Rad23 binds to Rpn1 or Rpn10 of the proteasomal 19S regulatory particle [72,73]. Binding to Rpn1 or Rpn10 positions Rad23 at the epicenter of the base, in close proximity to the entrance of the proteasomal core particle [74]. Similarly, the human homologues of Rad23, HR23A and HR23B, also bind to the proteasomal 19S subunit to present poly-ubiquitinated substrates by interacting with S5a (the human homologue of yeast Rpn10) [75,76]. These proteasome-interacting proteins bridge poly-ubiquitinated proteins and the proteasome by binding Ub-conjugates through their UBA domains which, interestingly, bind mono-Ub and poly-Ub but with district specificities [77]. Based on Rad23 UBA domains studies, UBA1 recognizes Lys63-linked Ub chains and has a major impact on Ub binding to Rad23. In contrast, UBA2 prefers Lys48-linked Ub chains, but affects Ub binding by Rad23 only in a minor manner because deletion of the UBA2 domain does not strongly affect Rad23 binding of poly-ubiquitinated proteins [77,78,79,80,81].
Rad23 also stabilizes proteasome substrates and inhibits the formation of Lys48-linked Ub chains [79,82,83]. Rhp23, the S. pombe homologue of Rad23, protects poly-ubiquitinated conjugates against deubiquitination by the enzyme Ub-specific protease Y (UBPY), which disassembles tetra-Ub chains via its UBA domain. Similarly, Rad23 reduces Rad4 multi-ubiquitination and stabilizes Rad4 levels. Thus, Rad23 and its homologues appear to function both in degradation and stabilization of ubiquitinated substrates.
3.2. Dsk2 and Ubiquilin Protein Families
Dsk2, which was initially identified as a protein involved in spindle pole body duplication [84], is another member of the UbL-UBA protein family characterized by an N-terminal UbL domain and a C-terminal UBA domain (Figure 2). However, its N-terminal UbL domain weakly interacts with the proteasome and binds with lower affinity than the interaction of Rad23 with the UIM domain of Rpn10 [56,72,85,86,87,88,89]. Dsk2 association with proteasomes has been only found in the absence of Rpn10 because free Rpn10 competes for Dsk2 binding to the proteasome [89]. In addition, the binding specificity of the Dsk2 UBA domain is quite unclear. Previous reports noted that Dsk2 binds through its UBA domain to poly-Ub conjugates with a preference for Lys48-linked Ub [87]. However, another group has proven recently that Dsk2 has no substantial preference toward Lys48-linked Ub and binds both Lys48-linked and Lys63-linked, purified Ub chains, as well as to mono-Ub [90]. Contradictory results obtained by these two groups could be related to usage of different experimental approaches. In the first approach, a series of lysine mutants of Ub (e.g., K29R, K48R, K63R) were made and the extracts of yeast cells were used, which could include additional factors affecting Dsk2 binding predisposition to poly-ubiquitinated substrates. The second study used free Lys48- and Lys63-linked poly-Ub chains to measure the binding affinities of isolated Dsk2 UBA (amino acids 300-373) to mono- and di-Ub chains. In addition to its unclear mechanism of action, Dsk2 protects poly-ubiquitinated conjugates against deubiquitination, as seen in Rad23, preventing the disassembly of tetra-Ub chains, and stabilizing Ub conjugates [87,89].
Mammalian ubiquilin proteins are the closest homologues to yeast Dsk2 protein. The human genome contains four ubiquilins (UBQLN1–4). While ubiquilin1 is ubiquitously expressed and ubiquilin2 and ubiquilin4 are expressed in most tissues, ubiquilin3 is only expressed in testis. All ubiquilins are highly conserved and share high degrees of sequence and domain structural homology to each other. Like yeast Dsk2, all ubiquilins harbor UbL domains at their N-termini and UBA domains at their C-termini. Located between these two domains are STI1 (stress-inducible heat shock chaperonin-binding motif) motifs, which may confer ubiquilins with chaperone-like functions (Figure 2) [91,92]. The best characterized member of the this family is ubiquilin1, which is also known as PLIC-1 (protein linking integrin associated protein with cytoskeleton 1). This cytosolic protein not only delivers poly-ubiquitinated proteins to the 26S proteasome [56,93] but also associates with aggregates or aggresomes [94] and delivers substrates to lysosomes for degradation via autophagy and chaperone-mediated autophagy (CMA) [95,96]. Additionally, its involvement in the ER-associated degradation (ERAD) pathway has also been documented [97]. Therefore, it is not a surprise that the UBA domain of ubiquilin1 shows similar binding affinity to Lys48-linked and Lys63-linked Ub chains [77].
3.3. Ddi1-Like Proteins
DNA Damage-Inducible 1 (Ddi1) protein also belongs to a family of shuttle proteins targeting poly-ubiquitinated cargo for proteasomal clearance, but differs from the other shuttling proteins in its proteolytic roles and its interacting partner. Like Rad23 and Dsk2, it usually contains an N-terminal UbL as well as a C-terminal UBA domain, which has been lost in mammals (but not in plants) (Figure 2).
It also has additional unique domains, such as conserved retroviral protease (RVP) fold domain, followed by a putative protein rich in Pro, Glu, Ser and Thr (PEST) region and Sso-binding domains (Sso-BD) [68,69]. Besides transferring ubiquitinated substrates to the proteasome for degradation [55,58,98,99], Ddi1 participates in many other cellular processes [68,81,98,100]. While the Ddi1 UBA domain forms a characteristic UBA-Ub complex, the Ddi1 UbL domain has unusual binding preferences, which may explain why, in higher eukaryotes, the UBA domain has been lost. In contrast to the Ub and UbL domains of Rad23 and Dsk2, the UbL domain of Ddi1 does not interact with UBA domains or the UIM of Rpn10 [101] and associates very weakly with Rpn1 [102]. Instead it binds Ub quite strongly by forming a unique interface mediated by hydrophobic contacts and by salt-bridges between oppositely-charged residues of the Ddi1 UbL domain and Ub [103].
Because the UbL domain of Ddi1 binds both Ub and the proteasome, it has a different mechanism of docking to substrates in comparison with Rad23 and Dsk2 proteins and may not act as a classic shuttle receptor. Instead, Ddi1 might assist Rad23 and Dsk2 rather than compete with them for targeting ubiquitinated substrates to the proteasome. For example, via the sequential dissociation of UbL from ubiquitinated substrates and its association with Rpn1, Ddi1 could increase the concentration of Ub conjugates in the proximity of the proteasomal 19S subunits and, therefore, expose poly-ubiquitinated proteins to the proteasome-associated Ub receptors, Rpn10 and Rpn13. Alternatively, via hetero-dimerization between Ddi1 and Rad23 through their UBA domains, a tandem (Ddi1-Rad23) shuttle could be formed [104]. Moreover, dimerization of Ddi1 through its RVP domain, which has been documented, might allow its simultaneous binding to an ubiquitinated substrate and to the proteasomal 19S subunit [103].
Loss of the UBA domain in the human Ddi1-like protein, DDI2, is partially restored by the presence of a novel UIM-like motif at the C-terminus of DDI2, which weakly but specifically binds mono-Ub [105]. Such a motif was not identified in human DDI1 [105].
4. Ub-Binding Receptors in Autophagy
Another group of shuttles plays a key role in both the recognition and the delivery of cytoplasmic substrates for autophagic clearance. In mammals, Ub tags serve as a general mechanism for labeling various substrates (from individual proteins through organelles to pathogens) for autophagic turnover. In yeast, recognition of autophagic cargo is not mediated by a common tag, and Cue5 is the only yeast receptor for Ub-dependent autophagy [106]. In contrast, in mammals, several autophagic receptors associate with poly-ubiquitinated proteins to deliver these substrates for autophagic clearance. These include TOLLIP, the mammalian homologue of Cue5 [106], SQSTM1, and NBR1, which are the best characterized autophagic receptors that recognize a broad range of substrates and are involved in almost every type of selective autophagy [27,29,30,31,107,108,109,110,111,112,113], OPTN, which is involved in the autophagic clearance of ubiquitinated aggregates, pathogens, and mitochondria [33,114,115,116], and, finally, NDP52, which is required for the autophagic clearance of ubiquitinated mitochondria and pathogens [117,118,119].
Unfortunately, little is known about autophagic receptors in general and about Ub-dependent receptors in plants. So far, based on domain architecture homology, AtNBR1/NtJoka2, hybrid homologues of human SQSTM1 and NBR1 have been described [65,66] (Figure 2).
4.1. CUET Proteins
CUE-domain targeting (Cue5-TOLLIP; CUET) proteins are probably ancient autophagic receptors involved in the clearance of cytotoxic protein aggregates [106]. CUETs are functionally conserved from yeast to mammals. This group of proteins is represented by yeast Cue5 and its human homologue, TOLLIP, and binds Ub via its CUE domain [106,120]. However, the domain organizations in TOLLIP and Cue5 are notably different (Figure 2). TOLLIP contains, in addition to a CUE domain, a large N-terminal extension harboring a TOM1 (target of myb1 homologue)-binding domain and a phospholipid-binding Ca+2-dependent membrane-targeting module (C2), which are postulated to be required for other non-autophagic functions [121,122]. TOLLIP plays several roles within the cells such as in protein traffic by the endocytotic pathway and in Toll-like receptor (TLR)-mediated innate immunity responses [122,123]. Also, in contrast to Cue5, which contains one C-terminally located ATG8-interacting motif (AIM), TOLLIP contains two functional LC3-interacting regions (LIRs) located within the C2 domain [106].
Interestingly, the CUE domains of both Cue5 and TOLLIP have no preferences toward Lys48-linked or Lys63-linked Ub chains and, therefore, might not distinguish between various types of Ub conjugates [106]. Because the CUE domains of Cue5 and TOLLIP bind mono-Ub and poly-Ub chains of various types, it is suggested that CUET receptors clear cells of protein aggregates [124,125], which contain diverse Ub linkages as a consequence of clustering various proteins ubiquitinated by different E3 ligases. In agreement with such a hypothesis, TOLLIP binds Ub-conjugated proteins better than does SQSTM1. The CUE domain of TOLLIP binds free Ub with similar affinity as does SQSTM1, but its binding to Lys48-linked and Lys63-linked poly-Ub chains is stronger than that of SQSTM1, especially those of a longer chain length [106]. In view of this ability to bind Ub, it is surprising that TOLLIP is recruited to aggregates independently of its recognition of Ub [106]. Further studies on TOLLIP might reveal the mechanism involved in TOLLIP targeting to aggregates.
Additionally, with regard to TOLLIP’s Ub-binding propensity, two contradictory results were obtained. In contrast to a previous report [120], a recent study found no evidence for Ub-binding activity for the C2 domain and has proven only the involvement of the CUE domain in Ub recruitment, as it is in Cue5 [106]. However, constructs of different length, containing the C2 domain, were used in these two experiments. While Mitra et al. [120] clearly showed that the isolated C2 domain of TOLLIP (amino acids 54–182) bound Ub with a similar affinity as other known Ub-binding domains [53]. Lu et al. [106] used the N-terminal (amino acids 1-180) region of TOLLIP containing the C2 domain. It is possible that the N-terminal region of TOLLIP, which precedes the C2 domain, has an inhibitory effect on Ub binding to the C2 region. This matter also needs further clarification.
4.2. SQSTM1
SQSTM1 was the first selective autophagy receptor discovered in mammals and is also the best characterized one [108,126,127]. It is a multi-functional protein consisting of an N-terminal Phox-BEM1 (PB1) domain, which is a central ZZ-type zinc finger domain, and a C-terminally located UBA domain. SQSTM1 also contains one LIR and a KEAP1 (Kelch-like ECH-associated protein 1)-interacting region (KIR), as well as a functional nuclear localization signal (NLS) and a nuclear export motif (NES) (Figure 2) [108,127,128,129,130]. Although SQSTM1 does not have a specific UbL region seen in the proteins of the UbL-UBA family, it does have a PB1 domain resembling the UbL domain. This fact, in combination with the presence of a UBA region in SQSTM1, may explain some of the properties that SQSTM1 shares with other UbL-UBA proteins. For example, it interacts via its UBA domain with poly-ubiquitinated proteins with preference for the Lys63-linked Ub chains and binds to the proteasome via its PB1 domain, which, like the UbL domain, forms a Ub-like, β-grasp fold, recognizable by the proteasomal components, S5a and Rpt1 [131,132]. The presence of both the NLS and NES motifs allows SQSTM1 to shuttle between the nucleus and cytoplasm [128]. This protein is implicated in autophagy and in several other pathways including DNA repair [133], the KEAP1-NRF2 signal transduction pathway [134], and in Wnt [135] or NFκB signaling [136]. However, SQSTM1 is mainly known as an autophagic receptor [27,29,31,108,109,134,137,138]. The PB1 domain of SQSTM1 has a high potential for homo-oligomerization, as well as for hetero-oligomerization with PB1-containing proteins, such as NBR1 [29]. The interaction of SQSTM1 and NBR1 organizes SQSTM1 into higher oligomers, which promotes SQSTM1 clustering with Ub-positive aggregates during aggrephagy, wherein its functional LIR motif strongly associates with mammalian LC3/GABARAP [108,139].
In addition to its role as an autophagic receptor, SQSTM1 also regulates the accumulation and autophagic clearance of protein aggregates. Recent studies show that SQSTM1 co-localizes with HDAC6 at ubiquitinated aggregates and by binding to the catalytic deacetylase domain 2 (DD2) of HDAC6, SQSTM1 negatively regulates HDAC6 deacetylase activity, promotes F-actin network assembly at the MTOC, and assists autophagosome-lysosome fusion [140].
4.3. NBR1
Like SQSTM1, NBR1 contains a PB1 domain at its N-terminus, which is followed by the ZZ domain and an UBA domain at its C-terminus. However, instead of the single LIR motif in SQSTM1, NBR1 contains two LIRs (one conforming to the typical consensus, W/F/YXXL/I/V, and the other being atypical) [30]. Besides these modules, NBR1 also contains two coiled-coil (CC) domains involved in NBR1 homo-oligomerization. This is a feature that allows its association with the microtubule network through microtubule-associated protein 1B (MAP1B) [141]. Additionally, NBR1 also has, preceding the UBA domain, a JUBA (juxta-UBA) α-helix that binds lipids [142]. The in silico analysis of the NBR1 protein sequence reveals two potential NESs, but no NLS (Figure 2).
Both NBR1 and SQSTM1 play critical roles in the recruitment of aggregate-prone proteins, long-lived proteins, or damaged organelles into autophagosomes and in the formation of protein aggregates. However, the occurrence of several distinct, dissimilar modules in NBR1 and SQSTM1 also confers on these proteins some additional unique properties. While SQSTM1 is considered as a main receptor for aggrephagy, and NBR1 significantly contributes, it is not essential [27,31]. The unique JUBA region that allows NBR1 to anchor directly to organelle membranes makes it a necessary and sufficient receptor for pexophagy (peroxisome degradation via autophagy) [111,142]. Moreover, the PB1 and UBA domains of NBR1 differ from those of SQSTM1 in oligomerization ability and interaction with Ub, respectively. The PB1 domain of SQSTM1 has the ability to polymerize [143,144,145] while this domain of NBR1 is unable to homo-oligomerize. Yet, it can still bind another PB1 domain (such as that in SQSTM1) [143,146]. Recently, the UBA domain of NBR1 was described to have a much higher affinity for Ub than that of SQSTM1 [147].
4.4. Plant NBR1-Like Proteins
NBR1 from Arabidopsis [66] and Joka2 from tobacco [65] behave as hybrid proteins that combine features of mammalian SQSTM1 and NBR1 and contain a similar PB1, ZZ, and UBA domain architecture [65,66,148]. However, instead of one C-terminally located UBA, plant receptors contain two, non-identical UBA domains, where only the second domain (UBA2) binds Ub in vitro [66] (Figure 2). Plant NBR1-like proteins target insoluble, detergent-resistant, poly-ubiquitinated protein aggregates accumulated during plant abiotic stresses and non-assembled particles of a cauliflower mosaic virus (CaMV) for their degradation via NBR1-mediated autophagy [149,150].
Both AtNBR1 and NtJoka2 are strongly aggregating proteins [65,66,148,149]. Like SQSTM1, but in contrast to mammalian NBR1, each of them possesses an acidic/basic PB1 surface that allows these NBR1-like proteins to self-oligomerize through their PB1 domain [66,148]. Thus, no CC regions analogous to those in mammalian NBR1 are necessary for oligomerization in the plant NBR1-like receptors. Consistent with the mechanistic data regarding cluster formation by SQSTM1, the PB1 domain in Joka2 is sufficient for its oligomerization, but the C-terminal region containing both UBA1 and UBA2 domains additionally promotes this process [148]. In addition, like all known autophagic receptors, plant NBR1-like proteins directly bind to ATG8 protein family members. Several potential AIMs are present within their sequences and at least one of them, located between the UBA domains, is functional [66].
Moreover, based on studies with Joka2, these proteins possess a functional NES and an uncharacterized NLS, which allows shuttling between the nucleus and the cytoplasm, analogous to SQSTM1 [148]. The role of NBR1-like proteins in the nucleus can only be hypothesized at present based on plausible functional conservation between plant NBR1-like proteins and SQSTM1. It is unclear whether these proteins like mammalian NBR1 contain a functional JUBA domain.
4.5. OPTN
Optineurin (OPTN) is another multi-domain protein that interacts with many partners and has versatile functions beside its role as an Ub-dependent autophagic receptor. Among its interactors are proteins involved in membrane and vesicle trafficking (Rab8, huntingtin, myosin VI), cellular morphogenesis, NF-κB regulation, signal transduction, transcription regulation, and cellular division control [151,152,153,154,155,156]. OPTN regulates autophagic degradation of mitochondria and cytosolic Salmonella enterica in processes called mitophagy and xenophagy, respectively, as well as of protein aggregates in both a Ub-dependent and Ub-independent manner [32,157].
OPTN involvement in autophagy depends on the activation of its LIR motif via the phosphorylation of a Ser residue preceding the core sequence, W/F/YXXL/I/V, by the TANK-binding kinase 1 (TBK1) [114,116]. OPTN interacts with linear and Lys63-linked poly-Ub chains via its UBAN domain [158] (Figure 2). However, OPTN can recognize protein aggregates not only in a Ub-dependent manner through its UBAN domain, which is required to recognize ubiquitinated Salmonella enterica [116] or mitochondria [114,115], but also through its C-terminal CC domain in a Ub-independent manner [32]. Moreover, a recent report has established that HECT domain and ankyrin repeat-containing E3 ubiquitin protein ligase 1 (HACE1)-mediated Lys27-linked and Lys48-linked poly-ubiquitylation of Lys193 in OPTN might activate autophagic removal of oxidative damaged proteins by promoting the formation of a SQSTM1-OPTN complex [159]. These data are consistent with the observation that OPTN can positively modulate protein aggregation [32].
In addition, to these roles in autophagy, OPTN also drives autophagosome maturation by interacting with myosin VI (MYO6), which is involved in the fusion of autophagosomes with endosomes [160].
Furthermore, it must be stressed that OPTN, like other autophagic receptors discussed in this scenario, can oligomerize to form 420 kDa homo-oligomers, which, based on the 67 kDa monomer size, were judged to be hexamers [161]. Unlike other autophagic receptors, which are degraded by autophagy, OPTN is normally degraded by the proteasome pathway [162].
5. PQC Pathway Choice Based on the Ub Code
Poly-Ub chains consisting of Lys48-linkages are synthesized by many E3 ligases and, therefore, are among the most abundant in the cell. Their level rapidly increases when proteasomes are inhibited, which points to their involvement in proteasomal degradation [163,164,165,166]. Moreover, a variety of E3 ligases, including UBR box N-recogins [167,168], gp78 - also known as AMFR (autocrine motility factor receptor) [169], E6AP (E6-associated protein) [170], and SCF (stem cell factor) [171], assemble Lys48-linked poly-Ub chains for proteasomal degradation, while others like Parkin [172], TRIM13 (tripartite motif-containing protein 13) [173], and CHIP (carboxyl terminus of Hsc70 interacting protein) [174] assemble Lys63-linked Ub chains on substrates for autophagic turnover. Therefore, in the Ub recognition code, it is well established that Lys48-linked Ub chains typically mark a protein for proteasomal degradation, while mono-ubiquitination and Lys63-linked Ub chains seem to play a role in autophagy [175,176]. Many early studies on Ub-binding receptors were in agreement with this hypothesis. For example, initial studies on the UBA domains from the UbL-UBA shuttles proposed that Dsk2/Dph1 from yeasts (in S. cerevisiae and S. pombe, respectively) and DSK2A-B from the Arabidopsis preferentially bind to Lys48-linked Ub chains, rather than to Lys63-linked Ub chains or mono-Ub [85,87,177]. Human HR23A also bind Lys48-linked Ub chains [83]. Similarly, the plant RAD23 family also has a strong preference for proteins marked by Lys48-linked Ub chains [178]. This led to the notion that the primary, Ub-based, proteasomal degron is the Lys48-linkage and UbL-UBA proteins such as yeast Dsk2, Rad23 and Ddi1, as well as their homologues in higher eukaryotes are proteasomal shuttling receptors.
Conversely, early studies on UBA domains of SQSTM1 and NBR1 showed that these proteins preferentially bind Lys63-linked Ub chains. The NBR1 UBA domain binds to Lys63-linked di-Ub about 60 times stronger than to mono-Ub [30]. Similar results have been found for the UBA domain of SQSTM1, which prefers binding to Lys63-linked Ub chains, but still has a lower affinity for Lys48-linked Ub chains [139,179,180]. On the basis of these experiments, it was proposed that Lys63-linked Ub chains mark proteins for autophagic clearance.
However, increasing evidence is accumulating to suggest that the Ub recognition code is inadequate to explain how a decision is made between proteasomal degradation and autophagic turnover as alternative PQC pathways. For example, further studies on the UBA domain of NBR1, which eliminated an artifact baused by dimerization of the glutathione-S-transferase tag fused to it, have shown that it lacks specificity for poly-Ub linkages [147]. Specifically, the UBA domain of NBR1 has no preference for Lys63-linked or Lys48-linked di-Ub and binds to each monomeric unit of Lys48-linked and Lys63-linked poly-Ub with similar affinities and via the same surface as its binding to mono-Ub [147]. It is especially intriguing in the context of this recent finding that Ub-positive inclusions do not show any linkage specificity, which demonstrates that Lys63-linked poly-Ub is not an exclusive and definitive label for autophagy [181].
New studies on yeast Dsk2 also require modifications of the previous views on the prevalent signals for proteasomal degradation. The Dsk2 UBA domain has high affinity for both mono-Ub and poly-Ub chains, but no substantial preferences for Lys48-linked or Lys63-linked Ub chains [90]. Similarly, the UBA domain of ubiquilin1 shows little or no binding selectivity toward a particular chain linkage, or between the two Ub moieties in the same chain [101].
Moreover, recently multiple Lys48-linked di-Ub conjugates have been proven to serve as stronger degrons for proteasomal degradation than tetrameric Lys48-linked poly-Ub chains, which were believed to be the most efficient proteasomal degron [182,183]. Additionally, recent studies indicate that other linkages could also be recognized by the proteasomal and autophagic machineries [34,175,184]. For instance, Lys11-linked Ub chains trigger proteasomal degradation of cell cycle regulators during mitosis [185,186]. In addition, but less frequently, Lys29-linked and Lys63-linked Ub chains mediate proteasomal degradation and reveal the plasticity in the recognition of Ub labels by degradation machineries [187,188,189,190,191]. These variations mean that the Ub code on certain substrates may not uniquely determine which degradation path they will follow. Almost any type of Ub chain: homogenous, heterogenous, linear, single, and multibranched chain, as well as mono-Ub and multi-mono-Ub label can be accepted by proteasomes [34]. For instance, SQSTM1, which delivers primarily ubiquitinated cargoes for autophagic degradation, also shuttles Lys63-linked poly-ubiquitinated substrates for proteasomal degradation. The UBA domain of SQSTM1 recognizes Lys63-linked poly-Ub chains of tau and mediates the interaction between Lys63-linked, poly-ubiquitinated tau [192] and the PSMC2 (proteasome 26S subunit, ATPase 2) homologous to yeast Rpt1 subunit of proteasomes, which is a docking site for shuttling factors. Similarly, SQSTM1 is involved in the proteasomal turnover of Lys63-linked, poly-ubiquitinated TrkA, also known as NTRK1 (neurotrophic receptor tyrosine kinase 1) [132]. In this regard, the function of SQSTM1 is similar to that of other proteins containing UBA and UbL domains, such as yeast Ddi1, Rad23, and Dsk2 (and their homologues in higher eukaryotes) in shuttling of substrates to the proteasome. However, instead of delivering proteins marked by Lys48-linked poly-Ub chains, substrates with Lys63-linked Ub chains are targeted to the proteasomes.
Recent studies on human and plant homologues of Dsk2 reiterate that shuttling of SQSTM1 between both degradation systems is not an exception among Ub-binding proteins. While human HR23 is still a major receptor for the proteasome pathway, ubiquilins function in several distinct protein degradation pathways by delivering substrates to either the proteasome, autophagy, or ERAD pathways [97]. In response to accumulation of aggregation-prone proteins, ubiquilin1 with its cargo is recruited by epidermal growth factor substrate 15 (EPS15) to the MTOC, where aggresomes form [94]. These results have been explained in terms of a model that proposes a dual role for ubiquilin1, whose binding to the UIM domains of the proteasomal subunit S5a and the EPS15 protein is mutually exclusive [94]. Thus, when there are low levels of protein aggregation, ubiquilin1 would promote the shuttling of poly-ubiquitinated proteins to the proteasome. However, when the level of protein aggregation is high and proteasomes may be running at capacity, ubiquilin1, via its UbL domain, can alternatively interact with EPS15. Therefore, this promotes aggresome formation. However, it is unknown if ubiquilin1-EPS15 interaction is evolutionarily conserved. Consistently, it has been reported that yeast Dsk2 facilitates the vacuole-mediated clearance of misfolded proteins by promoting inclusion body formation [193] and loss of the yeast homologue of EPS15, Pan1, enhances protein aggregate toxicity [194].
Ubiquilin2 and ubiquilin4 also have a role in autophagy. Their knockdown renders cells more susceptible to starvation-induced cell death while overexpression has a protective effect [96]. Ubiquilin2 co-localizes and co-immunoprecipitates with LC3 and assists in the maturation of autophagosomes to autolysosomes [95,96,195]. Additionally, ubiquilin4 directly associates with LC3 and, as an LC3-interacting partner, links the cargo-carrying ubiquilin1 to the autophagy machinery [196].
Other work has clearly indicated that plant DSK2, which associates with Lys48-linked Ub chains more strongly than with Lys63-linked Ub chains [177], when properly phosphorylated, serves as an autophagic receptor for bri1-EMS-suppressor 1 (BES1) protein due to the enhancement of its interaction with the autophagy protein ATG8 [197].
Lastly, recent results show that ubiquilin2 acts with the HSP70-HSP110 (heat shock 70 kDa protein—Heat shock 110 kDa protein) disaggregase machinery to deliver aggregated and misfolded proteins to the proteasome for their degradation [198] and, therefore, mediates autophagy-independent protein aggregate clearance by the proteasome and adds another layer of complexity in the fate of ubiquitinated proteins [198]. In the model proposed by these authors, under non-stressed conditions, ubiquilin2 is held inactive as homo-dimers or hetero-dimers, but the presence of HSP70 and ubiquitinated misfolded/aggregated proteins activates ubiquilin2, which then associates with HSP70 [198]. Next, HSP70/HSP110-dependent disaggregase activity pulls aggregated proteins apart, which allows ubiquilin2 to drive ubiquitinated misfolded proteins to the proteasomes for degradation.
Together, these results expose new complexities in the regulated proteolytic turnover of ubiquitinated substrates. Autophagic receptors shuttle between proteasome and autophagy pathways and proteasomal receptors might be tied to both degradation mechanisms. Since bi-directionality (towards proteasomes and autophagy) of Ub-binding shuttles appears to be a common mechanism that links the autophagy pathway and the UPS, previous models suggest that the Ub code alone determining this fate is not sufficient. It must be noted here that the Ub code indubitably plays an important role in the initial pre-commitment process. However, in light of recent data, the linkage of the accurate Ub label is insufficient to assign a protein substrate irreversibly to particular degradation pathway. These facts raise the question regarding which other factors may affect the PQC pathway choice.
6. PQC Pathway Choice Based on an Oligomeric State of the Receptor and Recognition of ATG8/LC3/GABARAP Proteins
To unravel the crucial determinants for the PQC pathway choice, a set of modular artificial receptors harboring identical Ub-binding modules, but with various oligomerization potentials, was created [90]. These modules either included or excluded an AIM/LIR and/or the UbL domain. Monomeric and dimeric forms of artificial receptors that contained the UbL domain (regardless of whether they possessed an AIM or not) supported efficient proteasomal degradation of soluble proteins but failed in clearing aggregated proteins. Conversely, oligomeric receptors containing an AIM (but not the UbL) supported autophagy-dependent degradation of aggregates but were impaired in the proteasomal clearance of soluble proteins. Thus, it has been clearly demonstrated that the PQC pathway choice can still be made at the level of the receptor after the initial substrate ubiquitination by competition between Ub-binding receptors harboring either proteasome-binding or ATG8-binding modules [90]. Therefore, a predisposition to organize higher oligomers and to bind ATG8 seems to be essential for autophagic degradation.
All known autophagic receptors involved in aggregate clearance, such as SQSTM1, NBR1, OPTN and others described above, can oligomerize and interact (directly or indirectly) with ATG8/LC3/GABARAP family members. For instance, SQSTM1 polymerizes via its PB1 domain into helical filaments and even assembles into higher order hetero-oligomers with different proteins like autophagy-linked FYVE protein (ALFY), a BEACH domain– and WD40-containing protein (WDR81), or Huntingtin (HTT), which additionally stimulate SQSTM1-mediated clustering of aggregated proteins and their autophagic turnover [26,137,199,200,201]. NBR1 self-oligomers via its CC1 domain and forms hetero-oligomers with SQSTM1 by PB1-PB1 interactions to render SQSTM1 clustering more efficient [27]. OPTN also oligomerizes and interacts with SQSTM1 [202].
Oligomerization of SQSTM1 is a factor that generates high-avidity interactions with Ub and the LC3/GABARAP proteins and, therefore, initiates autophagic turnover of substrates bound to SQSTM1. Oligomerization of SQSTM1 promotes the interaction with Ub and LC3B by drastically increasing the residence time of SQSTM1 on LC3B and Ub-coated aggregates [139], which may, perhaps, allow aggregates to be delivered by SQSTM1, together with the Ub-binding receptor, to autophagosomes and, subsequently, to lysosomes [139]. Consistently, deletion of the PB1 domain of SQSTM1, or mutations that interfere with its ability to multimerize, inhibit the recruitment of SQSTM1 to autophagosomes [138].
With regard to receptor binding to ATG8/LC3/GABARAP proteins, in some Ub-binding receptors, the AIM/LIR motif may be cryptic, and require activation by phosphorylation. Binding to ATG8/LC3/GABARAP proteins family members might also be indirect via other proteins that mediate receptor binding to the autophagosome membrane. For example, the function of ubiquilin1 as an autophagic carrier is mediated through ubiquilin4, which recognizes LC3 via two STI1 repeats lacking a canonical LIR [196].
7. PQC Pathway Choice Based on Other Factors
7.1. Chaperone Assistance in the Pathway Choice
The best example of the role of chaperones in modulating and regulating the PQC pathway choice comes from the studies on the Bcl2-associated athanogene (BAG) family of proteins in mammals, which are implicated in the coordination of proteasomal and autophagic activation. BAG1 is a well-known HSP70 chaperone that recognizes poly-ubiquitinated misfolded or unfolded substrates and initiates their sorting for proteasomal degradation [203,204]. However, in case of proteasome impairment or overload caused by stress or aging, another member of BAG family, BAG3, promotes turnover of poly-ubiquitinated proteins by autophagy [205,206,207,208]. BAG3 not only induces expression of LC3B and SQSTM1, but also directly interacts with SQSTM1, redirecting for autophagic degradation proteins labeled by Lys48-linked Ub chains that may originally have been destined for proteasomal clearance [209]. Moreover, BAG3, by interacting with the dynein motor and 14-3-3 proteins, actually induces the sequestration of poly-ubiquitinated proteins into aggresomes at the MTOC [210]. In summary, when cells switch their expression form BAG1 to BAG3, they also switch PQC systems from BAG1-mediated proteasomal degradation to BAG3-mediated autophagy.
Since all studies on the BAG1/BAG3 ratio and BAG3-mediated autophagy have come from the animal kingdom [206], it is worth asking if such modulation of the PQC system is also present in plants where the BAG protein family is conserved [211,212,213,214,215].
7.2. Conformational Changes of the Ub-Binding Receptors—The Intramolecular and Intermolecular Interactions
A number of proteins that contain both UbL and UBA domains, including the Ub-binding receptors involved in proteasomal degradation, form intramolecular interactions between the UbL and UBA domains. These domains from Rad23 and Dsk2 display intramolecular interactions even though the binding affinity was low [216]. Similar results have been obtained for human HR23A [75] and HR23B [76]. Such an interaction is possible due to adoption of an Ub-like fold by the UbL domain, which is recognized by the UBA domain. Studies on HR23A have revealed that UbL domain binding to the UBA domain and to proteasomal subunit S5a are mutually exclusive and HR23A loses its interdomain structure as a consequence of S5a binding [75]. At the same time, binding of the HR23A UBA domain to Ub also structurally alters the rest of the HR23A protein, wherein residues within the UbL domain that reside in the UBA contact surface shift. This suggests that binding of Ub to the UBA domain would prevent the intramolecular interaction with the UbL domain [217]. Based on these results, it seems reasonable that in the absence of proteasomal subunits, or Ub substrates that could interact with the UbL and UBA domains, respectively, intramolecular interaction between these domains can “lock” these proteins into a “closed” conformation. Because Dsk2 and Rad23 dimerize via their UBA domains [104,218], “closed” conformations would affect their homo-dimerization. However, not all UbL-UBA shuttles dimerize through their UBA domain. Ubiquilin1 mutants deleted for either their UbL or UBA domain retain the ability to interact with each other, which indicates that the central region is required for the interaction [219]. Similarly, Ddi1 can form homo-dimers even after the deletion of the UbL or UBA domains, which indicates that their dimerization is also mediated by the central region [104]. Therefore, it is an open question if adoption of such “closed” conformation would affect dimerization of these proteins.
Additionally, intermolecular interactions between these domains have also been detected. For instance, the UBA domain of one of the human Dsk2 homologues, ubiquilin2, can interact with the UbL domain of HR23Aas well as UBA2 domain of HR23A can interact with the UbL domain of ubiquilin2 [220]. Because intermolecular UbL-UBA binding should prevent the intramolecular UbL–UBA interaction in HR23A, it is possible that competition between intra-molecular and intermolecular UbL-UBA interactions occurs, which leads to the formation of shuttle monomers or multimers.
The NMR studies of the PB1 domain have shown that it creates an Ub-like, β-grasp fold, similar to the UbL domain [221]. Therefore, it has been postulated that the PB1 domain also directly interacts with its UBA [222,223]. Weak interaction between PB1 and UBA domains has been detected for the plant homologue of SQSTM1, Joka2 [148]. The Joka2 PB1-UBA interactions were only observed in aggregates, which suggests that clustering of Joka2 into higher order homo-oligomers requires PB1 oligomerization, as well as PB1-UBA interactions [148].
Thus, it appears that all proteins with UbL/PB1 and UBA domains possess the ability to dimerize/oligomerize through either the PB1 or UBA domains or the central region of the protein, and can form other intramolecular UbL/PB1-UBA interactions that could affect their predisposition to aggregation.
7.3. Post-Translational Modifications of the Ub-Binding Receptors
Post-translational modification is a common mechanism to modify protein properties. Additionally, when such modifications affect protein localization or function, it adds further flexibility to protein utility. Since Ub-binding protein shuttles cooperate with both proteasomal and autophagic PQC machineries, it can be expected that their involvement in these pathways should be regulated via various post-translational modification(s). Many shuttling receptors undergo specific modifications to properly respond to the cellular needs (Table 1). In this section, we discuss those modifications that might influence the decision-making process between autophagy and the UPS. For example, it has recently been reported that SQSTM1 is ubiquitinated at Lys7 within its PB1 domain by TRIM21, which negatively affects its oligomerization and sequestration activity [224]. Unfortunately, it is unknown if this has any effect on its involvement in proteasomal clearance (since PB1 binds Rpt1 and Rnp10 subunits of proteasome).

Table 1.
Ub-binding receptors involved in protein quality control (PQC) pathways.
Interestingly, SQSTM1 can be ubiquitinated within its UBA domain at the highly conserved Lys420 by KEAP1/CUL3 (Cullin-3). Modifications mediated by KEAP1/CUL3 increase SQSTM1 sequestering activity, association with LC3B, and its final degradation by affecting UBA dimerization [244]. It is unclear if this modification abrogates self-association of the UBA domain to increase Ub binding (simultaneous dimerization and Ub binding are mutually exclusive) or whether interaction of the UBA domain with other proteins promotes SQSTM1 oligomerization. However, it is clear that different ubiquitination statuses of SQSTM1 can positively or negatively affect its role as the autophagic receptor and, thereby, alter the fate of ubiquitinated substrates (for instance, TRIM21 ubiquitination would antagonize KEAP1/CUL3 mediated ubiquitination at the C-terminal UBA domain).
SQSTM1 is subject to phosphoregulation as well. Phosphorylation of Ser403 and Ser409 within the UBA domain of SQSTM1 destabilizes self-association of this domain and, therefore, positively regulates SQSTM1 activity [229,237]. It is suggested that phosphorylation at Ser403 increases SQSTM1 affinity for Ub by enabling the formation of additional polar contacts with Ub, analogous to polar contacts of the UBA domains of NBR1 and Dsk2, formed by Glu926 and Asp341, respectively, with the side chains of Lys6 and His68 in Ub. Because NBR1 and Dsk2 permanently possess negative residues upstream of the conserved MGF motif, which plays an integral role in Ub recognition, their binding to Ub is always stronger than that of the unphosphorylated UBA in SQSTM1 [147]. Intriguingly, around 17% of all known UBA domains have Ser residue in a position corresponding to Ser403 in SQSTM1, which suggests that phosphorylation might be a wide-ranging mechanism of attenuation of Ub-binding affinity in UBA domains and the subsequent activation of these proteins to recognize Ub conjugates.
Phosphoregulation of the AIM/LIR within Ub-binding receptors is another decisive post-translational modification that impacts PQC pathway choice by shuttling receptors. The canonical AIM/LIRs are represented by the W/F/F-X-X-L/I/V consensus, which is often adjacent to acidic or phosphorylated residues that form additional contact sites with members of the ATG8/LC3/GABARAP protein family [245,246]. Human SQSTM1 and NBR1, as well as its NBR1-like counterpart from A. thaliana, already contain negatively charged residue(s) just upstream of the core AIM motif, making them receptors already “primed” for autophagy [24]. However, about 25% of known AIM/LIR motifs harbor Ser or Thr residues at that position suggest that functionality of these AIM/LIRs might be regulated through phosphorylation. For instance, plant DSK2, when phosphorylated by the glycogen synthase 3 (GSK3)-like kinase, BIN2 (brassinosteroid insensitive 2), near its AIMs, targets BES1 for autophagic clearance. In other words, the targeting of BES1 for autophagic degradation is possible due to the DSK2-ATG8 interaction mediated by the activation of cryptic AIMs by BIN2 phosphorylation [197].
In contrast, phosphorylation of NBR1 by GSK3 at Thr586 within its non-canonical LIR inhibits its adaptor function in the formation of proteins aggregates and, therefore, prevents the aggregation of poly-ubiquitinated proteins and their subsequent selective degradation [242].
In summary, posttranslational modifications can modulate the functional status of Ub-binding shuttles and, therefore, can affect the pathway choice between the UPS and autophagy.
8. Suggested Model and Concluding Remarks
Proteasomal degradation and autophagy are two independent elements of the PQC system, but they act in a networked and interactive manner to maintain proteostasis. Multiple lines of evidence presented above suggest that Ub-binding shuttles can play roles as integration centers of both systems, which, thereby, coordinates the communication between the UPS and autophagy in the clearance of protein aggregates. The old model, focusing primarily on the Ub code, is incapable of explaining clearly the pathway chosen by a given ubiquitinated substrate. In view of new available data, we propose a model in which the Ub-binding receptors along with their associated regulators (interacting partners, chaperones, proteins conferring post-translational modifications) and the Ub code integrate the complex hierarchy of decisions regarding how unwanted proteins will be degraded.
In this model, both systems known as the UPS and autophagy share the ubiquitination process that label substrates. This is the first step where a pre-commitment decision can be made regarding which system will be used for protein clearance (Figure 1 and Figure 3). There is evidence demonstrating that labeling proteins with particular Ub codes already assigns this protein to a particular degradation system. Thus, the original model built on the conviction that the Ub code stores information about protein fate may still be correct, but refers mostly to optimal conditions and fully operating proteasomes. Because cells must handle constantly changing environments and respond to changes instantly, further “safe alternatives” from pre-commitment decisions are required. While the E2/E3 ligase complexes “write” the Ub code and attach them to the substrate, other deubiquitinating enzymes (DUBs) “erase” the code by removing Ub linkages (Figure 3). This mechanism of editing of previously written Ub codes enables the adjustment of Ub signaling to cellular needs.
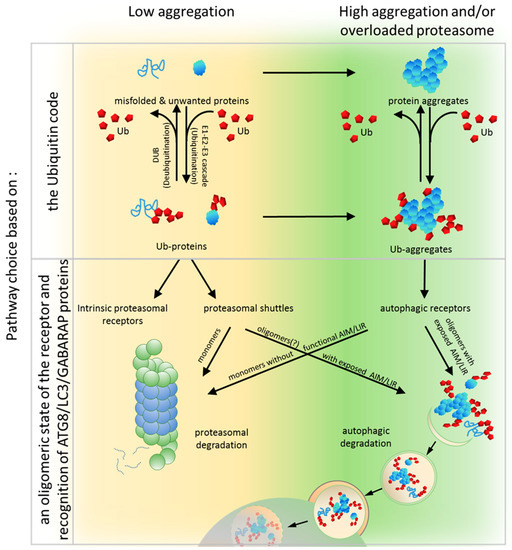
Figure 3.
A model for the substrate fate based on the Ub code and an oligomeric state of the receptor and recognition of ATG8/LC3/GABARAP (autophagy-related protein 8/microtubule-associated protein 1A/1B-light chain 3/GABA type A receptor-associated protein) proteins. The PQC network targeting misfolded or unwanted proteins via ubiquitination for degradation is hierarchically organized, but retains flexibility and coordinates the Ub-proteasome system (UPS) and autophagy pathways, according to cellular needs. In the low aggregation (undisturbed) condition, unwanted proteins initially become substrates for the UPS. During ubiquitination, particular Ub labels are attached to the target proteins via the E1–E2–E3 enzyme cascade. This process is reversed by deubiquitinases (DUBs). When the Ub code is written on the substrates, the cargos are then primarily recognized by intrinsic or shuttling proteasomal receptors characterized by higher affinity to Ub in comparison to the affinity of autophagic receptors to Ub. Simultaneous binding of shuttle receptors (probably in monomeric states) to ubiquitinated cargo and proteasomal proteins permit the transfer of the degradation substrates to the proteasome. However, in high aggregation conditions such as when proteasomes are overloaded or when misfolded or redundant proteins accumulate and form aggregates on which Ub is locally concentrated. Such aggregates are mainly recognized by autophagic receptors, which in their oligomeric states have high avidity for Ub and spontaneously coalesce with ubiquitinated proteins into larger clusters that further promote substrate aggregation and delivery to the autophagic machinery. In times of cellular need, shuttling receptors from the proteasomal or autophagic clearance pathways adapt to the alternative, but functional, degradation system to support cellular proteostasis. Proteasomal shuttles with exposed AIM/LIR motif and/or with a tendency to oligomerization can support protein turnover via autophagy by directly driving substrates to autophagic machinery or indirectly by sequestering ubiquitinated proteins in aggregates and stimulating cluster formation. Conversely, autophagic receptors in monomeric forms with non-functional AIM/LIR motifs can promote proteasomal degradation, if they are able to recognize and engage proteasomal subunits.
In principle, within the Ub code model, it is plausible that a code on a given protein could be erased and rewritten to shift the balance from one degradation pathway to another. However, the current evidence does not favor the rewriting of the Ub code as a major mechanism to redirect proteins to the alternative degradation pathway. Thus, after ubiquitination per se, additional decision-making steps must play a role in determining, for example, which Ub-binding adaptor proteins (such as receptors with PB1/UbL and UBA domains), will be used to recognize the Ub code on the target substrate, and which of the two PQC machineries these adaptors will engage with (Figure 3).
As we try to show in this manuscript, the role of these proteins in proteasomal and autophagic clearance is complex and has been an area of active debate and is under continued revision. A new appreciation is the role of protein modifications in regulating the function of Ub-binding shuttles and the effect of these on the PQC pathway choice. Numerous modifications within the UBA domain affect the affinities of these receptors for Ub or modulate the self-association of these receptors. Similarly, many post-translational modifications of the PB1 or UbL domain receptors impact their dimerization or interaction with proteasomal subunits. We also assume that both intra-molecular and inter-molecular interactions of PB1/UbL and UBA domains are part of the mechanism shared by these receptors to regulate their function by dynamically changing conformation from “closed“ to “open” or from monomeric to multimeric states and vice versa (Figure 4). Lastly, the interactions of these receptors with the autophagy machinery can also be regulated by phosphorylation of AIM/LIRs.
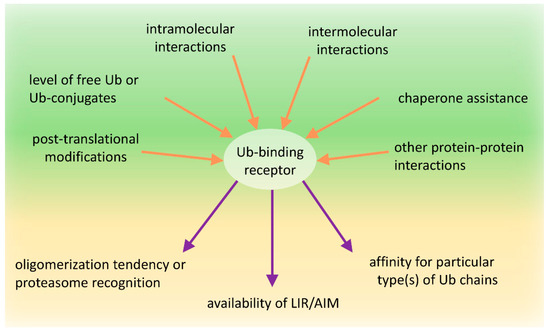
Figure 4.
Schematic representation of factors affecting conformational changes and function of Ub-binding receptors. The impact of shuttling Ub-binding receptors on the fate of ubiquitinated targets relies on the combined effect of several regulatory factors acting on those receptors and affecting their oligomerization tendency or proteasome recognition, availability of AIM/LIR motifs for ATG8/LC3/GABARAP-family member proteins decorating isolation membranes, and the receptor affinity for particular type(s) of Ub chains. Factors influencing these choices include post-translational modifications, intramolecular, intermolecular homo-oligomeric, and hetero-oligomeric interactions of each domain present in these receptors, chaperone assistance, levels of available free Ub and Ub-conjugates and effects of other proteins and molecules interacting with Ub-binding receptors and affecting their conformational changes and/or function.
It is plausible that, under un-induced conditions, the receptors with PB1/UbL and UBA domains could reside in a “closed” conformation where the UBA domain binds to the PB1/UbL domain. In such a state, the receptor may be shielded from binding ubiquitinated substrates as well as the proteasome or from oligomerizing and clustering into larger structures. However, because PB1/UbL binding to UBA is rather weak, more favorable conditions might activate the receptors and promote an “open” conformation. In such a case, the PB1 or UbL domains could bind the proteasome or other binding partners (either other proteins or another PB1 domain leading to oligomerization), which allows the UBA domain to homo-dimerize or to interact with ubiquitinated substrates, according to the ubiquitination/phosphorylation status of the UBA. While the molecular basis of the receptor conformation shifts remains unclear, the concentration of free Ub, ubiquitinated proteins, or the activity of particular E3 ligases and DUBs as well as coordinated phosphorylation/dephosphorylation are the most reasonable candidates for receptor conformation regulation (Figure 4).
Many reports show that ubiquitination or phosphorylation of certain residues affect receptor recognition by proteasome or its tendency to multimerization (Table 1). High concentrations of free mono-Ub and unanchored Ub chains inhibit clustering of SQSTM1, which might be related to the Ub-binding activity of the ZZ domain and might sterically inhibit oligomerization of SQSTM1 [27]. However, in contrast to Ub binding, the interaction of the N-arginylated chaperone, BiP (binding immunoglobulin protein), with the ZZ domain induces a conformational change of SQSTM1 exposing its PB1 and LIR, which accelerates its self-oligomerization and targeting to autophagosomes [247]. Since N-terminal arginylation of BiP is induced by accumulation of ubiquitinated proteins and aggregates, this serves as an interesting example of how cells control the crosstalk between the UPS and autophagy in decision-making process and react to accumulating proteins.
In conclusion, the clearance of unwanted and misfolded proteins is crucial for the maintenance of cellular proteostasis. Although the PQC network targeting ubiquitinated abnormal or unwanted proteins for degradation is hierarchically organized, it must retain flexibility and coordinate the two PQC pathways, according to the needs. Under normal conditions, unwanted proteins initially become substrates for the UPS and are primarily recognized by proteasomal receptors (such as Rad23 or Dsk2) characterized by higher affinity to Ub than that by autophagic receptors. Then, simultaneous binding of such receptors to proteasomal proteins via the UbL domain on the receptors and the substrates permits the transfer of the degradation substrate to the proteasome (Figure 3). However, when proteasomes are overloaded or malfunctioning, which could also be affected by stress, ubiquitinated proteins accumulate and form aggregates on which Ub is locally concentrated. Such structures can be recognized by autophagic receptors, such as SQSTM1, which, as a monomer, has low affinity, but as an oligomer, has high avidity to Ub and spontaneously coalesces with ubiquitinated proteins into larger clusters (Figure 3). Aggregation or aggresome formation can additionally be promoted by interaction with other oligomerizing and scaffolding proteins such as NBR1 [29,142], OPTN [32,115,116], or ALFY [199,200,248,249], which increase SQSTM1-mediated clustering [27,29,30,137]. In addition, the LIR motif sequence by itself, in contrast to its ability to bind to LC3B, promotes aggregation and delivery to the autophagic machinery [27]. However, in times of cellular need, shuttling receptors mainly assigned to the UPS or autophagy can adapt to the alternative such as a functional, degradation system in support of cellular proteostasis (Figure 3). We further hypothesize that the impact of shuttling Ub-binding receptors on the fate of ubiquitinated targets relies on the combined effect of several regulatory factors acting on those receptors: affinity to particular type(s) of Ub chains, intramolecular and intermolecular homo-oligomeric and hetero-oligomeric interactions of each domain present in these receptors, exposition of LIR/AIM, and post-translational modifications affecting protein conformational changes (Figure 4).
9. Future Perspectives
It has become clear that the UPS and autophagy are major PQC systems for the degradation of unwanted proteins in the cytoplasm. It has now been generally acknowledged that these two systems are highly interlinked and accept similar Ub labels on their targets. Moreover, based on recent studies, it appears that there is no single, specific signal that targets substrates exclusively to either the UPS or autophagy. The great majority of ubiquitin–UBA analyses have been done as in vitro studies and are often contradictory to the analyses of Ub linkages found in vivo, which include contributions of other cellular factors in Ub-linkage maintenance and selectivity. Therefore, it seems that revision of previous data and new careful analyses need to be done for receptor with UbL/PB1 and UBA domains to determine their predisposition to particular types of Ub conjugates in particular conditions. The physiological relevance of Ub linkages other than Lys48-linkages and Lys63-linkages or more complex structures, such as branched chains, remains open for additional research. We still do not know how dynamic and flexible Ub chains are and if their structure can be regulated to modulate recognition of particular Ub-binding receptors. Moreover, we are just beginning to understand how Ub-binding shuttles are regulated and how this regulation affects which degradation pathway they choose. Furthermore, an additional layer of complexity in the fate of ubiquitinated proteins comes from the fact that UbL fold can be found in numerous proteins that serve other unrelated functions within the cell. These important questions promise exciting times ahead for this field of research.
Author Contributions
K.Z-R. writing—original draft preparation. S.S. writing—review and editing.
Funding
This work was supported to an NIDDK grant (2 RO1 DK41737) to SS, who holds a Tata Chancellor’s Professorship in Molecular Biology at UC San Diego.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Schubert, U.; Anton, L.C.; Gibbs, J.; Norbury, C.C.; Yewdell, J.W.; Bennink, J.R. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 2000, 404, 770–774. [Google Scholar] [CrossRef]
- Dikic, I. Proteasomal and Autophagic Degradation Systems. Annu. Rev. Biochem. 2017, 86, 193–224. [Google Scholar] [CrossRef]
- Tanaka, K.; Matsuda, N. Proteostasis and neurodegeneration: The roles of proteasomal degradation and autophagy. Biochim. Biophys. Acta 2014, 1843, 197–204. [Google Scholar] [CrossRef]
- Scheffner, M.; Nuber, U.; Huibregtse, J.M. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature 1995, 373, 81–83. [Google Scholar] [CrossRef]
- da Fonseca, P.C.; Morris, E.P. Structure of the human 26S proteasome: Subunit radial displacements open the gate into the proteolytic core. J. Biol. Chem. 2008, 283, 23305–23314. [Google Scholar] [CrossRef]
- Budenholzer, L.; Cheng, C.L.; Li, Y.; Hochstrasser, M. Proteasome Structure and Assembly. J. Mol. Biol. 2017, 429, 3500–3524. [Google Scholar] [CrossRef]
- Bedford, L.; Paine, S.; Sheppard, P.W.; Mayer, R.J.; Roelofs, J. Assembly, structure, and function of the 26S proteasome. Trends Cell Biol. 2010, 20, 391–401. [Google Scholar] [CrossRef]
- Wang, X.; Su, H.; Ranek, M.J. Protein quality control and degradation in cardiomyocytes. J. Mol. Cell. Cardiol. 2008, 45, 11–27. [Google Scholar] [CrossRef]
- Verhoef, L.G.; Lindsten, K.; Masucci, M.G.; Dantuma, N.P. Aggregate formation inhibits proteasomal degradation of polyglutamine proteins. Hum. Mol. Genet. 2002, 11, 2689–2700. [Google Scholar] [CrossRef]
- Johnston, J.A.; Ward, C.L.; Kopito, R.R. Aggresomes: A cellular response to misfolded proteins. J. Cell Biol. 1998, 143, 1883–1898. [Google Scholar] [CrossRef]
- Taylor, J.P.; Tanaka, F.; Robitschek, J.; Sandoval, C.M.; Taye, A.; Markovic-Plese, S.; Fischbeck, K.H. Aggresomes protect cells by enhancing the degradation of toxic polyglutamine-containing protein. Hum. Mol. Genet. 2003, 12, 749–757. [Google Scholar] [CrossRef]
- Suraweera, A.; Munch, C.; Hanssum, A.; Bertolotti, A. Failure of amino acid homeostasis causes cell death following proteasome inhibition. Mol. Cell 2012, 48, 242–253. [Google Scholar] [CrossRef]
- Pandey, U.B.; Nie, Z.; Batlevi, Y.; McCray, B.A.; Ritson, G.P.; Nedelsky, N.B.; Schwartz, S.L.; DiProspero, N.A.; Knight, M.A.; Schuldiner, O.; et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature 2007, 447, 859–863. [Google Scholar] [CrossRef]
- Sugawara, K.; Suzuki, N.N.; Fujioka, Y.; Mizushima, N.; Ohsumi, Y.; Inagaki, F. The crystal structure of microtubule-associated protein light chain 3, a mammalian homologue of Saccharomyces cerevisiae Atg8. Genes Cells 2004, 9, 611–618. [Google Scholar] [CrossRef]
- Schwarten, M.; Stoldt, M.; Mohrluder, J.; Willbold, D. Solution structure of Atg8 reveals conformational polymorphism of the N-terminal domain. Biochem. Biophys. Res. Commun. 2010, 395, 426–431. [Google Scholar] [CrossRef]
- Paz, Y.; Elazar, Z.; Fass, D. Structure of GATE-16, membrane transport modulator and mammalian ortholog of autophagocytosis factor Aut7p. J. Biol. Chem. 2000, 275, 25445–25450. [Google Scholar] [CrossRef]
- Kumeta, H.; Watanabe, M.; Nakatogawa, H.; Yamaguchi, M.; Ogura, K.; Adachi, W.; Fujioka, Y.; Noda, N.N.; Ohsumi, Y.; Inagaki, F. The NMR structure of the autophagy-related protein Atg8. J. Biomol. NMR 2010, 47, 237–241. [Google Scholar] [CrossRef]
- Kouno, T.; Mizuguchi, M.; Tanida, I.; Ueno, T.; Kanematsu, T.; Mori, Y.; Shinoda, H.; Hirata, M.; Kominami, E.; Kawano, K. Solution structure of microtubule-associated protein light chain 3 and identification of its functional subdomains. J. Biol. Chem. 2005, 280, 24610–24617. [Google Scholar] [CrossRef]
- Hu, C.; Zhang, X.; Teng, Y.B.; Hu, H.X.; Li, W.F. Structure of autophagy-related protein Atg8 from the silkworm Bombyx mori. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010, 66, 787–790. [Google Scholar] [CrossRef]
- Coyle, J.E.; Qamar, S.; Rajashankar, K.R.; Nikolov, D.B. Structure of GABARAP in two conformations: Implications for GABA(A) receptor localization and tubulin binding. Neuron 2002, 33, 63–74. [Google Scholar] [CrossRef]
- Feng, Y.; He, D.; Yao, Z.; Klionsky, D.J. The machinery of macroautophagy. Cell Res. 2014, 24, 24–41. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Osawa, T.; Fujioka, Y.; Noda, N.N. Structural biology of the core autophagy machinery. Curr. Opin. Struct. Biol. 2017, 43, 10–17. [Google Scholar] [CrossRef]
- Yang, Z.; Klionsky, D.J. Mammalian autophagy: Core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 2010, 22, 124–131. [Google Scholar] [CrossRef]
- Behrends, C.; Fulda, S. Receptor proteins in selective autophagy. Int. J. Cell Biol. 2012, 2012, 673290. [Google Scholar] [CrossRef]
- Lamark, T.; Johansen, T. Aggrephagy: Selective disposal of protein aggregates by macroautophagy. Int. J. Cell Biol. 2012, 2012, 736905. [Google Scholar] [CrossRef]
- Liu, X.; Li, Y.; Wang, X.; Xing, R.; Liu, K.; Gan, Q.; Tang, C.; Gao, Z.; Jian, Y.; Luo, S.; et al. The BEACH-containing protein WDR81 coordinates p62 and LC3C to promote aggrephagy. J. Cell Biol. 2017, 216, 1301–1320. [Google Scholar] [CrossRef] [PubMed]
- Zaffagnini, G.; Savova, A.; Danieli, A.; Romanov, J.; Tremel, S.; Ebner, M.; Peterbauer, T.; Sztacho, M.; Trapannone, R.; Tarafder, A.K.; et al. p62 filaments capture and present ubiquitinated cargos for autophagy. EMBO J. 2018, 37. [Google Scholar] [CrossRef]
- Brown, A.I.; Rutenberg, A.D. Cluster coarsening on drops exhibits strong and sudden size-selectivity. Soft Matter 2015, 11, 3786–3793. [Google Scholar] [CrossRef]
- Kirkin, V.; Lamark, T.; Johansen, T.; Dikic, I. NBR1 cooperates with p62 in selective autophagy of ubiquitinated targets. Autophagy 2009, 5, 732–733. [Google Scholar] [CrossRef]
- Kirkin, V.; Lamark, T.; Sou, Y.S.; Bjorkoy, G.; Nunn, J.L.; Bruun, J.A.; Shvets, E.; McEwan, D.G.; Clausen, T.H.; Wild, P.; et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell 2009, 33, 505–516. [Google Scholar] [CrossRef]
- Lamark, T.; Kirkin, V.; Dikic, I.; Johansen, T. NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle 2009, 8, 1986–1990. [Google Scholar] [CrossRef] [PubMed]
- Korac, J.; Schaeffer, V.; Kovacevic, I.; Clement, A.M.; Jungblut, B.; Behl, C.; Terzic, J.; Dikic, I. Ubiquitin-independent function of optineurin in autophagic clearance of protein aggregates. J. Cell. Sci. 2013, 126, 580–592. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.C.; Li, H.Y.; Chen, G.C.; Chern, Y.; Tu, P.H. Mutations in the ubiquitin-binding domain of OPTN/optineurin interfere with autophagy-mediated degradation of misfolded proteins by a dominant-negative mechanism. Autophagy 2015, 11, 685–700. [Google Scholar] [CrossRef] [PubMed]
- Kravtsova-Ivantsiv, Y.; Sommer, T.; Ciechanover, A. The lysine48-based polyubiquitin chain proteasomal signal: Not a single child anymore. Angew. Chem. Int. Ed. Engl. 2013, 52, 192–198. [Google Scholar] [CrossRef] [PubMed]
- Ciechanover, A.; Kwon, Y.T. Degradation of misfolded proteins in neurodegenerative diseases: Therapeutic targets and strategies. Exp. Mol. Med. 2015, 47, e147. [Google Scholar] [CrossRef] [PubMed]
- Ciechanover, A.; Kwon, Y.T. Protein Quality Control by Molecular Chaperones in Neurodegeneration. Front. Neurosci. 2017, 11, 185. [Google Scholar] [CrossRef] [PubMed]
- Ciechanover, A. The unravelling of the ubiquitin system. Nat. Rev. Mol. Cell Biol. 2015, 16, 322–324. [Google Scholar] [CrossRef] [PubMed]
- Zientara-Rytter, K.; Sirko, A. To deliver or to degrade - an interplay of the ubiquitin-proteasome system, autophagy and vesicular transport in plants. FEBS J. 2016, 283, 3534–3555. [Google Scholar] [CrossRef] [PubMed]
- Alexander, A.; Walker, C.L. Differential localization of ATM is correlated with activation of distinct downstream signaling pathways. Cell Cycle 2010, 9, 3685–3686. [Google Scholar] [CrossRef]
- Ji, C.H.; Kwon, Y.T. Crosstalk and interplay between the ubiquitin-proteasome system and autophagy. Mol. Cells 2017, 40, 441–449. [Google Scholar] [CrossRef]
- Wang, Y.; Qin, Z.H. Coordination of autophagy with other cellular activities. Acta Pharmacol. Sin. 2013, 34, 585–594. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, X. The interplay between autophagy and the ubiquitin-proteasome system in cardiac proteotoxicity. Biochim. Biophys. Acta 2015, 1852, 188–194. [Google Scholar] [CrossRef] [PubMed]
- Lilienbaum, A. Relationship between the proteasomal system and autophagy. Int. J. Biochem. Mol. Biol. 2013, 4, 1–26. [Google Scholar] [PubMed]
- Komander, D.; Rape, M. The ubiquitin code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef] [PubMed]
- Rajalingam, K.; Dikic, I. SnapShot: Expanding the ubiquitin code. Cell 2016, 164. [Google Scholar] [CrossRef] [PubMed]
- Woelk, T.; Sigismund, S.; Penengo, L.; Polo, S. The ubiquitination code: A signalling problem. Cell Div. 2007, 2, 11. [Google Scholar] [CrossRef] [PubMed]
- Husnjak, K.; Dikic, I. Ubiquitin-binding proteins: Decoders of ubiquitin-mediated cellular functions. Annu. Rev. Biochem. 2012, 81, 291–322. [Google Scholar] [CrossRef]
- Madura, K. Rad23 and Rpn10: Perennial wallflowers join the melee. Trends Biochem. Sci. 2004, 29, 637–640. [Google Scholar] [CrossRef]
- Clarke, D.J.; Mondesert, G.; Segal, M.; Bertolaet, B.L.; Jensen, S.; Wolff, M.; Henze, M.; Reed, S.I. Dosage suppressors of pds1 implicate ubiquitin-associated domains in checkpoint control. Mol. Cell. Biol. 2001, 21, 1997–2007. [Google Scholar] [CrossRef]
- Elsasser, S.; Chandler-Militello, D.; Muller, B.; Hanna, J.; Finley, D. Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J. Biol. Chem. 2004, 279, 26817–26822. [Google Scholar] [CrossRef]
- Gomez, T.A.; Kolawa, N.; Gee, M.; Sweredoski, M.J.; Deshaies, R.J. Identification of a functional docking site in the Rpn1 LRR domain for the UBA-UBL domain protein Ddi1. BMC Biol. 2011, 9, 33. [Google Scholar] [CrossRef]
- Hartmann-Petersen, R.; Gordon, C. Integral UBL domain proteins: A family of proteasome interacting proteins. Semin. Cell Dev. Biol. 2004, 15, 247–259. [Google Scholar] [CrossRef] [PubMed]
- Hicke, L.; Schubert, H.L.; Hill, C.P. Ubiquitin-binding domains. Nat. Rev. Mol. Cell Biol. 2005, 6, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Husnjak, K.; Elsasser, S.; Zhang, N.; Chen, X.; Randles, L.; Shi, Y.; Hofmann, K.; Walters, K.J.; Finley, D.; Dikic, I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 2008, 453, 481–488. [Google Scholar] [CrossRef] [PubMed]
- Kaplun, L.; Tzirkin, R.; Bakhrat, A.; Shabek, N.; Ivantsiv, Y.; Raveh, D. The DNA damage-inducible UbL-UbA protein Ddi1 participates in Mec1-mediated degradation of HO endonuclease. Mol. Cell. Biol. 2005, 25, 5355–5362. [Google Scholar] [CrossRef]
- Kleijnen, M.F.; Shih, A.H.; Zhou, P.; Kumar, S.; Soccio, R.E.; Kedersha, N.L.; Gill, G.; Howley, P.M. The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol. Cell 2000, 6, 409–419. [Google Scholar] [CrossRef]
- Lambertson, D.; Chen, L.; Madura, K. Pleiotropic defects caused by loss of the proteasome-interacting factors Rad23 and Rpn10 of Saccharomyces cerevisiae. Genetics 1999, 153, 69–79. [Google Scholar] [PubMed]
- Saeki, Y.; Saitoh, A.; Toh-e, A.; Yokosawa, H. Ubiquitin-like proteins and Rpn10 play cooperative roles in ubiquitin-dependent proteolysis. Biochem. Biophys. Res. Commun. 2002, 293, 986–992. [Google Scholar] [CrossRef]
- Elsasser, S.; Finley, D. Delivery of ubiquitinated substrates to protein-unfolding machines. Nat. Cell Biol. 2005, 7, 742–749. [Google Scholar] [CrossRef]
- Saeki, Y. Ubiquitin recognition by the proteasome. J. Biochem. 2017, 161, 113–124. [Google Scholar] [CrossRef]
- Walters, K.J.; Kleijnen, M.F.; Goh, A.M.; Wagner, G.; Howley, P.M. Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a. Biochemistry 2002, 41, 1767–1777. [Google Scholar] [CrossRef]
- Mueller, T.D.; Feigon, J. Solution structures of UBA domains reveal a conserved hydrophobic surface for protein-protein interactions. J. Mol. Biol. 2002, 319, 1243–1255. [Google Scholar] [CrossRef]
- Dieckmann, T.; Withers-Ward, E.S.; Jarosinski, M.A.; Liu, C.F.; Chen, I.S.; Feigon, J. Structure of a human DNA repair protein UBA domain that interacts with HIV-1 Vpr. Nat. Struct. Biol. 1998, 5, 1042–1047. [Google Scholar] [CrossRef] [PubMed]
- Mueller, T.D.; Kamionka, M.; Feigon, J. Specificity of the interaction between ubiquitin-associated domains and ubiquitin. J. Biol. Chem. 2004, 279, 11926–11936. [Google Scholar] [CrossRef] [PubMed]
- Zientara-Rytter, K.; Lukomska, J.; Moniuszko, G.; Gwozdecki, R.; Surowiecki, P.; Lewandowska, M.; Liszewska, F.; Wawrzynska, A.; Sirko, A. Identification and functional analysis of Joka2, a tobacco member of the family of selective autophagy cargo receptors. Autophagy 2011, 7, 1145–1158. [Google Scholar] [CrossRef] [PubMed]
- Svenning, S.; Lamark, T.; Krause, K.; Johansen, T. Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 2011, 7, 993–1010. [Google Scholar] [CrossRef] [PubMed]
- Shaid, S.; Brandts, C.H.; Serve, H.; Dikic, I. Ubiquitination and selective autophagy. Cell Death Differ. 2013, 20, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Gabriely, G.; Kama, R.; Gelin-Licht, R.; Gerst, J.E. Different domains of the UBL-UBA ubiquitin receptor, Ddi1/Vsm1, are involved in its multiple cellular roles. Mol. Biol. Cell 2008, 19, 3625–3637. [Google Scholar] [CrossRef]
- Sirkis, R.; Gerst, J.E.; Fass, D. Ddi1, a eukaryotic protein with the retroviral protease fold. J. Mol. Biol. 2006, 364, 376–387. [Google Scholar] [CrossRef]
- Watkins, J.F.; Sung, P.; Prakash, L.; Prakash, S. The Saccharomyces cerevisiae DNA repair gene RAD23 encodes a nuclear protein containing a ubiquitin-like domain required for biological function. Mol. Cell. Biol. 1993, 13, 7757–7765. [Google Scholar] [CrossRef]
- Dantuma, N.P.; Heinen, C.; Hoogstraten, D. The ubiquitin receptor Rad23: At the crossroads of nucleotide excision repair and proteasomal degradation. DNA Repair (Amst) 2009, 8, 449–460. [Google Scholar] [CrossRef] [PubMed]
- Elsasser, S.; Gali, R.R.; Schwickart, M.; Larsen, C.N.; Leggett, D.S.; Muller, B.; Feng, M.T.; Tubing, F.; Dittmar, G.A.; Finley, D. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat. Cell Biol. 2002, 4, 725–730. [Google Scholar] [CrossRef] [PubMed]
- Saeki, Y.; Sone, T.; Toh-e, A.; Yokosawa, H. Identification of ubiquitin-like protein-binding subunits of the 26S proteasome. Biochem. Biophys. Res. Commun. 2002, 296, 813–819. [Google Scholar] [CrossRef]
- Rosenzweig, R.; Osmulski, P.A.; Gaczynska, M.; Glickman, M.H. The central unit within the 19S regulatory particle of the proteasome. Nat. Struct. Mol. Biol. 2008, 15, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Walters, K.J.; Lech, P.J.; Goh, A.M.; Wang, Q.; Howley, P.M. DNA-repair protein hHR23a alters its protein structure upon binding proteasomal subunit S5a. Proc. Natl. Acad. Sci. USA 2003, 100, 12694–12699. [Google Scholar] [CrossRef] [PubMed]
- Ryu, K.S.; Lee, K.J.; Bae, S.H.; Kim, B.K.; Kim, K.A.; Choi, B.S. Binding surface mapping of intra- and interdomain interactions among hHR23B, ubiquitin, and polyubiquitin binding site 2 of S5a. J. Biol. Chem. 2003, 278, 36621–36627. [Google Scholar] [CrossRef] [PubMed]
- Raasi, S.; Varadan, R.; Fushman, D.; Pickart, C.M. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat. Struct. Mol. Biol. 2005, 12, 708–714. [Google Scholar] [CrossRef]
- Rao, H.; Sastry, A. Recognition of specific ubiquitin conjugates is important for the proteolytic functions of the ubiquitin-associated domain proteins Dsk2 and Rad23. J. Biol. Chem. 2002, 277, 11691–11695. [Google Scholar] [CrossRef]
- Chen, L.; Shinde, U.; Ortolan, T.G.; Madura, K. Ubiquitin-associated (UBA) domains in Rad23 bind ubiquitin and promote inhibition of multi-ubiquitin chain assembly. EMBO Rep. 2001, 2, 933–938. [Google Scholar] [CrossRef]
- Chen, L.; Madura, K. Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol. Cell. Biol. 2002, 22, 4902–4913. [Google Scholar] [CrossRef]
- Bertolaet, B.L.; Clarke, D.J.; Wolff, M.; Watson, M.H.; Henze, M.; Divita, G.; Reed, S.I. UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nat. Struct. Biol. 2001, 8, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Ortolan, T.G.; Tongaonkar, P.; Lambertson, D.; Chen, L.; Schauber, C.; Madura, K. The DNA repair protein Rad23 is a negative regulator of multi-ubiquitin chain assembly. Nat. Cell Biol. 2000, 2, 601–608. [Google Scholar] [CrossRef]
- Raasi, S.; Pickart, C.M. Rad23 ubiquitin-associated domains (UBA) inhibit 26 S proteasome-catalyzed proteolysis by sequestering lysine 48-linked polyubiquitin chains. J. Biol. Chem. 2003, 278, 8951–8959. [Google Scholar] [CrossRef] [PubMed]
- Biggins, S.; Ivanovska, I.; Rose, M.D. Yeast ubiquitin-like genes are involved in duplication of the microtubule organizing center. J. Cell Biol. 1996, 133, 1331–1346. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, C.R.; Seeger, M.; Hartmann-Petersen, R.; Stone, M.; Wallace, M.; Semple, C.; Gordon, C. Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat. Cell Biol. 2001, 3, 939–943. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Madura, K. Evidence for distinct functions for human DNA repair factors hHR23A and hHR23B. FEBS Lett. 2006, 580, 3401–3408. [Google Scholar] [CrossRef]
- Funakoshi, M.; Sasaki, T.; Nishimoto, T.; Kobayashi, H. Budding yeast Dsk2p is a polyubiquitin-binding protein that can interact with the proteasome. Proc. Natl. Acad. Sci. USA 2002, 99, 745–750. [Google Scholar] [CrossRef]
- Tanaka, K.; Funakoshi, M.; Inoue, K.; Kobayashi, H. Identification of two isoforms of Dsk2-related protein XDRP1 in Xenopus eggs. Biochem. Biophys. Res. Commun. 2006, 350, 768–773. [Google Scholar] [CrossRef]
- Matiuhin, Y.; Kirkpatrick, D.S.; Ziv, I.; Kim, W.; Dakshinamurthy, A.; Kleifeld, O.; Gygi, S.P.; Reis, N.; Glickman, M.H. Extraproteasomal Rpn10 restricts access of the polyubiquitin-binding protein Dsk2 to proteasome. Mol. Cell 2008, 32, 415–425. [Google Scholar] [CrossRef]
- Lu, K.; den Brave, F.; Jentsch, S. Receptor oligomerization guides pathway choice between proteasomal and autophagic degradation. Nat. Cell Biol. 2017, 19, 732–739. [Google Scholar] [CrossRef]
- Zhao, G.; Zhou, X.; Wang, L.; Li, G.; Kisker, C.; Lennarz, W.J.; Schindelin, H. Structure of the mouse peptide N-glycanase-HR23 complex suggests co-evolution of the endoplasmic reticulum-associated degradation and DNA repair pathways. J. Biol. Chem. 2006, 281, 13751–13761. [Google Scholar] [CrossRef] [PubMed]
- El Ayadi, A.; Stieren, E.S.; Barral, J.M.; Boehning, D. Ubiquilin-1 and protein quality control in Alzheimer disease. Prion 2013, 7, 164–169. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Su, V.; Kurata, W.E.; Jin, C.; Lau, A.F. A novel connexin43-interacting protein, CIP75, which belongs to the UbL-UBA protein family, regulates the turnover of connexin43. J. Biol. Chem. 2008, 283, 5748–5759. [Google Scholar] [CrossRef] [PubMed]
- Heir, R.; Ablasou, C.; Dumontier, E.; Elliott, M.; Fagotto-Kaufmann, C.; Bedford, F.K. The UBL domain of PLIC-1 regulates aggresome formation. EMBO Rep. 2006, 7, 1252–1258. [Google Scholar] [CrossRef] [PubMed]
- Rothenberg, C.; Srinivasan, D.; Mah, L.; Kaushik, S.; Peterhoff, C.M.; Ugolino, J.; Fang, S.; Cuervo, A.M.; Nixon, R.A.; Monteiro, M.J. Ubiquilin functions in autophagy and is degraded by chaperone-mediated autophagy. Hum. Mol. Genet. 2010, 19, 3219–3232. [Google Scholar] [CrossRef]
- N’Diaye, E.N.; Kajihara, K.K.; Hsieh, I.; Morisaki, H.; Debnath, J.; Brown, E.J. PLIC proteins or ubiquilins regulate autophagy-dependent cell survival during nutrient starvation. EMBO Rep. 2009, 10, 173–179. [Google Scholar] [CrossRef]
- Lee, D.Y.; Brown, E.J. Ubiquilins in the crosstalk among proteolytic pathways. Biol. Chem. 2012, 393, 441–447. [Google Scholar] [CrossRef]
- Ivantsiv, Y.; Kaplun, L.; Tzirkin-Goldin, R.; Shabek, N.; Raveh, D. Unique role for the UbL-UbA protein Ddi1 in turnover of SCFUfo1 complexes. Mol. Cell. Biol. 2006, 26, 1579–1588. [Google Scholar] [CrossRef]
- Voloshin, O.; Bakhrat, A.; Herrmann, S.; Raveh, D. Transfer of Ho endonuclease and Ufo1 to the proteasome by the UbL-UbA shuttle protein, Ddi1, analysed by complex formation in vitro. PLoS ONE 2012, 7, e39210. [Google Scholar] [CrossRef]
- Kaplun, L.; Ivantsiv, Y.; Kornitzer, D.; Raveh, D. Functions of the DNA damage response pathway target HO endonuclease of yeast for degradation via the ubiquitin-26S proteasome system. Proc. Natl. Acad. Sci. USA 2000, 97, 10077–10082. [Google Scholar] [CrossRef]
- Zhang, D.; Raasi, S.; Fushman, D. Affinity makes the difference: Nonselective interaction of the UBA domain of Ubiquilin-1 with monomeric ubiquitin and polyubiquitin chains. J. Mol. Biol. 2008, 377, 162–180. [Google Scholar] [CrossRef] [PubMed]
- Rosenzweig, R.; Bronner, V.; Zhang, D.; Fushman, D.; Glickman, M.H. Rpn1 and Rpn2 coordinate ubiquitin processing factors at proteasome. J. Biol. Chem. 2012, 287, 14659–14671. [Google Scholar] [CrossRef] [PubMed]
- Nowicka, U.; Zhang, D.; Walker, O.; Krutauz, D.; Castaneda, C.A.; Chaturvedi, A.; Chen, T.Y.; Reis, N.; Glickman, M.H.; Fushman, D. DNA-damage-inducible 1 protein (Ddi1) contains an uncharacteristic ubiquitin-like domain that binds ubiquitin. Structure 2015, 23, 542–557. [Google Scholar] [CrossRef]
- Bertolaet, B.L.; Clarke, D.J.; Wolff, M.; Watson, M.H.; Henze, M.; Divita, G.; Reed, S.I. UBA domains mediate protein-protein interactions between two DNA damage-inducible proteins. J. Mol. Biol. 2001, 313, 955–963. [Google Scholar] [CrossRef] [PubMed]
- Siva, M.; Svoboda, M.; Veverka, V.; Trempe, J.F.; Hofmann, K.; Kozisek, M.; Hexnerova, R.; Sedlak, F.; Belza, J.; Brynda, J.; et al. Human DNA-damage-inducible 2 protein is structurally and functionally distinct from its yeast ortholog. Sci. Rep. 2016, 6, 30443. [Google Scholar] [CrossRef]
- Lu, K.; Psakhye, I.; Jentsch, S. Autophagic clearance of polyQ proteins mediated by ubiquitin-Atg8 adaptors of the conserved CUET protein family. Cell 2014, 158, 549–563. [Google Scholar] [CrossRef] [PubMed]
- Waters, S.; Marchbank, K.; Solomon, E.; Whitehouse, C.; Gautel, M. Interactions with LC3 and polyubiquitin chains link nbr1 to autophagic protein turnover. FEBS Lett. 2009, 583, 1846–1852. [Google Scholar] [CrossRef]
- Pankiv, S.; Clausen, T.H.; Lamark, T.; Brech, A.; Bruun, J.A.; Outzen, H.; Overvatn, A.; Bjorkoy, G.; Johansen, T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007, 282, 24131–24145. [Google Scholar] [CrossRef]
- Geisler, S.; Holmstrom, K.M.; Skujat, D.; Fiesel, F.C.; Rothfuss, O.C.; Kahle, P.J.; Springer, W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 2010, 12, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Ishimura, R.; Tanaka, K.; Komatsu, M. Dissection of the role of p62/Sqstm1 in activation of Nrf2 during xenophagy. FEBS Lett. 2014, 588, 822–828. [Google Scholar] [CrossRef]
- Deosaran, E.; Larsen, K.B.; Hua, R.; Sargent, G.; Wang, Y.; Kim, S.; Lamark, T.; Jauregui, M.; Law, K.; Lippincott-Schwartz, J.; et al. NBR1 acts as an autophagy receptor for peroxisomes. J. Cell. Sci. 2013, 126, 939–952. [Google Scholar] [CrossRef] [PubMed]
- Isakson, P.; Lystad, A.H.; Breen, K.; Koster, G.; Stenmark, H.; Simonsen, A. TRAF6 mediates ubiquitination of KIF23/MKLP1 and is required for midbody ring degradation by selective autophagy. Autophagy 2013, 9, 1955–1964. [Google Scholar] [CrossRef]
- Narendra, D.; Kane, L.A.; Hauser, D.N.; Fearnley, I.M.; Youle, R.J. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy 2010, 6, 1090–1106. [Google Scholar] [CrossRef] [PubMed]
- Richter, B.; Sliter, D.A.; Herhaus, L.; Stolz, A.; Wang, C.; Beli, P.; Zaffagnini, G.; Wild, P.; Martens, S.; Wagner, S.A.; et al. Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc. Natl. Acad. Sci. USA 2016, 113, 4039–4044. [Google Scholar] [CrossRef] [PubMed]
- Wong, Y.C.; Holzbaur, E.L. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc. Natl. Acad. Sci. USA 2014, 111, E4439–E4448. [Google Scholar] [CrossRef]
- Wild, P.; Farhan, H.; McEwan, D.G.; Wagner, S.; Rogov, V.V.; Brady, N.R.; Richter, B.; Korac, J.; Waidmann, O.; Choudhary, C.; et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 2011, 333, 228–233. [Google Scholar] [CrossRef] [PubMed]
- Verlhac, P.; Gregoire, I.P.; Azocar, O.; Petkova, D.S.; Baguet, J.; Viret, C.; Faure, M. Autophagy receptor NDP52 regulates pathogen-containing autophagosome maturation. Cell Host. Microbe 2015, 17, 515–525. [Google Scholar] [CrossRef]
- Lazarou, M.; Sliter, D.A.; Kane, L.A.; Sarraf, S.A.; Wang, C.; Burman, J.L.; Sideris, D.P.; Fogel, A.I.; Youle, R.J. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 2015, 524, 309–314. [Google Scholar] [CrossRef]
- von Muhlinen, N.; Thurston, T.; Ryzhakov, G.; Bloor, S.; Randow, F. NDP52, a novel autophagy receptor for ubiquitin-decorated cytosolic bacteria. Autophagy 2010, 6, 288–289. [Google Scholar] [CrossRef]
- Mitra, S.; Traughber, C.A.; Brannon, M.K.; Gomez, S.; Capelluto, D.G. Ubiquitin interacts with the Tollip C2 and CUE domains and inhibits binding of Tollip to phosphoinositides. J. Biol. Chem. 2013, 288, 25780–25791. [Google Scholar] [CrossRef]
- Ankem, G.; Mitra, S.; Sun, F.; Moreno, A.C.; Chutvirasakul, B.; Azurmendi, H.F.; Li, L.; Capelluto, D.G. The C2 domain of Tollip, a Toll-like receptor signalling regulator, exhibits broad preference for phosphoinositides. Biochem. J. 2011, 435, 597–608. [Google Scholar] [CrossRef] [PubMed]
- Capelluto, D.G. Tollip: A multitasking protein in innate immunity and protein trafficking. Microbes Infect. 2012, 14, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Visvikis, O.; Boyer, L.; Torrino, S.; Doye, A.; Lemonnier, M.; Lores, P.; Rolando, M.; Flatau, G.; Mettouchi, A.; Bouvard, D.; et al. Escherichia coli producing CNF1 toxin hijacks Tollip to trigger Rac1-dependent cell invasion. Traffic 2011, 12, 579–590. [Google Scholar] [CrossRef]
- Doi, H.; Mitsui, K.; Kurosawa, M.; Machida, Y.; Kuroiwa, Y.; Nukina, N. Identification of ubiquitin-interacting proteins in purified polyglutamine aggregates. FEBS Lett. 2004, 571, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Oguro, A.; Kubota, H.; Shimizu, M.; Ishiura, S.; Atomi, Y. Protective role of the ubiquitin binding protein Tollip against the toxicity of polyglutamine-expansion proteins. Neurosci. Lett. 2011, 503, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, M.; Waguri, S.; Koike, M.; Sou, Y.S.; Ueno, T.; Hara, T.; Mizushima, N.; Iwata, J.; Ezaki, J.; Murata, S.; et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 2007, 131, 1149–1163. [Google Scholar] [CrossRef] [PubMed]
- Bjorkoy, G.; Lamark, T.; Brech, A.; Outzen, H.; Perander, M.; Overvatn, A.; Stenmark, H.; Johansen, T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 2005, 171, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Pankiv, S.; Lamark, T.; Bruun, J.A.; Overvatn, A.; Bjorkoy, G.; Johansen, T. Nucleocytoplasmic shuttling of p62/SQSTM1 and its role in recruitment of nuclear polyubiquitinated proteins to promyelocytic leukemia bodies. J. Biol. Chem. 2010, 285, 5941–5953. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Lamark, T.; Sjottem, E.; Larsen, K.B.; Awuh, J.A.; Overvatn, A.; McMahon, M.; Hayes, J.D.; Johansen, T. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem. 2010, 285, 22576–22591. [Google Scholar] [CrossRef]
- Lin, X.; Li, S.; Zhao, Y.; Ma, X.; Zhang, K.; He, X.; Wang, Z. Interaction domains of p62: A bridge between p62 and selective autophagy. DNA Cell Biol. 2013, 32, 220–227. [Google Scholar] [CrossRef]
- Seibenhener, M.L.; Babu, J.R.; Geetha, T.; Wong, H.C.; Krishna, N.R.; Wooten, M.W. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol. Cell. Biol. 2004, 24, 8055–8068. [Google Scholar] [CrossRef] [PubMed]
- Geetha, T.; Seibenhener, M.L.; Chen, L.; Madura, K.; Wooten, M.W. p62 serves as a shuttling factor for TrkA interaction with the proteasome. Biochem. Biophys. Res. Commun. 2008, 374, 33–37. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, N.; Zhang, L.; Li, R.; Fu, W.; Ma, K.; Li, X.; Wang, L.; Wang, J.; Zhang, H.; et al. Autophagy regulates chromatin ubiquitination in DNA damage response through elimination of SQSTM1/p62. Mol. Cell 2016, 63, 34–48. [Google Scholar] [CrossRef] [PubMed]
- Katsuragi, Y.; Ichimura, Y.; Komatsu, M. p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J. 2015, 282, 4672–4678. [Google Scholar] [CrossRef] [PubMed]
- Puvirajesinghe, T.M.; Bertucci, F.; Jain, A.; Scerbo, P.; Belotti, E.; Audebert, S.; Sebbagh, M.; Lopez, M.; Brech, A.; Finetti, P.; et al. Identification of p62/SQSTM1 as a component of non-canonical Wnt VANGL2-JNK signalling in breast cancer. Nat. Commun. 2016, 7, 10318. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Qiang, L.; Sample, A.; Shah, P.; He, Y.Y. NF-kappaB signaling activation induced by chloroquine requires autophagosome, p62 protein, and c-Jun N-terminal Kinase (JNK) signaling and promotes tumor cell resistance. J. Biol. Chem. 2017, 292, 3379–3388. [Google Scholar] [CrossRef] [PubMed]
- Clausen, T.H.; Lamark, T.; Isakson, P.; Finley, K.; Larsen, K.B.; Brech, A.; Overvatn, A.; Stenmark, H.; Bjorkoy, G.; Simonsen, A.; et al. p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy 2010, 6, 330–344. [Google Scholar] [CrossRef]
- Itakura, E.; Mizushima, N. p62 Targeting to the autophagosome formation site requires self-oligomerization but not LC3 binding. J. Cell Biol. 2011, 192, 17–27. [Google Scholar] [CrossRef]
- Wurzer, B.; Zaffagnini, G.; Fracchiolla, D.; Turco, E.; Abert, C.; Romanov, J.; Martens, S. Oligomerization of p62 allows for selection of ubiquitinated cargo and isolation membrane during selective autophagy. Elife 2015, 4, e08941. [Google Scholar] [CrossRef]
- Yan, J.; Seibenhener, M.L.; Calderilla-Barbosa, L.; Diaz-Meco, M.T.; Moscat, J.; Jiang, J.; Wooten, M.W.; Wooten, M.C. SQSTM1/p62 interacts with HDAC6 and regulates deacetylase activity. PLoS ONE 2013, 8, e76016. [Google Scholar] [CrossRef]
- Marchbank, K.; Waters, S.; Roberts, R.G.; Solomon, E.; Whitehouse, C.A. MAP1B Interaction with the FW domain of the autophagic receptor Nbr1 facilitates its association to the microtubule network. Int. J. Cell Biol. 2012, 2012, 208014. [Google Scholar] [CrossRef] [PubMed]
- Mardakheh, F.K.; Auciello, G.; Dafforn, T.R.; Rappoport, J.Z.; Heath, J.K. Nbr1 is a novel inhibitor of ligand-mediated receptor tyrosine kinase degradation. Mol. Cell. Biol. 2010, 30, 5672–5685. [Google Scholar] [CrossRef] [PubMed]
- Lamark, T.; Perander, M.; Outzen, H.; Kristiansen, K.; Overvatn, A.; Michaelsen, E.; Bjorkoy, G.; Johansen, T. Interaction codes within the family of mammalian Phox and Bem1p domain-containing proteins. J. Biol. Chem. 2003, 278, 34568–34581. [Google Scholar] [CrossRef] [PubMed]
- Christian, F.; Krause, E.; Houslay, M.D.; Baillie, G.S. PKA phosphorylation of p62/SQSTM1 regulates PB1 domain interaction partner binding. Biochim. Biophys. Acta 2014, 1843, 2765–2774. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.I.; Gill, D.J.; Perisic, O.; Quinn, M.T.; Williams, R.L. PB1 domain-mediated heterodimerization in NADPH oxidase and signaling complexes of atypical protein kinase C with Par6 and p62. Mol. Cell 2003, 12, 39–50. [Google Scholar] [CrossRef]
- Muller, S.; Kursula, I.; Zou, P.; Wilmanns, M. Crystal structure of the PB1 domain of NBR1. FEBS Lett. 2006, 580, 341–344. [Google Scholar] [CrossRef]
- Walinda, E.; Morimoto, D.; Sugase, K.; Konuma, T.; Tochio, H.; Shirakawa, M. Solution structure of the ubiquitin-associated (UBA) domain of human autophagy receptor NBR1 and its interaction with ubiquitin and polyubiquitin. J. Biol. Chem. 2014, 289, 13890–13902. [Google Scholar] [CrossRef]
- Zientara-Rytter, K.; Sirko, A. Significant role of PB1 and UBA domains in multimerization of Joka2, a selective autophagy cargo receptor from tobacco. Front. Plant Sci. 2014, 5, 13. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, J.; Cheng, Y.; Chi, Y.J.; Fan, B.; Yu, J.Q.; Chen, Z. NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet. 2013, 9, e1003196. [Google Scholar] [CrossRef]
- Hafren, A.; Macia, J.L.; Love, A.J.; Milner, J.J.; Drucker, M.; Hofius, D. Selective autophagy limits cauliflower mosaic virus infection by NBR1-mediated targeting of viral capsid protein and particles. Proc. Natl. Acad. Sci. USA 2017, 114, E2026–E2035. [Google Scholar] [CrossRef]
- Kachaner, D.; Filipe, J.; Laplantine, E.; Bauch, A.; Bennett, K.L.; Superti-Furga, G.; Israel, A.; Weil, R. Plk1-dependent phosphorylation of optineurin provides a negative feedback mechanism for mitotic progression. Mol. Cell 2012, 45, 553–566. [Google Scholar] [CrossRef] [PubMed]
- del Toro, D.; Alberch, J.; Lazaro-Dieguez, F.; Martin-Ibanez, R.; Xifro, X.; Egea, G.; Canals, J.M. Mutant huntingtin impairs post-Golgi trafficking to lysosomes by delocalizing optineurin/Rab8 complex from the Golgi apparatus. Mol. Biol. Cell 2009, 20, 1478–1492. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Wu, C.J.; Zhao, Y.; Ashwell, J.D. Optineurin negatively regulates TNFalpha- induced NF-kappaB activation by competing with NEMO for ubiquitinated RIP. Curr. Biol. 2007, 17, 1438–1443. [Google Scholar] [CrossRef] [PubMed]
- Anborgh, P.H.; Godin, C.; Pampillo, M.; Dhami, G.K.; Dale, L.B.; Cregan, S.P.; Truant, R.; Ferguson, S.S. Inhibition of metabotropic glutamate receptor signaling by the huntingtin-binding protein optineurin. J. Biol. Chem. 2005, 280, 34840–34848. [Google Scholar] [CrossRef]
- Sahlender, D.A.; Roberts, R.C.; Arden, S.D.; Spudich, G.; Taylor, M.J.; Luzio, J.P.; Kendrick-Jones, J.; Buss, F. Optineurin links myosin VI to the Golgi complex and is involved in Golgi organization and exocytosis. J. Cell Biol. 2005, 169, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Hattula, K.; Peranen, J. FIP-2, a coiled-coil protein, links Huntingtin to Rab8 and modulates cellular morphogenesis. Curr. Biol. 2000, 10, 1603–1606. [Google Scholar] [CrossRef]
- Slowicka, K.; Vereecke, L.; van Loo, G. Cellular functions of Optineurin in health and disease. Trends Immunol. 2016, 37, 621–633. [Google Scholar] [CrossRef]
- Gleason, C.E.; Ordureau, A.; Gourlay, R.; Arthur, J.S.; Cohen, P. Polyubiquitin binding to optineurin is required for optimal activation of TANK-binding kinase 1 and production of interferon beta. J. Biol. Chem. 2011, 286, 35663–35674. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Chen, P.; Gao, H.; Gu, Y.; Yang, J.; Peng, H.; Xu, X.; Wang, H.; Yang, M.; Liu, X.; et al. Ubiquitylation of autophagy receptor Optineurin by HACE1 activates selective autophagy for tumor suppression. Cancer Cell 2014, 26, 106–120. [Google Scholar] [CrossRef] [PubMed]
- Tumbarello, D.A.; Waxse, B.J.; Arden, S.D.; Bright, N.A.; Kendrick-Jones, J.; Buss, F. Autophagy receptors link myosin VI to autophagosomes to mediate Tom1-dependent autophagosome maturation and fusion with the lysosome. Nat. Cell Biol. 2012, 14, 1024–1035. [Google Scholar] [CrossRef] [PubMed]
- Ying, H.; Shen, X.; Park, B.; Yue, B.Y. Posttranslational modifications, localization, and protein interactions of optineurin, the product of a glaucoma gene. PLoS ONE 2010, 5, e9168. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Ying, H.; Qiu, Y.; Park, J.S.; Shyam, R.; Chi, Z.L.; Iwata, T.; Yue, B.Y. Processing of optineurin in neuronal cells. J. Biol. Chem. 2011, 286, 3618–3629. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, S.E.; Riley, B.E.; Shaler, T.A.; Trevino, R.S.; Becker, C.H.; Schulman, H.; Kopito, R.R. Protein standard absolute quantification (PSAQ) method for the measurement of cellular ubiquitin pools. Nat. Methods 2011, 8, 691–696. [Google Scholar] [CrossRef]
- Kim, W.; Bennett, E.J.; Huttlin, E.L.; Guo, A.; Li, J.; Possemato, A.; Sowa, M.E.; Rad, R.; Rush, J.; Comb, M.J.; et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 2011, 44, 325–340. [Google Scholar] [CrossRef]
- Peng, J.; Schwartz, D.; Elias, J.E.; Thoreen, C.C.; Cheng, D.; Marsischky, G.; Roelofs, J.; Finley, D.; Gygi, S.P. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 2003, 21, 921–926. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Duong, D.M.; Seyfried, N.T.; Cheng, D.; Xie, Y.; Robert, J.; Rush, J.; Hochstrasser, M.; Finley, D.; Peng, J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 2009, 137, 133–145. [Google Scholar] [CrossRef]
- Kwon, Y.T.; Reiss, Y.; Fried, V.A.; Hershko, A.; Yoon, J.K.; Gonda, D.K.; Sangan, P.; Copeland, N.G.; Jenkins, N.A.; Varshavsky, A. The mouse and human genes encoding the recognition component of the N-end rule pathway. Proc. Natl. Acad. Sci. USA 1998, 95, 7898–7903. [Google Scholar] [CrossRef] [PubMed]
- Tasaki, T.; Mulder, L.C.; Iwamatsu, A.; Lee, M.J.; Davydov, I.V.; Varshavsky, A.; Muesing, M.; Kwon, Y.T. A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol. Cell. Biol. 2005, 25, 7120–7136. [Google Scholar] [CrossRef]
- Liu, W.; Shang, Y.; Li, W. gp78 elongates of polyubiquitin chains from the distal end through the cooperation of its G2BR and CUE domains. Sci. Rep. 2014, 4, 7138. [Google Scholar] [CrossRef]
- Kim, H.C.; Huibregtse, J.M. Polyubiquitination by HECT E3s and the determinants of chain type specificity. Mol. Cell. Biol. 2009, 29, 3307–3318. [Google Scholar] [CrossRef]
- Suber, T.; Wei, J.; Jacko, A.M.; Nikolli, I.; Zhao, Y.; Zhao, J.; Mallampalli, R.K. SCF(FBXO17) E3 ligase modulates inflammation by regulating proteasomal degradation of glycogen synthase kinase-3beta in lung epithelia. J. Biol. Chem. 2017, 292, 7452–7461. [Google Scholar] [CrossRef]
- McKeon, J.E.; Sha, D.; Li, L.; Chin, L.S. Parkin-mediated K63-polyubiquitination targets ubiquitin C-terminal hydrolase L1 for degradation by the autophagy-lysosome system. Cell. Mol. Life Sci. 2015, 72, 1811–1824. [Google Scholar] [CrossRef] [PubMed]
- Tomar, D.; Prajapati, P.; Sripada, L.; Singh, K.; Singh, R.; Singh, A.K.; Singh, R. TRIM13 regulates caspase-8 ubiquitination, translocation to autophagosomes and activation during ER stress induced cell death. Biochim. Biophys. Acta 2013, 1833, 3134–3144. [Google Scholar] [CrossRef]
- Ferreira, J.V.; Soares, A.R.; Ramalho, J.S.; Pereira, P.; Girao, H. K63 linked ubiquitin chain formation is a signal for HIF1A degradation by chaperone-mediated autophagy. Sci. Rep. 2015, 5, 10210. [Google Scholar] [CrossRef] [PubMed]
- Ciechanover, A.; Stanhill, A. The complexity of recognition of ubiquitinated substrates by the 26S proteasome. Biochim. Biophys. Acta 2014, 1843, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.M.; Wong, E.S.; Kirkpatrick, D.S.; Pletnikova, O.; Ko, H.S.; Tay, S.P.; Ho, M.W.; Troncoso, J.; Gygi, S.P.; Lee, M.K.; et al. Lysine 63-linked ubiquitination promotes the formation and autophagic clearance of protein inclusions associated with neurodegenerative diseases. Hum. Mol. Genet. 2008, 17, 431–439. [Google Scholar] [CrossRef]
- Lin, Y.L.; Sung, S.C.; Tsai, H.L.; Yu, T.T.; Radjacommare, R.; Usharani, R.; Fatimababy, A.S.; Lin, H.Y.; Wang, Y.Y.; Fu, H. The defective proteasome but not substrate recognition function is responsible for the null phenotypes of the Arabidopsis proteasome subunit RPN10. Plant Cell 2011, 23, 2754–2773. [Google Scholar] [CrossRef]
- Farmer, L.M.; Book, A.J.; Lee, K.H.; Lin, Y.L.; Fu, H.; Vierstra, R.D. The RAD23 family provides an essential connection between the 26S proteasome and ubiquitylated proteins in Arabidopsis. Plant Cell 2010, 22, 124–142. [Google Scholar] [CrossRef]
- Wooten, M.W.; Geetha, T.; Babu, J.R.; Seibenhener, M.L.; Peng, J.; Cox, N.; Diaz-Meco, M.T.; Moscat, J. Essential role of sequestosome 1/p62 in regulating accumulation of Lys63-ubiquitinated proteins. J. Biol. Chem. 2008, 283, 6783–6789. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Gallagher, T.R.; Cavey, J.R.; Sheppard, P.W.; Ralston, S.H.; Layfield, R.; Searle, M.S. Ubiquitin recognition by the ubiquitin-associated domain of p62 involves a novel conformational switch. J. Biol. Chem. 2008, 283, 5427–5440. [Google Scholar] [CrossRef]
- Riley, B.E.; Kaiser, S.E.; Shaler, T.A.; Ng, A.C.; Hara, T.; Hipp, M.S.; Lage, K.; Xavier, R.J.; Ryu, K.Y.; Taguchi, K.; et al. Ubiquitin accumulation in autophagy-deficient mice is dependent on the Nrf2-mediated stress response pathway: A potential role for protein aggregation in autophagic substrate selection. J. Cell Biol. 2010, 191, 537–552. [Google Scholar] [CrossRef]
- Thrower, J.S.; Hoffman, L.; Rechsteiner, M.; Pickart, C.M. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000, 19, 94–102. [Google Scholar] [CrossRef]
- Chau, V.; Tobias, J.W.; Bachmair, A.; Marriott, D.; Ecker, D.J.; Gonda, D.K.; Varshavsky, A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 1989, 243, 1576–1583. [Google Scholar] [CrossRef]
- Schreiber, A.; Peter, M. Substrate recognition in selective autophagy and the ubiquitin-proteasome system. Biochim. Biophys. Acta 2014, 1843, 163–181. [Google Scholar] [CrossRef]
- Jin, L.; Williamson, A.; Banerjee, S.; Philipp, I.; Rape, M. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell 2008, 133, 653–665. [Google Scholar] [CrossRef]
- Matsumoto, M.L.; Wickliffe, K.E.; Dong, K.C.; Yu, C.; Bosanac, I.; Bustos, D.; Phu, L.; Kirkpatrick, D.S.; Hymowitz, S.G.; Rape, M.; et al. K11-linked polyubiquitination in cell cycle control revealed by a K11 linkage-specific antibody. Mol. Cell 2010, 39, 477–484. [Google Scholar] [CrossRef]
- Kim, H.T.; Kim, K.P.; Lledias, F.; Kisselev, A.F.; Scaglione, K.M.; Skowyra, D.; Gygi, S.P.; Goldberg, A.L. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J. Biol. Chem. 2007, 282, 17375–17386. [Google Scholar] [CrossRef]
- Kirkpatrick, D.S.; Hathaway, N.A.; Hanna, J.; Elsasser, S.; Rush, J.; Finley, D.; King, R.W.; Gygi, S.P. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat. Cell Biol. 2006, 8, 700–710. [Google Scholar] [CrossRef]
- Johnson, E.S.; Ma, P.C.; Ota, I.M.; Varshavsky, A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 1995, 270, 17442–17456. [Google Scholar] [CrossRef]
- Koegl, M.; Hoppe, T.; Schlenker, S.; Ulrich, H.D.; Mayer, T.U.; Jentsch, S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 1999, 96, 635–644. [Google Scholar] [CrossRef]
- Saeki, Y.; Kudo, T.; Sone, T.; Kikuchi, Y.; Yokosawa, H.; Toh-e, A.; Tanaka, K. Lysine 63-linked polyubiquitin chain may serve as a targeting signal for the 26S proteasome. EMBO J. 2009, 28, 359–371. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, S.; Zheng, H. The cargo receptor SQSTM1 ameliorates neurofibrillary tangle pathology and spreading through selective targeting of pathological MAPT (microtubule associated protein tau). Autophagy 2018, 1–16. [Google Scholar] [CrossRef]
- Chuang, K.H.; Liang, F.; Higgins, R.; Wang, Y. Ubiquilin/Dsk2 promotes inclusion body formation and vacuole (lysosome)-mediated disposal of mutated huntingtin. Mol. Biol. Cell 2016, 27, 2025–2036. [Google Scholar] [CrossRef]
- Meriin, A.B.; Zhang, X.; Miliaras, N.B.; Kazantsev, A.; Chernoff, Y.O.; McCaffery, J.M.; Wendland, B.; Sherman, M.Y. Aggregation of expanded polyglutamine domain in yeast leads to defects in endocytosis. Mol. Cell. Biol. 2003, 23, 7554–7565. [Google Scholar] [CrossRef]
- Rothenberg, C.; Monteiro, M.J. Ubiquilin at a crossroads in protein degradation pathways. Autophagy 2010, 6, 979–980. [Google Scholar] [CrossRef]
- Lee, D.Y.; Arnott, D.; Brown, E.J. Ubiquilin4 is an adaptor protein that recruits Ubiquilin1 to the autophagy machinery. EMBO Rep. 2013, 14, 373–381. [Google Scholar] [CrossRef]
- Nolan, T.M.; Brennan, B.; Yang, M.; Chen, J.; Zhang, M.; Li, Z.; Wang, X.; Bassham, D.C.; Walley, J.; Yin, Y. Selective autophagy of BES1 mediated by DSK2 balances plant growth and survival. Dev. Cell 2017, 41, 33–46.e37. [Google Scholar] [CrossRef]
- Hjerpe, R.; Bett, J.S.; Keuss, M.J.; Solovyova, A.; McWilliams, T.G.; Johnson, C.; Sahu, I.; Varghese, J.; Wood, N.; Wightman, M.; et al. UBQLN2 mediates autophagy-independent protein aggregate clearance by the proteasome. Cell 2016, 166, 935–949. [Google Scholar] [CrossRef]
- Filimonenko, M.; Isakson, P.; Finley, K.D.; Anderson, M.; Jeong, H.; Melia, T.J.; Bartlett, B.J.; Myers, K.M.; Birkeland, H.C.; Lamark, T.; et al. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Mol. Cell 2010, 38, 265–279. [Google Scholar] [CrossRef]
- Han, H.; Wei, W.; Duan, W.; Guo, Y.; Li, Y.; Wang, J.; Bi, Y.; Li, C. Autophagy-linked FYVE protein (Alfy) promotes autophagic removal of misfolded proteins involved in amyotrophic lateral sclerosis (ALS). In Vitro Cell. Dev. Biol. Anim. 2015, 51, 249–263. [Google Scholar] [CrossRef]
- Rui, Y.N.; Xu, Z.; Patel, B.; Chen, Z.; Chen, D.; Tito, A.; David, G.; Sun, Y.; Stimming, E.F.; Bellen, H.J.; et al. Huntingtin functions as a scaffold for selective macroautophagy. Nat. Cell Biol. 2015, 17, 262–275. [Google Scholar] [CrossRef]
- Gao, J.; Ohtsubo, M.; Hotta, Y.; Minoshima, S. Oligomerization of optineurin and its oxidative stress- or E50K mutation-driven covalent cross-linking: Possible relationship with glaucoma pathology. PLoS ONE 2014, 9, e101206. [Google Scholar] [CrossRef]
- Elliott, E.; Tsvetkov, P.; Ginzburg, I. BAG-1 associates with Hsc70.Tau complex and regulates the proteasomal degradation of Tau protein. J. Biol. Chem. 2007, 282, 37276–37284. [Google Scholar] [CrossRef]
- Luders, J.; Demand, J.; Hohfeld, J. The ubiquitin-related BAG-1 provides a link between the molecular chaperones Hsc70/Hsp70 and the proteasome. J. Biol. Chem. 2000, 275, 4613–4617. [Google Scholar] [CrossRef]
- Behl, C. Breaking BAG: The co-chaperone BAG3 in health and disease. Trends Pharmacol. Sci. 2016, 37, 672–688. [Google Scholar] [CrossRef]
- Gamerdinger, M.; Hajieva, P.; Kaya, A.M.; Wolfrum, U.; Hartl, F.U.; Behl, C. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 2009, 28, 889–901. [Google Scholar] [CrossRef]
- Carra, S.; Seguin, S.J.; Landry, J. HspB8 and Bag3: A new chaperone complex targeting misfolded proteins to macroautophagy. Autophagy 2008, 4, 237–239. [Google Scholar] [CrossRef]
- Carra, S.; Seguin, S.J.; Lambert, H.; Landry, J. HspB8 chaperone activity toward poly(Q)-containing proteins depends on its association with Bag3, a stimulator of macroautophagy. J. Biol. Chem. 2008, 283, 1437–1444. [Google Scholar] [CrossRef]
- Minoia, M.; Boncoraglio, A.; Vinet, J.; Morelli, F.F.; Brunsting, J.F.; Poletti, A.; Krom, S.; Reits, E.; Kampinga, H.H.; Carra, S. BAG3 induces the sequestration of proteasomal clients into cytoplasmic puncta: Implications for a proteasome-to-autophagy switch. Autophagy 2014, 10, 1603–1621. [Google Scholar] [CrossRef]
- Gamerdinger, M.; Kaya, A.M.; Wolfrum, U.; Clement, A.M.; Behl, C. BAG3 mediates chaperone-based aggresome-targeting and selective autophagy of misfolded proteins. EMBO Rep. 2011, 12, 149–156. [Google Scholar] [CrossRef]
- Doukhanina, E.V.; Chen, S.; van der Zalm, E.; Godzik, A.; Reed, J.; Dickman, M.B. Identification and functional characterization of the BAG protein family in Arabidopsis thaliana. J. Biol. Chem. 2006, 281, 18793–18801. [Google Scholar] [CrossRef] [PubMed]
- Kabbage, M.; Dickman, M.B. The BAG proteins: A ubiquitous family of chaperone regulators. Cell. Mol. Life Sci. 2008, 65, 1390–1402. [Google Scholar] [CrossRef] [PubMed]
- Kabbage, M.; Kessens, R.; Dickman, M.B. A plant Bcl-2-associated athanogene is proteolytically activated to confer fungal resistance. Microb. Cell 2016, 3, 224–226. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Dickman, M. Processing of AtBAG6 triggers autophagy and fungal resistance. Plant Signal. Behav. 2016, 11, e1175699. [Google Scholar] [CrossRef]
- Sebti, S.; Prebois, C.; Perez-Gracia, E.; Bauvy, C.; Desmots, F.; Pirot, N.; Gongora, C.; Bach, A.S.; Hubberstey, A.V.; Palissot, V.; et al. BAG6/BAT3 modulates autophagy by affecting EP300/p300 intracellular localization. Autophagy 2014, 10, 1341–1342. [Google Scholar] [CrossRef] [PubMed]
- Lowe, E.D.; Hasan, N.; Trempe, J.F.; Fonso, L.; Noble, M.E.; Endicott, J.A.; Johnson, L.N.; Brown, N.R. Structures of the Dsk2 UBL and UBA domains and their complex. Acta Crystallogr. D Biol. Crystallogr. 2006, 62, 177–188. [Google Scholar] [CrossRef]
- Wang, Q.; Goh, A.M.; Howley, P.M.; Walters, K.J. Ubiquitin recognition by the DNA repair protein hHR23a. Biochemistry 2003, 42, 13529–13535. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, T.; Funakoshi, M.; Endicott, J.A.; Kobayashi, H. Budding yeast Dsk2 protein forms a homodimer via its C-terminal UBA domain. Biochem. Biophys. Res. Commun. 2005, 336, 530–535. [Google Scholar] [CrossRef]
- Ford, D.L.; Monteiro, M.J. Dimerization of ubiquilin is dependent upon the central region of the protein: Evidence that the monomer, but not the dimer, is involved in binding presenilins. Biochem. J. 2006, 399, 397–404. [Google Scholar] [CrossRef]
- Kang, Y.; Zhang, N.; Koepp, D.M.; Walters, K.J. Ubiquitin receptor proteins hHR23a and hPLIC2 interact. J. Mol. Biol. 2007, 365, 1093–1101. [Google Scholar] [CrossRef]
- Hirano, Y.; Yoshinaga, S.; Ogura, K.; Yokochi, M.; Noda, Y.; Sumimoto, H.; Inagaki, F. Solution structure of atypical protein kinase C PB1 domain and its mode of interaction with ZIP/p62 and MEK5. J. Biol. Chem. 2004, 279, 31883–31890. [Google Scholar] [CrossRef] [PubMed]
- Su, V.; Lau, A.F. Ubiquitin-like and ubiquitin-associated domain proteins: Significance in proteasomal degradation. Cell. Mol. Life Sci. 2009, 66, 2819–2833. [Google Scholar] [CrossRef]
- Isogai, S.; Morimoto, D.; Arita, K.; Unzai, S.; Tenno, T.; Hasegawa, J.; Sou, Y.S.; Komatsu, M.; Tanaka, K.; Shirakawa, M.; et al. Crystal structure of the ubiquitin-associated (UBA) domain of p62 and its interaction with ubiquitin. J. Biol. Chem. 2011, 286, 31864–31874. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.A.; Sun, Y.; Jiang, Y.P.; Bott, A.J.; Jaber, N.; Dou, Z.; Yang, B.; Chen, J.S.; Catanzaro, J.M.; Du, C.; et al. TRIM21 ubiquitylates SQSTM1/p62 and suppresses protein sequestration to regulate redox homeostasis. Mol. Cell 2016, 61, 720–733. [Google Scholar] [CrossRef] [PubMed]
- Liang, R.Y.; Chen, L.; Ko, B.T.; Shen, Y.H.; Li, Y.T.; Chen, B.R.; Lin, K.T.; Madura, K.; Chuang, S.M. Rad23 interaction with the proteasome is regulated by phosphorylation of its ubiquitin-like (UbL) domain. J. Mol. Biol. 2014, 426, 4049–4060. [Google Scholar] [CrossRef]
- Godderz, D.; Giovannucci, T.A.; Lalakova, J.; Menendez-Benito, V.; Dantuma, N.P. The deubiquitylating enzyme Ubp12 regulates Rad23-dependent proteasomal degradation. J. Cell. Sci. 2017, 130, 3336–3346. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Ghosh, S. Negative regulation of toll-like receptor-mediated signaling by Tollip. J. Biol. Chem. 2002, 277, 7059–7065. [Google Scholar] [CrossRef] [PubMed]
- Song, P.; Li, S.; Wu, H.; Gao, R.; Rao, G.; Wang, D.; Chen, Z.; Ma, B.; Wang, H.; Sui, N.; et al. Parkin promotes proteasomal degradation of p62: Implication of selective vulnerability of neuronal cells in the pathogenesis of Parkinson’s disease. Protein Cell 2016, 7, 114–129. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, G.; Wada, K.; Okuno, M.; Kurosawa, M.; Nukina, N. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol. Cell 2011, 44, 279–289. [Google Scholar] [CrossRef]
- Heath, R.J.; Goel, G.; Baxt, L.A.; Rush, J.S.; Mohanan, V.; Paulus, G.L.C.; Jani, V.; Lassen, K.G.; Xavier, R.J. RNF166 determines recruitment of adaptor proteins during antibacterial autophagy. Cell. Rep. 2016, 17, 2183–2194. [Google Scholar] [CrossRef]
- Kalogeropulou, A.F.; Zhao, J.; Bolliger, M.F.; Memou, A.; Narasimha, S.; Molitor, T.P.; Wilson, W.H.; Rideout, H.J.; Nichols, R.J. P62/SQSTM1 is a novel leucine-rich repeat kinase 2 (LRRK2) substrate that enhances neuronal toxicity. Biochem. J. 2018, 475, 1271–1293. [Google Scholar] [CrossRef] [PubMed]
- Linares, J.F.; Amanchy, R.; Greis, K.; Diaz-Meco, M.T.; Moscat, J. Phosphorylation of p62 by cdk1 controls the timely transit of cells through mitosis and tumor cell proliferation. Mol. Cell. Biol. 2011, 31, 105–117. [Google Scholar] [CrossRef]
- Ha, S.; Jeong, S.H.; Yi, K.; Chung, K.M.; Hong, C.J.; Kim, S.W.; Kim, E.K.; Yu, S.W. Phosphorylation of p62 by AMP-activated protein kinase mediates autophagic cell death in adult hippocampal neural stem cells. J. Biol. Chem. 2017, 292, 13795–13808. [Google Scholar] [CrossRef]
- Watanabe, Y.; Tsujimura, A.; Taguchi, K.; Tanaka, M. HSF1 stress response pathway regulates autophagy receptor SQSTM1/p62-associated proteostasis. Autophagy 2017, 13, 133–148. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, K.; Simmons, A.N.; Kajino-Sakamoto, R.; Tsuji, Y.; Ninomiya-Tsuji, J. TAK1 regulates the Nrf2 antioxidant system through modulating p62/SQSTM1. Antioxid. Redox Signal. 2016, 25, 953–964. [Google Scholar] [CrossRef] [PubMed]
- Ichimura, Y.; Waguri, S.; Sou, Y.S.; Kageyama, S.; Hasegawa, J.; Ishimura, R.; Saito, T.; Yang, Y.; Kouno, T.; Fukutomi, T.; et al. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol. Cell 2013, 51, 618–631. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Lachenmayer, M.L.; Wu, S.; Liu, W.; Kundu, M.; Wang, R.; Komatsu, M.; Oh, Y.J.; Zhao, Y.; Yue, Z. Proteotoxic stress induces phosphorylation of p62/SQSTM1 by ULK1 to regulate selective autophagic clearance of protein aggregates. PLoS Genet. 2015, 11, e1004987. [Google Scholar] [CrossRef] [PubMed]
- Pilli, M.; Arko-Mensah, J.; Ponpuak, M.; Roberts, E.; Master, S.; Mandell, M.A.; Dupont, N.; Ornatowski, W.; Jiang, S.; Bradfute, S.B.; et al. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity 2012, 37, 223–234. [Google Scholar] [CrossRef]
- Lee, Y.; Chou, T.F.; Pittman, S.K.; Keith, A.L.; Razani, B.; Weihl, C.C. Keap1/Cullin3 modulates p62/SQSTM1 activity via UBA domain ubiquitination. Cell. Rep. 2017, 19, 188–202. [Google Scholar] [CrossRef]
- Jongsma, M.L.; Berlin, I.; Wijdeven, R.H.; Janssen, L.; Janssen, G.M.; Garstka, M.A.; Janssen, H.; Mensink, M.; van Veelen, P.A.; Spaapen, R.M.; et al. An ER-associated pathway defines endosomal architecture for controlled cargo transport. Cell 2016, 166, 152–166. [Google Scholar] [CrossRef]
- Lin, Q.; Dai, Q.; Meng, H.; Sun, A.; Wei, J.; Peng, K.; Childress, C.; Chen, M.; Shao, G.; Yang, W. The HECT E3 ubiquitin ligase NEDD4 interacts with and ubiquitylates SQSTM1 for inclusion body autophagy. J. Cell. Sci. 2017, 130, 3839–3850. [Google Scholar] [CrossRef] [PubMed]
- Nicot, A.S.; Lo Verso, F.; Ratti, F.; Pilot-Storck, F.; Streichenberger, N.; Sandri, M.; Schaeffer, L.; Goillot, E. Phosphorylation of NBR1 by GSK3 modulates protein aggregation. Autophagy 2014, 10, 1036–1053. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.M.; Ordureau, A.; Paulo, J.A.; Rinehart, J.; Harper, J.W. The PINK1-PARKIN mitochondrial ubiquitylation pathway drives a program of OPTN/NDP52 recruitment and TBK1 activation to promote mitophagy. Mol. Cell 2015, 60, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Chou, T.F.; Pittman, S.K.; Keith, A.L.; Razani, B.; Weihl, C.C. Keap1/Cullin3 modulates p62/SQSTM1 activity via UBA domain ubiquitination. Cell. Rep. 2017, 20, 1994. [Google Scholar] [CrossRef]
- Jacomin, A.C.; Samavedam, S.; Promponas, V.; Nezis, I.P. iLIR database: A web resource for LIR motif-containing proteins in eukaryotes. Autophagy 2016, 12, 1945–1953. [Google Scholar] [CrossRef] [PubMed]
- Birgisdottir, A.B.; Lamark, T.; Johansen, T. The LIR motif—Crucial for selective autophagy. J. Cell. Sci. 2013, 126, 3237–3247. [Google Scholar] [CrossRef] [PubMed]
- Cha-Molstad, H.; Sung, K.S.; Hwang, J.; Kim, K.A.; Yu, J.E.; Yoo, Y.D.; Jang, J.M.; Han, D.H.; Molstad, M.; Kim, J.G.; et al. Amino-terminal arginylation targets endoplasmic reticulum chaperone BiP for autophagy through p62 binding. Nat. Cell Biol. 2015, 17, 917–929. [Google Scholar] [CrossRef]
- Isakson, P.; Holland, P.; Simonsen, A. The role of ALFY in selective autophagy. Cell Death Differ. 2013, 20, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Simonsen, A.; Birkeland, H.C.; Gillooly, D.J.; Mizushima, N.; Kuma, A.; Yoshimori, T.; Slagsvold, T.; Brech, A.; Stenmark, H. Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. J. Cell. Sci. 2004, 117, 4239–4251. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).