Receptor Tyrosine Kinase Ubiquitination and De-Ubiquitination in Signal Transduction and Receptor Trafficking
Abstract
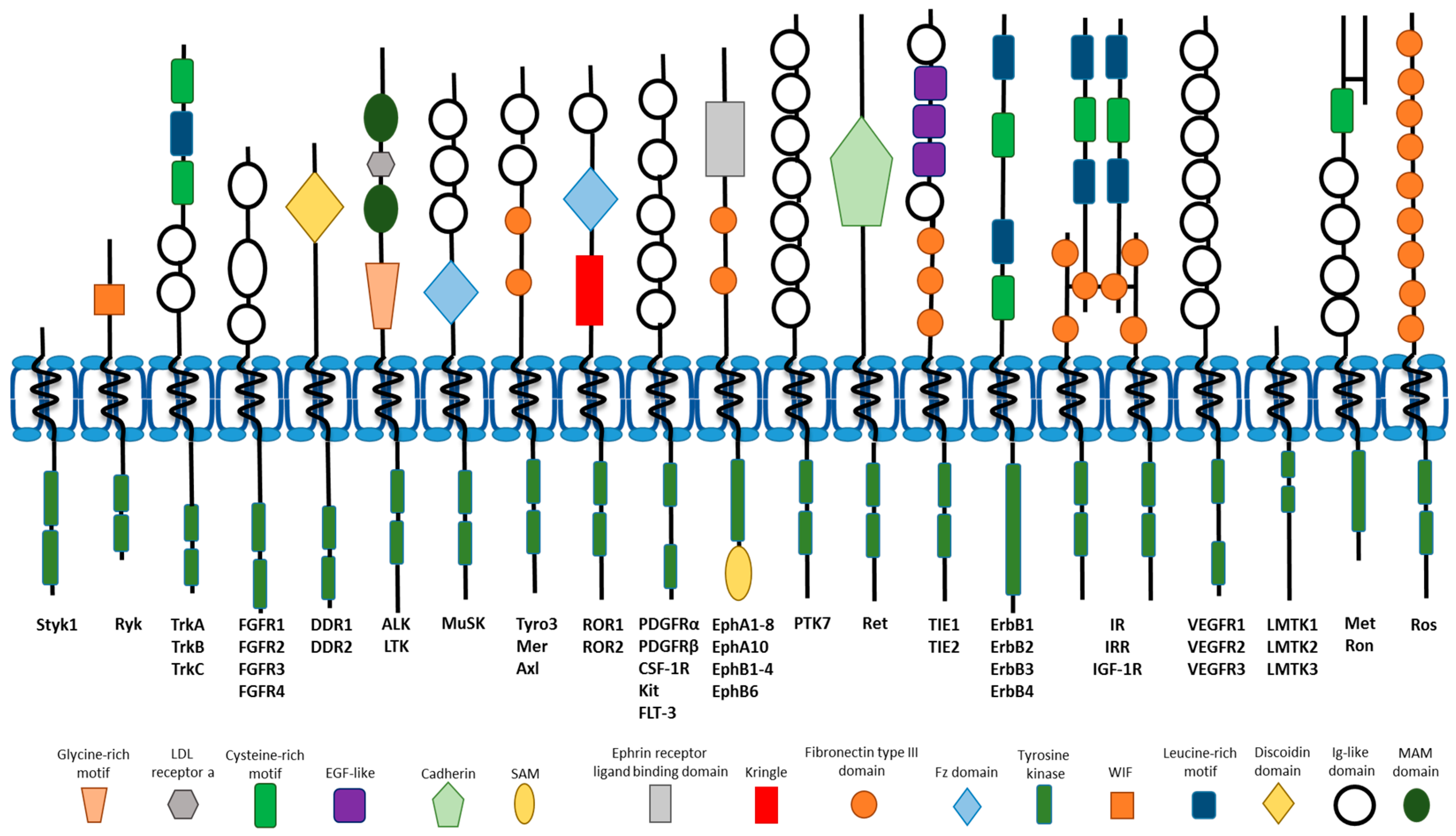
1. Introduction
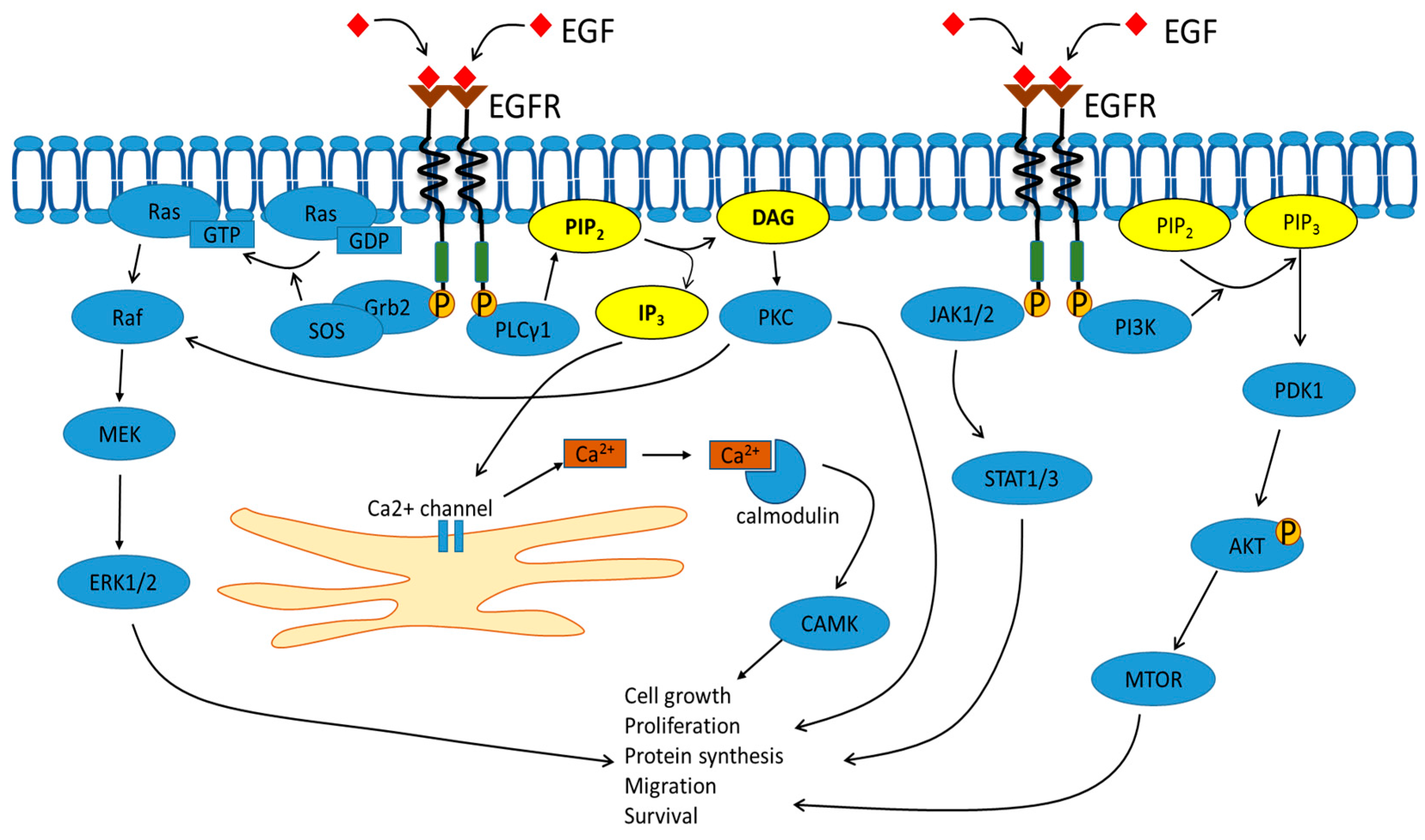
2. Signal Transduction
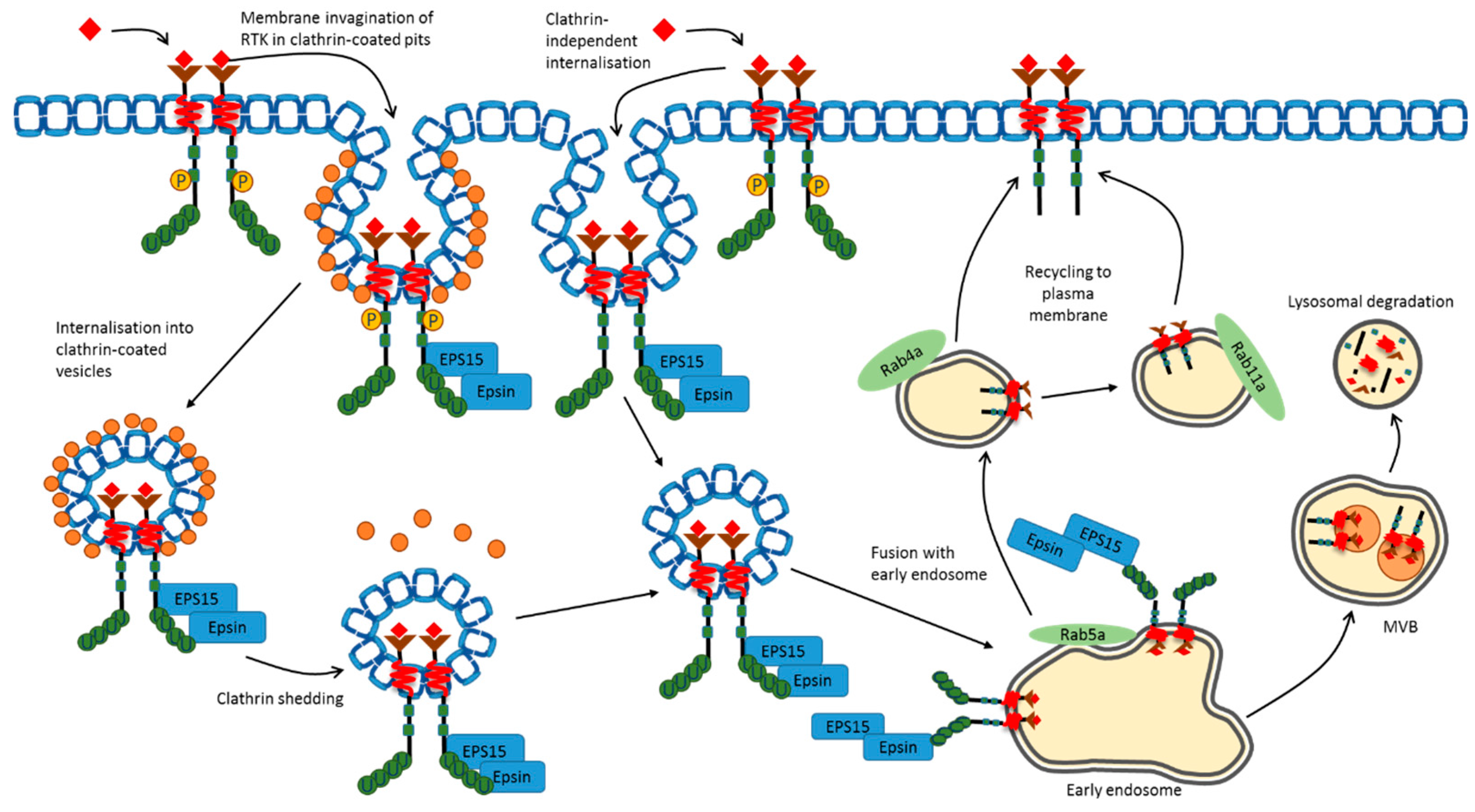
3. Plasma Membrane Endocytosis
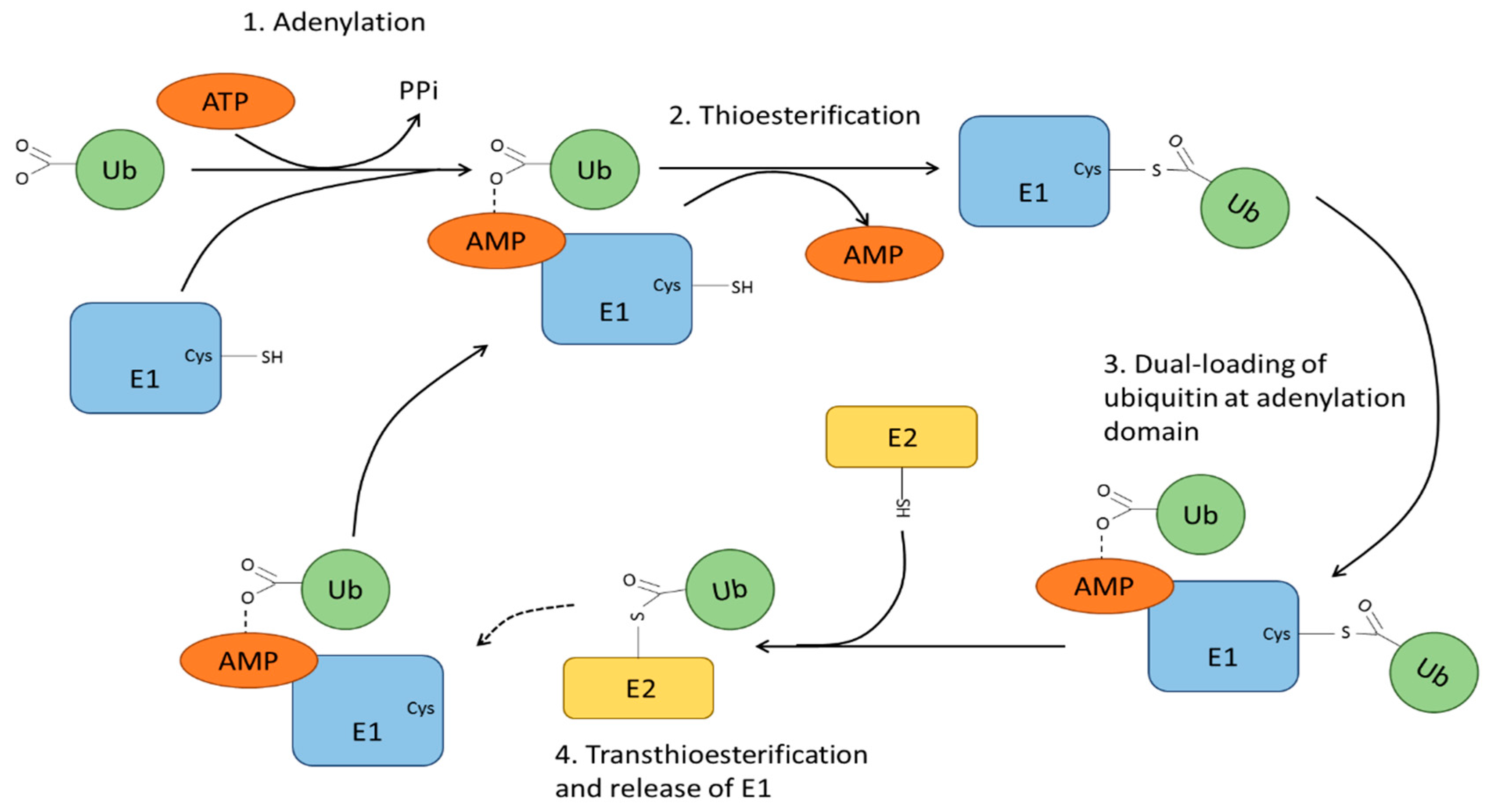
4. Ubiquitin Modification and Ubiquitin Ligases
5. Plasma Membrane RTK Ubiquitination and Sorting
6. RTK Ubiquitination, Recycling and Proteolysis
7. RTK Targeting by E3 Ubiquitin Ligases
8. RTK De-Ubiquitination
9. Conclusions
Acknowledgements
Author Contributions
Conflicts of Interest
References
- Robinson, D.R.; Wu, Y.M.; Lin, S.F. The protein tyrosine kinase family of the human genome. Oncogene 2000, 19, 5548–5557. [Google Scholar] [CrossRef] [PubMed]
- Blume-Jensen, P.; Hunter, T. Oncogenic kinase signaling. Nature 2001, 411, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Lemmon, M.A.; Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 2010, 141, 1117–1134. [Google Scholar] [CrossRef] [PubMed]
- Marino-Buslje, C.; Mizuguchi, K.; Siddle, K.; Blundell, T.L. A third fibronectin type iii domain in the extracellular region of the insulin receptor family. FEBS Lett. 1998, 441, 331–336. [Google Scholar] [CrossRef]
- Tuzi, N.L.; Gullick, W.J. Eph, the largest known family of putative growth factor receptors. Br. J. Cancer 1994, 69, 417–421. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Garrett, T.P.; McKern, N.M.; Lou, M.; Frenkel, M.J.; Bentley, J.D.; Lovrecz, G.O.; Elleman, T.C.; Cosgrove, L.J.; Ward, C.W. Crystal structure of the first three domains of the type-1 insulin-like growth factor receptor. Nature 1998, 394, 395–399. [Google Scholar] [CrossRef] [PubMed]
- Ward, C.W.; Hoyne, P.A.; Flegg, R.H. Insulin and epidermal growth factor receptors contain the cysteine repeat motif found in the tumor necrosis factor receptor. Proteins 1995, 22, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Grassot, J.; Gouy, M.; Perriere, G.; Mouchiroud, G. Origin and molecular evolution of receptor tyrosine kinases with immunoglobulin-like domains. Mol. Biol. Evol. 2006, 23, 1232–1241. [Google Scholar] [CrossRef] [PubMed]
- Windisch, J.M.; Auer, B.; Marksteiner, R.; Lang, M.E.; Schneider, R. Specific neurotrophin binding to leucine-rich motif peptides of trka and trkb. FEBS Lett. 1995, 374, 125–129. [Google Scholar] [CrossRef]
- Reshetnyak, A.V.; Murray, P.B.; Shi, X.; Mo, E.S.; Mohanty, J.; Tome, F.; Bai, H.; Gunel, M.; Lax, I.; Schlessinger, J. Augmentor alpha and beta (fam150) are ligands of the receptor tyrosine kinases alk and ltk: Hierarchy and specificity of ligand-receptor interactions. Proc. Natl. Acad. Sci. USA 2015, 112, 15862–15867. [Google Scholar] [CrossRef] [PubMed]
- Manni, S.; Kisko, K.; Schleier, T.; Missimer, J.; Ballmer-Hofer, K. Functional and structural characterization of the kinase insert and the carboxy terminal domain in vegf receptor 2 activation. FASEB J. 2014, 28, 4914–4923. [Google Scholar] [CrossRef] [PubMed]
- Sarabipour, S.; Hristova, K. Mechanism of fgf receptor dimerization and activation. Nat. Commun. 2016, 7, 10262. [Google Scholar] [CrossRef] [PubMed]
- Jahangiri, A.; Nguyen, A.; Chandra, A.; Sidorov, M.K.; Yagnik, G.; Rick, J.; Han, S.W.; Chen, W.; Flanigan, P.M.; Schneidman-Duhovny, D.; et al. Cross-activating c-met/beta1 integrin complex drives metastasis and invasive resistance in cancer. Proc. Natl. Acad. Sci. USA 2017, 114, E8685–E8694. [Google Scholar] [PubMed]
- Weiss, F.U.; Daub, H.; Ullrich, A. Novel mechanisms of rtk signal generation. Curr. Opin. Genet. Dev. 1997, 7, 80–86. [Google Scholar] [CrossRef]
- Hubbard, S.R.; Miller, W.T. Receptor tyrosine kinases: Mechanisms of activation and signaling. Curr. Opin. Cell Biol. 2007, 19, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Volinsky, N.; Kholodenko, B.N. Complexity of receptor tyrosine kinase signal processing. Cold Spring Harbor Perspect. Biol. 2013, 5, a009043. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, M.Y.; Yang, S.; Raymond-Stinz, M.A.; Edwards, J.S.; Wilson, B.S. Spatio-temporal modeling of signaling protein recruitment to egfr. BMC Syst. Biol. 2010, 4, 57. [Google Scholar] [CrossRef] [PubMed]
- Ott, C.M.; Lingappa, V.R. Integral membrane protein biosynthesis: Why topology is hard to predict. J. Cell Sci. 2002, 115, 2003–2009. [Google Scholar] [PubMed]
- Scharaw, S.; Iskar, M.; Ori, A.; Boncompain, G.; Laketa, V.; Poser, I.; Lundberg, E.; Perez, F.; Beck, M.; Bork, P.; et al. The endosomal transcriptional regulator rnf11 integrates degradation and transport of egfr. J. Cell Biol. 2016, 215, 543–558. [Google Scholar] [CrossRef] [PubMed]
- Crepaldi, T.; Pollack, A.L.; Prat, M.; Zborek, A.; Mostov, K.; Comoglio, P.M. Targeting of the sf/hgf receptor to the basolateral domain of polarized epithelial cells. J. Cell Biol. 1994, 125, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Hobert, M.E.; Kil, S.J.; Medof, M.E.; Carlin, C.R. The cytoplasmic juxtamembrane domain of the epidermal growth factor receptor contains a novel autonomous basolateral sorting determinant. J. Biol. Chem. 1997, 272, 32901–32909. [Google Scholar] [CrossRef] [PubMed]
- Mittar, S.; Ulyatt, C.; Howell, G.J.; Bruns, A.F.; Zachary, I.; Walker, J.H.; Ponnambalam, S. Vegfr1 receptor tyrosine kinase localization to the golgi apparatus is calcium-dependent. Exp. Cell Res. 2009, 315, 877–889. [Google Scholar] [CrossRef] [PubMed]
- Manickam, V.; Tiwari, A.; Jung, J.J.; Bhattacharya, R.; Goel, A.; Mukhopadhyay, D.; Choudhury, A. Regulation of vascular endothelial growth factor receptor 2 trafficking and angiogenesis by golgi localized t-snare syntaxin 6. Blood 2011, 117, 1425–1435. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, A.; Jung, J.J.; Inamdar, S.M.; Nihalani, D.; Choudhury, A. The myosin motor myo1c is required for vegfr2 delivery to the cell surface and for angiogenic signaling. Am. J. Physiol. Heart Circ. Physiol. 2013, 304, H687–H696. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.H.; Nakajima, Y.; Geyer, M.; Wary, K.K.; Ushio-Fukai, M.; Komarova, Y.; Malik, A.B. Kif13b regulates angiogenesis through golgi to plasma membrane trafficking of vegfr2. J. Cell Sci. 2014, 127, 4518–4530. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Zhao, L.; Li, X.; Ji, Y.S.; Li, N.; Xu, X.F.; Chen, Z.Y. Syntaxin 8 modulates the post-synthetic trafficking of the trka receptor and inflammatory pain transmission. J. Biol. Chem. 2014, 289, 19556–19569. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Shen, J.; Hsu, J.L.; Han, Z.; Hsu, M.C.; Yang, C.C.; Kuo, H.P.; Wang, Y.N.; Yamaguchi, H.; Miller, S.A.; et al. Syntaxin 6-mediated golgi translocation plays an important role in nuclear functions of egfr through microtubule-dependent trafficking. Oncogene 2014, 33, 756–770. [Google Scholar] [CrossRef] [PubMed]
- Warren, C.M.; Ziyad, S.; Briot, A.; Der, A.; Iruela-Arispe, M.L. A ligand-independent vegfr2 signaling pathway limits angiogenic responses in diabetes. Sci. Signal. 2014, 7, ra1. [Google Scholar] [CrossRef] [PubMed]
- Maghsoudlou, A.; Meyer, R.D.; Rezazadeh, K.; Arafa, E.; Pudney, J.; Hartsough, E.; Rahimi, N. Rnf121 inhibits angiogenic growth factor signaling by restricting cell surface expression of vegfr-2. Traffic 2016, 17, 289–300. [Google Scholar] [CrossRef] [PubMed]
- Jastrzebski, K.; Zdzalik-Bielecka, D.; Maminska, A.; Kalaidzidis, Y.; Hellberg, C.; Miaczynska, M. Multiple routes of endocytic internalization of pdgfrbeta contribute to pdgf-induced stat3 signaling. J. Cell Sci. 2017, 130, 577–589. [Google Scholar] [CrossRef] [PubMed]
- Vieira, A.V.; Lamaze, C.; Schmid, S.L. Control of egf receptor signaling by clathrin-mediated endocytosis. Science 1996, 274, 2086–2089. [Google Scholar] [CrossRef] [PubMed]
- Sigismund, S.; Argenzio, E.; Tosoni, D.; Cavallaro, E.; Polo, S.; Di Fiore, P.P. Clathrin-mediated internalization is essential for sustained egfr signaling but dispensable for degradation. Dev. Cell 2008, 15, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Zhou, Y.; Ozawa, T.; Okizono, R.; Banba, A.; Yamamura, T.; Oga, E.; Muraguchi, A.; Sakurai, H. Ligand-activated epidermal growth factor receptor (egfr) signaling governs endocytic trafficking of unliganded receptor monomers by non-canonical phosphorylation. J. Biol. Chem. 2018, 293, 2288–2301. [Google Scholar] [CrossRef] [PubMed]
- Ewan, L.C.; Jopling, H.M.; Jia, H.; Mittar, S.; Bagherzadeh, A.; Howell, G.J.; Walker, J.H.; Zachary, I.C.; Ponnambalam, S. Intrinsic tyrosine kinase activity is required for vascular endothelial growth factor receptor 2 ubiquitination, sorting and degradation in endothelial cells. Traffic 2006, 7, 1270–1282. [Google Scholar] [CrossRef] [PubMed]
- Lampugnani, M.G.; Orsenigo, F.; Gagliani, M.C.; Tacchetti, C.; Dejana, E. Vascular endothelial cadherin controls vegfr-2 internalization and signaling from intracellular compartments. J. Cell Biol. 2006, 174, 593–604. [Google Scholar] [CrossRef] [PubMed]
- Bruns, A.F.; Herbert, S.P.; Odell, A.F.; Jopling, H.M.; Hooper, N.M.; Zachary, I.C.; Walker, J.H.; Ponnambalam, S. Ligand-stimulated vegfr2 signaling is regulated by co-ordinated trafficking and proteolysis. Traffic 2010, 11, 161–174. [Google Scholar] [CrossRef] [PubMed]
- Fearnley, G.W.; Smith, G.A.; Abdul-Zani, I.; Yuldasheva, N.; Mughal, N.A.; Homer-Vanniasinkam, S.; Kearney, M.T.; Zachary, I.C.; Tomlinson, D.C.; Harrison, M.A.; et al. Vegf-a isoforms program differential vegfr2 signal transduction, trafficking and proteolysis. Biol. Open 2016, 5, 571–583. [Google Scholar] [CrossRef] [PubMed]
- Delos Santos, R.C.; Bautista, S.; Lucarelli, S.; Bone, L.N.; Dayam, R.M.; Abousawan, J.; Botelho, R.J.; Antonescu, C.N. Selective regulation of clathrin-mediated epidermal growth factor receptor signaling and endocytosis by phospholipase c and calcium. Mol. Biol. Cell 2017, 28, 2802–2818. [Google Scholar] [CrossRef] [PubMed]
- Bogdanovic, E.; Coombs, N.; Dumont, D.J. Oligomerized tie2 localizes to clathrin-coated pits in response to angiopoietin-1. Histochem. Cell Biol. 2009, 132, 225–237. [Google Scholar] [CrossRef] [PubMed]
- Wilde, A.; Beattie, E.C.; Lem, L.; Riethof, D.A.; Liu, S.H.; Mobley, W.C.; Soriano, P.; Brodsky, F.M. Egf receptor signaling stimulates src kinase phosphorylation of clathrin, influencing clathrin redistribution and egf uptake. Cell 1999, 96, 677–687. [Google Scholar] [CrossRef]
- Goh, L.K.; Huang, F.; Kim, W.; Gygi, S.; Sorkin, A. Multiple mechanisms collectively regulate clathrin-mediated endocytosis of the epidermal growth factor receptor. J. Cell Biol. 2010, 189, 871–883. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Huang, F.; Marusyk, A.; Sorkin, A. Grb2 regulates internalization of egf receptors through clathrin-coated pits. Mol. Biol. Cell 2003, 14, 858–870. [Google Scholar] [CrossRef] [PubMed]
- Ioannou, M.S.; Kulasekaran, G.; Fotouhi, M.; Morein, J.J.; Han, C.; Tse, S.; Nossova, N.; Han, T.; Mannard, E.; McPherson, P.S. Intersectin-s interaction with dennd2b facilitates recycling of epidermal growth factor receptor. EMBO Rep. 2017, 18, 2119–2130. [Google Scholar] [CrossRef] [PubMed]
- Puri, C.; Tosoni, D.; Comai, R.; Rabellino, A.; Segat, D.; Caneva, F.; Luzzi, P.; Di Fiore, P.P.; Tacchetti, C. Relationships between egfr signaling-competent and endocytosis-competent membrane microdomains. Mol. Biol. Cell 2005, 16, 2704–2718. [Google Scholar] [CrossRef] [PubMed]
- Rappoport, J.Z.; Simon, S.M. Endocytic trafficking of activated egfr is ap-2 dependent and occurs through preformed clathrin spots. J. Cell Sci. 2009, 122, 1301–1305. [Google Scholar] [CrossRef] [PubMed]
- Sigismund, S.; Woelk, T.; Puri, C.; Maspero, E.; Tacchetti, C.; Transidico, P.; Di Fiore, P.P.; Polo, S. Clathrin-independent endocytosis of ubiquitinated cargos. Proc. Natl. Acad. Sci. USA 2005, 102, 2760–2765. [Google Scholar] [CrossRef] [PubMed]
- Caldieri, G.; Barbieri, E.; Nappo, G.; Raimondi, A.; Bonora, M.; Conte, A.; Verhoef, L.; Confalonieri, S.; Malabarba, M.G.; Bianchi, F.; et al. Reticulon 3-dependent er-pm contact sites control egfr nonclathrin endocytosis. Science 2017, 356, 617–624. [Google Scholar] [CrossRef] [PubMed]
- Basagiannis, D.; Zografou, S.; Murphy, C.; Fotsis, T.; Morbidelli, L.; Ziche, M.; Bleck, C.; Mercer, J.; Christoforidis, S. Vegf induces signaling and angiogenesis by directing vegfr2 internalisation through macropinocytosis. J. Cell Sci. 2016, 129, 4091–4104. [Google Scholar] [PubMed]
- Yamabhai, M.; Anderson, R.G. Second cysteine-rich region of epidermal growth factor receptor contains targeting information for caveolae/rafts. J. Biol. Chem. 2002, 277, 24843–24846. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, S.; Ushio-Fukai, M.; Zuo, L.; Tojo, T.; Dikalov, S.; Patrushev, N.A.; Alexander, R.W. Novel role of arf6 in vascular endothelial growth factor-induced signaling and angiogenesis. Circ. Res. 2005, 96, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T.; Lu, C.; Ida, L.; Yanagisawa, K.; Usukura, J.; Cheng, J.; Hotta, N.; Shimada, Y.; Isomura, H.; Suzuki, M.; et al. Ror1 sustains caveolae and survival signaling as a scaffold of cavin-1 and caveolin-1. Nat. Commun. 2016, 7, 10060. [Google Scholar] [CrossRef] [PubMed]
- Mastick, C.C.; Brady, M.J.; Saltiel, A.R. Insulin stimulates the tyrosine phosphorylation of caveolin. J. Cell Biol. 1995, 129, 1523–1531. [Google Scholar] [CrossRef] [PubMed]
- Labrecque, L.; Royal, I.; Surprenant, D.S.; Patterson, C.; Gingras, D.; Beliveau, R. Regulation of vascular endothelial growth factor receptor-2 activity by caveolin-1 and plasma membrane cholesterol. Mol. Biol. Cell 2003, 14, 334–347. [Google Scholar] [CrossRef] [PubMed]
- Orlichenko, L.; Huang, B.; Krueger, E.; McNiven, M.A. Epithelial growth factor-induced phosphorylation of caveolin 1 at tyrosine 14 stimulates caveolae formation in epithelial cells. J. Biol. Chem. 2006, 281, 4570–4579. [Google Scholar] [CrossRef] [PubMed]
- Verma, N.; Keinan, O.; Selitrennik, M.; Karn, T.; Filipits, M.; Lev, S. Pyk2 sustains endosomal-derived receptor signaling and enhances epithelial-to-mesenchymal transition. Nat. Commun. 2015, 6, 6064. [Google Scholar] [CrossRef] [PubMed]
- Taub, N.; Teis, D.; Ebner, H.L.; Hess, M.W.; Huber, L.A. Late endosomal traffic of the epidermal growth factor receptor ensures spatial and temporal fidelity of mitogen-activated protein kinase signaling. Mol. Biol. Cell 2007, 18, 4698–4710. [Google Scholar] [CrossRef] [PubMed]
- Laviolette, L.A.; Mermoud, J.; Calvo, I.A.; Olson, N.; Boukhali, M.; Steinlein, O.K.; Roider, E.; Sattler, E.C.; Huang, D.; Teh, B.T.; et al. Negative regulation of egfr signaling by the human folliculin tumour suppressor protein. Nat. Commun. 2017, 8, 15866. [Google Scholar] [CrossRef] [PubMed]
- Kermorgant, S.; Parker, P.J. Receptor trafficking controls weak signal delivery: A strategy used by c-met for stat3 nuclear accumulation. J. Cell Biol. 2008, 182, 855–863. [Google Scholar] [CrossRef] [PubMed]
- Menard, L.; Parker, P.J.; Kermorgant, S. Receptor tyrosine kinase c-met controls the cytoskeleton from different endosomes via different pathways. Nat. Commun. 2014, 5, 3907. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Pennock, S.D.; Chen, X.; Kazlauskas, A.; Wang, Z. Platelet-derived growth factor receptor-mediated signal transduction from endosomes. J. Biol. Chem. 2004, 279, 8038–8046. [Google Scholar] [CrossRef] [PubMed]
- Kofler, N.; Corti, F.; Rivera-Molina, F.; Deng, Y.; Toomre, D.; Simons, M. The rab-effector protein rabep2 regulates endosomal trafficking to mediate vascular endothelial growth factor receptor-2 (vegfr2)-dependent signaling. J. Biol. Chem. 2018. [Google Scholar] [CrossRef] [PubMed]
- Acconcia, F.; Sigismund, S.; Polo, S. Ubiquitin in trafficking: The network at work. Exp. Cell Res. 2009, 315, 1610–1618. [Google Scholar] [CrossRef] [PubMed]
- Girnita, L.; Takahashi, S.I.; Crudden, C.; Fukushima, T.; Worrall, C.; Furuta, H.; Yoshihara, H.; Hakuno, F.; Girnita, A. Chapter seven-when phosphorylation encounters ubiquitination: A balanced perspective on igf-1r signaling. Prog. Mol. Biol. Transl. Sci. 2016, 141, 277–311. [Google Scholar] [PubMed]
- Smith, G.A.; Tomlinson, D.C.; Harrison, M.A.; Ponnambalam, S. Chapter eight-ubiquitin-mediated regulation of cellular responses to vascular endothelial growth factors. Prog. Mol. Biol. Transl. Sci. 2016, 141, 313–338. [Google Scholar] [PubMed]
- Smith, G.A.; Fearnley, G.W.; Abdul-Zani, I.; Wheatcroft, S.B.; Tomlinson, D.C.; Harrison, M.A.; Ponnambalam, S. Ubiquitination of basal vegfr2 regulates signal transduction and endothelial function. Biol. Open 2017, 6, 1404–1415. [Google Scholar] [CrossRef] [PubMed]
- Sorkin, A.; Goh, L.K. Endocytosis and intracellular trafficking of erbbs. Exp. Cell Res. 2008, 314, 3093–3106. [Google Scholar] [CrossRef] [PubMed]
- Gucwa, A.L.; Brown, D.A. Uim domain-dependent recruitment of the endocytic adaptor protein eps15 to ubiquitin-enriched endosomes. BMC Cell Biol. 2014, 15, 34. [Google Scholar] [CrossRef] [PubMed]
- Lund, P.K.; Moats-Staats, B.M.; Simmons, J.G.; Hoyt, E.; D’Ercole, A.J.; Martin, F.; Van Wyk, J.J. Nucleotide sequence analysis of a cdna encoding human ubiquitin reveals that ubiquitin is synthesized as a precursor. J. Biol. Chem. 1985, 260, 7609–7613. [Google Scholar] [PubMed]
- Wiborg, O.; Pedersen, M.S.; Wind, A.; Berglund, L.E.; Marcker, K.A.; Vuust, J. The human ubiquitin multigene family: Some genes contain multiple directly repeated ubiquitin coding sequences. EMBO J. 1985, 4, 755–759. [Google Scholar] [PubMed]
- McGrath, J.P.; Jentsch, S.; Varshavsky, A. Uba 1: An essential yeast gene encoding ubiquitin-activating enzyme. EMBO J. 1991, 10, 227–236. [Google Scholar] [PubMed]
- Pelzer, C.; Kassner, I.; Matentzoglu, K.; Singh, R.K.; Wollscheid, H.P.; Scheffner, M.; Schmidtke, G.; Groettrup, M. Ube1l2, a novel e1 enzyme specific for ubiquitin. J. Biol. Chem. 2007, 282, 23010–23014. [Google Scholar] [CrossRef] [PubMed]
- Desterro, J.M.; Rodriguez, M.S.; Kemp, G.D.; Hay, R.T. Identification of the enzyme required for activation of the small ubiquitin-like protein sumo-1. J. Biol. Chem. 1999, 274, 10618–10624. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Yeh, E.T. Identification of the activating and conjugating enzymes of the nedd8 conjugation pathway. J. Biol. Chem. 1999, 274, 12036–12042. [Google Scholar] [CrossRef] [PubMed]
- Van der Veen, A.G.; Schorpp, K.; Schlieker, C.; Buti, L.; Damon, J.R.; Spooner, E.; Ploegh, H.L.; Jentsch, S. Role of the ubiquitin-like protein urm1 as a noncanonical lysine-directed protein modifier. Proc. Natl. Acad. Sci. USA 2011, 108, 1763–1770. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, M.; Chiba, T.; Tatsumi, K.; Iemura, S.; Tanida, I.; Okazaki, N.; Ueno, T.; Kominami, E.; Natsume, T.; Tanaka, K. A novel protein-conjugating system for ufm1, a ubiquitin-fold modifier. EMBO J. 2004, 23, 1977–1986. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Krug, R.M. Influenza b virus ns1 protein inhibits conjugation of the interferon (ifn)-induced ubiquitin-like isg15 protein. EMBO J. 2001, 20, 362–371. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.T.; Miller, D.W.; Mathew, R.; Cassell, R.; Holton, J.M.; Roussel, M.F.; Schulman, B.A. A unique e1-e2 interaction required for optimal conjugation of the ubiquitin-like protein nedd8. Nat. Struct. Mol. Biol. 2004, 11, 927–935. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Noda, T.; Yoshimori, T.; Tanaka, Y.; Ishii, T.; George, M.D.; Klionsky, D.J.; Ohsumi, M.; Ohsumi, Y. A protein conjugation system essential for autophagy. Nature 1998, 395, 395–398. [Google Scholar] [CrossRef] [PubMed]
- Oweis, W.; Padala, P.; Hassouna, F.; Cohen-Kfir, E.; Gibbs, D.R.; Todd, E.A.; Berndsen, C.E.; Wiener, R. Trans-binding mechanism of ubiquitin-like protein activation revealed by a uba5-ufm1 complex. Cell Rep. 2016, 16, 3113–3120. [Google Scholar] [CrossRef] [PubMed]
- Mashahreh, B.; Hassouna, F.; Soudah, N.; Cohen-Kfir, E.; Strulovich, R.; Haitin, Y.; Wiener, R. Trans-binding of ufm1 to uba5 stimulates uba5 homodimerization and atp binding. FASEB J. 2018, fj201701057R. [Google Scholar] [CrossRef] [PubMed]
- Cook, B.W.; Shaw, G.S. Architecture of the catalytic hpn motif is conserved in all e2 conjugating enzymes. Biochem. J. 2012, 445, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.Y.; Hanlon, M.; Eddins, M.; Tsui, C.; Rogers, R.S.; Jensen, J.P.; Matunis, M.J.; Weissman, A.M.; Wolberger, C.; Pickart, C.M. A conserved catalytic residue in the ubiquitin-conjugating enzyme family. EMBO J. 2003, 22, 5241–5250. [Google Scholar] [CrossRef] [PubMed]
- Morreale, F.E.; Walden, H. Types of ubiquitin ligases. Cell 2016, 165, 248. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Bengtson, M.H.; Ulbrich, A.; Matsuda, A.; Reddy, V.A.; Orth, A.; Chanda, S.K.; Batalov, S.; Joazeiro, C.A. Genome-wide and functional annotation of human e3 ubiquitin ligases identifies mulan, a mitochondrial e3 that regulates the organelle’s dynamics and signaling. PLoS ONE 2008, 3, e1487. [Google Scholar] [CrossRef] [PubMed]
- Huibregtse, J.M.; Scheffner, M.; Beaudenon, S.; Howley, P.M. A family of proteins structurally and functionally related to the e6-ap ubiquitin-protein ligase. Proc. Natl. Acad. Sci. USA 1995, 92, 5249. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.C.; Steffen, A.M.; Oldham, M.L.; Chen, J.; Huibregtse, J.M. Structure and function of a hect domain ubiquitin-binding site. EMBO Rep. 2011, 12, 334–341. [Google Scholar] [CrossRef] [PubMed]
- Hatakeyama, S.; Nakayama, K.I. U-box proteins as a new family of ubiquitin ligases. Biochem. Biophys. Res. Commun. 2003, 302, 635–645. [Google Scholar] [CrossRef]
- Kamadurai, H.B.; Souphron, J.; Scott, D.C.; Duda, D.M.; Miller, D.J.; Stringer, D.; Piper, R.C.; Schulman, B.A. Insights into ubiquitin transfer cascades from a structure of a ubch5b approximately ubiquitin-hect(nedd4l) complex. Mol. Cell 2009, 36, 1095–1102. [Google Scholar] [CrossRef] [PubMed]
- Deshaies, R.J.; Joazeiro, C.A. Ring domain e3 ubiquitin ligases. Ann. Rev. Biochem. 2009, 78, 399–434. [Google Scholar] [CrossRef] [PubMed]
- Pruneda, J.N.; Littlefield, P.J.; Soss, S.E.; Nordquist, K.A.; Chazin, W.J.; Brzovic, P.S.; Klevit, R.E. Structure of an e3:E2~ub complex reveals an allosteric mechanism shared among ring/u-box ligases. Mol. Cell 2012, 47, 933–942. [Google Scholar] [CrossRef] [PubMed]
- Kirisako, T.; Kamei, K.; Murata, S.; Kato, M.; Fukumoto, H.; Kanie, M.; Sano, S.; Tokunaga, F.; Tanaka, K.; Iwai, K. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 2006, 25, 4877–4887. [Google Scholar] [CrossRef] [PubMed]
- Guzzo, C.M.; Berndsen, C.E.; Zhu, J.; Gupta, V.; Datta, A.; Greenberg, R.A.; Wolberger, C.; Matunis, M.J. Rnf4-dependent hybrid sumo-ubiquitin chains are signals for rap80 and thereby mediate the recruitment of brca1 to sites of DNA damage. Sci. Signal. 2012, 5, ra88. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, C.K.; Galanty, Y.; Sczaniecka-Clift, M.; Coates, J.; Jhujh, S.; Demir, M.; Cornwell, M.; Beli, P.; Jackson, S.P. Systematic e2 screening reveals a ube2d-rnf138-ctip axis promoting DNA repair. Nat. Cell Biol. 2015, 17, 1458–1470. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Liang, J.; Lu, Y.; Xu, L.; Miao, S.; Lu, L.Y.; Song, W.; Wang, L. Ubiquitylation of rad51d mediated by e3 ligase rnf138 promotes the homologous recombination repair pathway. PLoS ONE 2016, 11, e0155476. [Google Scholar] [CrossRef] [PubMed]
- Uckelmann, M.; Sixma, T.K. Histone ubiquitination in the DNA damage response. DNA Repair 2017, 56, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Wijnhoven, P.; Konietzny, R.; Blackford, A.N.; Travers, J.; Kessler, B.M.; Nishi, R.; Jackson, S.P. Usp4 auto-deubiquitylation promotes homologous recombination. Mol. Cell 2015, 60, 362–373. [Google Scholar] [CrossRef] [PubMed]
- Beli, P.; Jackson, S.P. Ubiquitin regulates dissociation of DNA repair factors from chromatin. Oncotarget 2015, 6, 14727–14728. [Google Scholar] [CrossRef] [PubMed]
- Baranes-Bachar, K.; Levy-Barda, A.; Oehler, J.; Reid, D.A.; Soria-Bretones, I.; Voss, T.C.; Chung, D.; Park, Y.; Liu, C.; Yoon, J.B.; et al. The ubiquitin e3/e4 ligase ube4a adjusts protein ubiquitylation and accumulation at sites of DNA damage, facilitating double-strand break repair. Mol. Cell 2018, 69, 866–878. [Google Scholar] [CrossRef] [PubMed]
- Elia, A.E.; Boardman, A.P.; Wang, D.C.; Huttlin, E.L.; Everley, R.A.; Dephoure, N.; Zhou, C.; Koren, I.; Gygi, S.P.; Elledge, S.J. Quantitative proteomic atlas of ubiquitination and acetylation in the DNA damage response. Mol. Cell 2015, 59, 867–881. [Google Scholar] [CrossRef] [PubMed]
- Gatti, M.; Pinato, S.; Maiolica, A.; Rocchio, F.; Prato, M.G.; Aebersold, R.; Penengo, L. Rnf168 promotes noncanonical k27 ubiquitination to signal DNA damage. Cell Rep. 2015, 10, 226–238. [Google Scholar] [CrossRef] [PubMed]
- Meyer, H.J.; Rape, M. Enhanced protein degradation by branched ubiquitin chains. Cell 2014, 157, 910–921. [Google Scholar] [CrossRef] [PubMed]
- Fei, C.; Li, Z.; Li, C.; Chen, Y.; Chen, Z.; He, X.; Mao, L.; Wang, X.; Zeng, R.; Li, L. Smurf1-mediated lys29-linked nonproteolytic polyubiquitination of axin negatively regulates wnt/beta-catenin signaling. Mol. Cell. Biol. 2013, 33, 4095–4105. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Lee, B.H.; King, R.W.; Finley, D.; Kirschner, M.W. Substrate degradation by the proteasome: A single-molecule kinetic analysis. Science 2015, 348, 1250834. [Google Scholar] [CrossRef] [PubMed]
- Nathan, J.A.; Kim, H.T.; Ting, L.; Gygi, S.P.; Goldberg, A.L. Why do cellular proteins linked to k63-polyubiquitin chains not associate with proteasomes? EMBO J. 2013, 32, 552–565. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.S.; Yang-Yen, H.F.; Suen, C.S.; Hwang, M.J.; Yen, J.J. Cbl-mediated k63-linked ubiquitination of jak2 enhances jak2 phosphorylation and signal transduction. Sci. Rep. 2017, 7, 4613. [Google Scholar] [CrossRef] [PubMed]
- Ohtake, F.; Saeki, Y.; Ishido, S.; Kanno, J.; Tanaka, K. The k48-k63 branched ubiquitin chain regulates nf-kappab signaling. Mol. Cell 2016, 64, 251–266. [Google Scholar] [CrossRef] [PubMed]
- Ohtake, F.; Tsuchiya, H.; Saeki, Y.; Tanaka, K. K63 ubiquitylation triggers proteasomal degradation by seeding branched ubiquitin chains. Proc. Natl. Acad. Sci. USA 2018, 115, E1401–E1408. [Google Scholar] [CrossRef] [PubMed]
- Fiil, B.K.; Damgaard, R.B.; Wagner, S.A.; Keusekotten, K.; Fritsch, M.; Bekker-Jensen, S.; Mailand, N.; Choudhary, C.; Komander, D.; Gyrd-Hansen, M. Otulin restricts met1-linked ubiquitination to control innate immune signaling. Mol. Cell 2013, 50, 818–830. [Google Scholar] [CrossRef] [PubMed]
- Dziedzic, S.A.; Su, Z.; Jean Barrett, V.; Najafov, A.; Mookhtiar, A.K.; Amin, P.; Pan, H.; Sun, L.; Zhu, H.; Ma, A.; et al. Abin-1 regulates ripk1 activation by linking met1 ubiquitylation with lys63 deubiquitylation in tnf-rsc. Nat. Cell Biol. 2018, 20, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Alkan, Z.; Duong, F.L.; Hawkes, W.C. Selenoprotein w controls epidermal growth factor receptor surface expression, activation and degradation via receptor ubiquitination. Biochim. Biophys. Acta 2015, 1853, 1087–1095. [Google Scholar] [CrossRef] [PubMed]
- Haugsten, E.M.; Zakrzewska, M.; Brech, A.; Pust, S.; Olsnes, S.; Sandvig, K.; Wesche, J. Clathrin- and dynamin-independent endocytosis of fgfr3—Implications for signaling. PLoS ONE 2011, 6, e21708. [Google Scholar] [CrossRef] [PubMed]
- Lanahan, A.A.; Hermans, K.; Claes, F.; Kerley-Hamilton, J.S.; Zhuang, Z.W.; Giordano, F.J.; Carmeliet, P.; Simons, M. Vegf receptor 2 endocytic trafficking regulates arterial morphogenesis. Dev. Cell 2010, 18, 713–724. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Je, H.S.; Young, P.; Gross, J.; Lu, B.; Feng, G. Regulation of synaptic growth and maturation by a synapse-associated e3 ubiquitin ligase at the neuromuscular junction. J. Cell Biol. 2007, 177, 1077–1089. [Google Scholar] [CrossRef] [PubMed]
- Arevalo, J.C.; Waite, J.; Rajagopal, R.; Beyna, M.; Chen, Z.Y.; Lee, F.S.; Chao, M.V. Cell survival through trk neurotrophin receptors is differentially regulated by ubiquitination. Neuron 2006, 50, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Geetha, T.; Jiang, J.; Wooten, M.W. Lysine 63 polyubiquitination of the nerve growth factor receptor trka directs internalization and signaling. Mol. Cell 2005, 20, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Geetha, T.; Wooten, M.W. Trka receptor endolysosomal degradation is both ubiquitin and proteasome dependent. Traffic 2008, 9, 1146–1156. [Google Scholar] [CrossRef] [PubMed]
- Haglund, K.; Sigismund, S.; Polo, S.; Szymkiewicz, I.; Di Fiore, P.P.; Dikic, I. Multiple monoubiquitination of rtks is sufficient for their endocytosis and degradation. Nat. Cell Biol. 2003, 5, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Zeng, X.; Kim, W.; Balasubramani, M.; Fortian, A.; Gygi, S.P.; Yates, N.A.; Sorkin, A. Lysine 63-linked polyubiquitination is required for egf receptor degradation. Proc. Natl. Acad. Sci. USA 2013, 110, 15722–15727. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, K.; Falquet, L. A ubiquitin-interacting motif conserved in components of the proteasomal and lysosomal protein degradation systems. Trends Biochem. Sci. 2001, 26, 347–350. [Google Scholar] [CrossRef]
- Polo, S.; Sigismund, S.; Faretta, M.; Guidi, M.; Capua, M.R.; Bossi, G.; Chen, H.; De Camilli, P.; Di Fiore, P.P. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature 2002, 416, 451–455. [Google Scholar] [CrossRef] [PubMed]
- Barriere, H.; Nemes, C.; Lechardeur, D.; Khan-Mohammad, M.; Fruh, K.; Lukacs, G.L. Molecular basis of oligoubiquitin-dependent internalization of membrane proteins in mammalian cells. Traffic 2006, 7, 282–297. [Google Scholar] [CrossRef] [PubMed]
- Baldys, A.; Raymond, J.R. Critical role of escrt machinery in egfr recycling. Biochemistry 2009, 48, 9321–9323. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Letourneau, D.; Holleran, B.; Leduc, R.; Lavigne, P.; Lavoie, C. Galphas protein binds ubiquitin to regulate epidermal growth factor receptor endosomal sorting. Proc. Natl. Acad. Sci. USA 2017, 114, 13477–13482. [Google Scholar] [CrossRef] [PubMed]
- Deshar, R.; Cho, E.B.; Yoon, S.K.; Yoon, J.B. Cc2d1a and cc2d1b regulate degradation and signaling of egfr and tlr4. Biochem. Biophys. Res. Commun. 2016, 480, 280–287. [Google Scholar] [CrossRef] [PubMed]
- Soubeyran, P.; Kowanetz, K.; Szymkiewicz, I.; Langdon, W.Y.; Dikic, I. Cbl-cin85-endophilin complex mediates ligand-induced downregulation of egf receptors. Nature 2002, 416, 183–187. [Google Scholar] [CrossRef] [PubMed]
- Petrelli, A.; Gilestro, G.F.; Lanzardo, S.; Comoglio, P.M.; Migone, N.; Giordano, S. The endophilin-cin85-cbl complex mediates ligand-dependent downregulation of c-met. Nature 2002, 416, 187–190. [Google Scholar] [CrossRef] [PubMed]
- Diesenberg, K.; Beerbaum, M.; Fink, U.; Schmieder, P.; Krauss, M. Sept9 negatively regulates ubiquitin-dependent downregulation of egfr. J. Cell Sci. 2015, 128, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Umebayashi, K.; Stenmark, H.; Yoshimori, T. Ubc4/5 and c-cbl continue to ubiquitinate egf receptor after internalization to facilitate polyubiquitination and degradation. Mol. Biol. Cell 2008, 19, 3454–3462. [Google Scholar] [CrossRef] [PubMed]
- Malerod, L.; Stuffers, S.; Brech, A.; Stenmark, H. Vps22/eap30 in escrt-ii mediates endosomal sorting of growth factor and chemokine receptors destined for lysosomal degradation. Traffic 2007, 8, 1617–1629. [Google Scholar] [CrossRef] [PubMed]
- Longva, K.E.; Blystad, F.D.; Stang, E.; Larsen, A.M.; Johannessen, L.E.; Madshus, I.H. Ubiquitination and proteasomal activity is required for transport of the egf receptor to inner membranes of multivesicular bodies. J. Cell Biol. 2002, 156, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Chi, S.; Cao, H.; Wang, Y.; McNiven, M.A. Recycling of the epidermal growth factor receptor is mediated by a novel form of the clathrin adaptor protein eps15. J. Biol. Chem. 2011, 286, 35196–35208. [Google Scholar] [CrossRef] [PubMed]
- Jopling, H.M.; Odell, A.F.; Pellet-Many, C.; Latham, A.M.; Frankel, P.; Sivaprasadarao, A.; Walker, J.H.; Zachary, I.C.; Ponnambalam, S. Endosome-to-plasma membrane recycling of vegfr2 receptor tyrosine kinase regulates endothelial function and blood vessel formation. Cells 2014, 3, 363–385. [Google Scholar] [CrossRef] [PubMed]
- Hellberg, C.; Schmees, C.; Karlsson, S.; Ahgren, A.; Heldin, C.H. Activation of protein kinase c alpha is necessary for sorting the pdgf beta-receptor to rab4a-dependent recycling. Mol. Biol. Cell 2009, 20, 2856–2863. [Google Scholar] [CrossRef] [PubMed]
- Romanelli, R.J.; LeBeau, A.P.; Fulmer, C.G.; Lazzarino, D.A.; Hochberg, A.; Wood, T.L. Insulin-like growth factor type-i receptor internalization and recycling mediate the sustained phosphorylation of akt. J. Biol. Chem. 2007, 282, 22513–22524. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Lavigne, P.; Lavoie, C. Gga3 mediates trka endocytic recycling to promote sustained akt phosphorylation and cell survival. Mol. Biol. Cell 2015, 26, 4412–4426. [Google Scholar] [CrossRef] [PubMed]
- Luiskandl, S.; Woller, B.; Schlauf, M.; Schmid, J.A.; Herbst, R. Endosomal trafficking of the receptor tyrosine kinase musk proceeds via clathrin-dependent pathways, arf6 and actin. FEBS J. 2013, 280, 3281–3297. [Google Scholar] [CrossRef] [PubMed]
- Mihai, C.; Chotani, M.; Elton, T.S.; Agarwal, G. Mapping of ddr1 distribution and oligomerization on the cell surface by fret microscopy. J. Mol. Biol. 2009, 385, 432–445. [Google Scholar] [CrossRef] [PubMed]
- Haugsten, E.M.; Malecki, J.; Bjorklund, S.M.; Olsnes, S.; Wesche, J. Ubiquitination of fibroblast growth factor receptor 1 is required for its intracellular sorting but not for its endocytosis. Mol. Biol. Cell 2008, 19, 3390–3403. [Google Scholar] [CrossRef] [PubMed]
- Decker, S.J. Epidermal growth factor and transforming growth factor-alpha induce differential processing of the epidermal growth factor receptor. Biochem. Biophys. Res. Commun. 1990, 166, 615–621. [Google Scholar] [CrossRef]
- Stern, K.A.; Place, T.L.; Lill, N.L. Egf and amphiregulin differentially regulate cbl recruitment to endosomes and egf receptor fate. Biochem. J. 2008, 410, 585–594. [Google Scholar] [CrossRef] [PubMed]
- Baldys, A.; Gooz, M.; Morinelli, T.A.; Lee, M.H.; Raymond, J.R., Jr.; Luttrell, L.M.; Raymond, J.R., Sr. Essential role of c-cbl in amphiregulin-induced recycling and signaling of the endogenous epidermal growth factor receptor. Biochemistry 2009, 48, 1462–1473. [Google Scholar] [CrossRef] [PubMed]
- Roepstorff, K.; Grandal, M.V.; Henriksen, L.; Knudsen, S.L.; Lerdrup, M.; Grovdal, L.; Willumsen, B.M.; van Deurs, B. Differential effects of egfr ligands on endocytic sorting of the receptor. Traffic 2009, 10, 1115–1127. [Google Scholar] [CrossRef] [PubMed]
- Sehat, B.; Tofigh, A.; Lin, Y.; Trocme, E.; Liljedahl, U.; Lagergren, J.; Larsson, O. Sumoylation mediates the nuclear translocation and signaling of the igf-1 receptor. Sci. Signal. 2010, 3, ra10. [Google Scholar] [CrossRef] [PubMed]
- Knittle, A.M.; Helkkula, M.; Johnson, M.S.; Sundvall, M.; Elenius, K. Sumoylation regulates nuclear accumulation and signaling activity of the soluble intracellular domain of the erbb4 receptor tyrosine kinase. J. Biol. Chem. 2017, 292, 19890–19904. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Lin, Y.; Badin, M.; Vasilcanu, D.; Stromberg, T.; Jernberg-Wiklund, H.; Sehat, B.; Larsson, O. Over-accumulation of nuclear igf-1 receptor in tumor cells requires elevated expression of the receptor and the sumo-conjugating enzyme ubc9. Biochem. Biophys. Res. Commun. 2011, 404, 667–671. [Google Scholar] [CrossRef] [PubMed]
- Visser Smit, G.D.; Place, T.L.; Cole, S.L.; Clausen, K.A.; Vemuganti, S.; Zhang, G.; Koland, J.G.; Lill, N.L. Cbl controls egfr fate by regulating early endosome fusion. Sci. Signal. 2009, 2, ra86. [Google Scholar] [CrossRef] [PubMed]
- Ravid, T.; Heidinger, J.M.; Gee, P.; Khan, E.M.; Goldkorn, T. C-cbl-mediated ubiquitinylation is required for epidermal growth factor receptor exit from the early endosomes. J. Biol. Chem. 2004, 279, 37153–37162. [Google Scholar] [CrossRef] [PubMed]
- Severe, N.; Miraoui, H.; Marie, P.J. The casitas b lineage lymphoma (cbl) mutant g306e enhances osteogenic differentiation in human mesenchymal stromal cells in part by decreased cbl-mediated platelet-derived growth factor receptor alpha and fibroblast growth factor receptor 2 ubiquitination. J. Biol. Chem. 2011, 286, 24443–24450. [Google Scholar] [CrossRef] [PubMed]
- Reddi, A.L.; Ying, G.; Duan, L.; Chen, G.; Dimri, M.; Douillard, P.; Druker, B.J.; Naramura, M.; Band, V.; Band, H. Binding of cbl to a phospholipase cgamma1-docking site on platelet-derived growth factor receptor beta provides a dual mechanism of negative regulation. J. Biol. Chem. 2007, 282, 29336–29347. [Google Scholar] [CrossRef] [PubMed]
- Rorsman, C.; Tsioumpekou, M.; Heldin, C.H.; Lennartsson, J. The ubiquitin ligases c-cbl and cbl-b negatively regulate platelet-derived growth factor (pdgf) bb-induced chemotaxis by affecting pdgf receptor beta (pdgfrbeta) internalization and signaling. J. Biol. Chem. 2016, 291, 11608–11618. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Shimokawa, N.; Esmaeili-Mahani, S.; Morita, A.; Masuda, H.; Iwasaki, T.; Tamura, J.; Haglund, K.; Koibuchi, N. Ligand-induced downregulation of trka is partly regulated through ubiquitination by cbl. FEBS Lett. 2011, 585, 1741–1747. [Google Scholar] [CrossRef] [PubMed]
- Hyndman, B.D.; Crupi, M.J.F.; Peng, S.; Bone, L.N.; Rekab, A.N.; Lian, E.Y.; Wagner, S.M.; Antonescu, C.N.; Mulligan, L.M. Differential recruitment of e3 ubiquitin ligase complexes regulates ret isoform internalization. J. Cell Sci. 2017, 130, 3282–3296. [Google Scholar] [CrossRef] [PubMed]
- Sharfe, N.; Freywald, A.; Toro, A.; Roifman, C.M. Ephrin-a1 induces c-cbl phosphorylation and epha receptor down-regulation in t cells. J. Immunol. 2003, 170, 6024–6032. [Google Scholar] [CrossRef] [PubMed]
- Fasen, K.; Cerretti, D.P.; Huynh-Do, U. Ligand binding induces cbl-dependent ephb1 receptor degradation through the lysosomal pathway. Traffic 2008, 9, 251–266. [Google Scholar] [CrossRef] [PubMed]
- Duval, M.; Bedard-Goulet, S.; Delisle, C.; Gratton, J.P. Vascular endothelial growth factor-dependent down-regulation of flk-1/kdr involves cbl-mediated ubiquitination. Consequences on nitric oxide production from endothelial cells. J. Biol. Chem. 2003, 278, 20091–20097. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S.; Sawano, A.; Nojima, Y.; Shibuya, M.; Maru, Y. The c-cbl/cd2ap complex regulates vegf-induced endocytosis and degradation of flt-1 (vegfr-1). FASEB J. 2004, 18, 929–931. [Google Scholar] [CrossRef] [PubMed]
- Parks, E.E.; Ceresa, B.P. Cell surface epidermal growth factor receptors increase src and c-cbl activity and receptor ubiquitylation. J. Biol. Chem. 2014, 289, 25537–25545. [Google Scholar] [CrossRef] [PubMed]
- Emdal, K.B.; Pedersen, A.K.; Bekker-Jensen, D.B.; Tsafou, K.P.; Horn, H.; Lindner, S.; Schulte, J.H.; Eggert, A.; Jensen, L.J.; Francavilla, C.; et al. Temporal proteomics of ngf-trka signaling identifies an inhibitory role for the e3 ligase cbl-b in neuroblastoma cell differentiation. Sci. Signal. 2015, 8, ra40. [Google Scholar] [CrossRef] [PubMed]
- Sirisaengtaksin, N.; Gireud, M.; Yan, Q.; Kubota, Y.; Meza, D.; Waymire, J.C.; Zage, P.E.; Bean, A.J. Ube4b protein couples ubiquitination and sorting machineries to enable epidermal growth factor receptor (egfr) degradation. J. Biol. Chem. 2014, 289, 3026–3039. [Google Scholar] [CrossRef] [PubMed]
- Gschweitl, M.; Ulbricht, A.; Barnes, C.A.; Enchev, R.I.; Stoffel-Studer, I.; Meyer-Schaller, N.; Huotari, J.; Yamauchi, Y.; Greber, U.F.; Helenius, A.; et al. A spopl/cullin-3 ubiquitin ligase complex regulates endocytic trafficking by targeting eps15 at endosomes. eLife 2016, 5, e13841. [Google Scholar] [CrossRef] [PubMed]
- Song, R.; Peng, W.; Zhang, Y.; Lv, F.; Wu, H.K.; Guo, J.; Cao, Y.; Pi, Y.; Zhang, X.; Jin, L.; et al. Central role of e3 ubiquitin ligase mg53 in insulin resistance and metabolic disorders. Nature 2013, 494, 375–379. [Google Scholar] [CrossRef] [PubMed]
- Nagarajan, A.; Petersen, M.C.; Nasiri, A.R.; Butrico, G.; Fung, A.; Ruan, H.B.; Kursawe, R.; Caprio, S.; Thibodeau, J.; Bourgeois-Daigneault, M.C.; et al. March1 regulates insulin sensitivity by controlling cell surface insulin receptor levels. Nat. Commun. 2016, 7, 12639. [Google Scholar] [CrossRef] [PubMed]
- Sehat, B.; Andersson, S.; Girnita, L.; Larsson, O. Identification of c-cbl as a new ligase for insulin-like growth factor-i receptor with distinct roles from mdm2 in receptor ubiquitination and endocytosis. Cancer Res. 2008, 68, 5669–5677. [Google Scholar] [CrossRef] [PubMed]
- Vecchione, A.; Marchese, A.; Henry, P.; Rotin, D.; Morrione, A. The grb10/nedd4 complex regulates ligand-induced ubiquitination and stability of the insulin-like growth factor i receptor. Mol. Cell. Biol. 2003, 23, 3363–3372. [Google Scholar] [CrossRef] [PubMed]
- Girnita, L.; Girnita, A.; Larsson, O. Mdm2-dependent ubiquitination and degradation of the insulin-like growth factor 1 receptor. Proc. Natl. Acad. Sci. USA 2003, 100, 8247–8252. [Google Scholar] [CrossRef] [PubMed]
- Monami, G.; Emiliozzi, V.; Morrione, A. Grb10/nedd4-mediated multiubiquitination of the insulin-like growth factor receptor regulates receptor internalization. J. Cell. Physiol. 2008, 216, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Murdaca, J.; Treins, C.; Monthouel-Kartmann, M.N.; Pontier-Bres, R.; Kumar, S.; Van Obberghen, E.; Giorgetti-Peraldi, S. Grb10 prevents nedd4-mediated vascular endothelial growth factor receptor-2 degradation. J. Biol. Chem. 2004, 279, 26754–26761. [Google Scholar] [CrossRef] [PubMed]
- Hasseine, L.K.; Murdaca, J.; Suavet, F.; Longnus, S.; Giorgetti-Peraldi, S.; Van Obberghen, E. Hrs is a positive regulator of vegf and insulin signaling. Exp. Cell Res. 2007, 313, 1927–1942. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Scheel, H.; Hofmann, K.; Komander, D. Dissection of usp catalytic domains reveals five common insertion points. Mol. Biosyst. 2009, 5, 1797–1808. [Google Scholar] [CrossRef] [PubMed]
- Mevissen, T.E.; Hospenthal, M.K.; Geurink, P.P.; Elliott, P.R.; Akutsu, M.; Arnaudo, N.; Ekkebus, R.; Kulathu, Y.; Wauer, T.; El Oualid, F.; et al. Otu deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell 2013, 154, 169–184. [Google Scholar] [CrossRef] [PubMed]
- Seki, T.; Gong, L.; Williams, A.J.; Sakai, N.; Todi, S.V.; Paulson, H.L. Josd1, a membrane-targeted deubiquitinating enzyme, is activated by ubiquitination and regulates membrane dynamics, cell motility, and endocytosis. J. Biol. Chem. 2013, 288, 17145–17155. [Google Scholar] [CrossRef] [PubMed]
- Abdul Rehman, S.A.; Kristariyanto, Y.A.; Choi, S.Y.; Nkosi, P.J.; Weidlich, S.; Labib, K.; Hofmann, K.; Kulathu, Y. Mindy-1 is a member of an evolutionarily conserved and structurally distinct new family of deubiquitinating enzymes. Mol. Cell 2016, 63, 146–155. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, N.; Niu, J.; Saad, Y.; Kumar, S.; Sirakova, T.; Becerra, E.; Li, X.; Kolattukudy, P.E. Transcription factors stat6 and klf4 implement macrophage polarization via the dual catalytic powers of mcpip. J. Immunol. 2015, 194, 6011–6023. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Saad, Y.; Lei, T.; Wang, J.; Qi, D.; Yang, Q.; Kolattukudy, P.E.; Fu, M. Mcp-induced protein 1 deubiquitinates traf proteins and negatively regulates jnk and nf-kappab signaling. J. Exp. Med. 2010, 207, 2959–2973. [Google Scholar] [CrossRef] [PubMed]
- Komander, D.; Clague, M.J.; Urbe, S. Breaking the chains: Structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009, 10, 550–563. [Google Scholar] [CrossRef] [PubMed]
- Grou, C.P.; Pinto, M.P.; Mendes, A.V.; Domingues, P.; Azevedo, J.E. The de novo synthesis of ubiquitin: Identification of deubiquitinases acting on ubiquitin precursors. Sci. Rep. 2015, 5, 12836. [Google Scholar] [CrossRef] [PubMed]
- Komander, D.; Lord, C.J.; Scheel, H.; Swift, S.; Hofmann, K.; Ashworth, A.; Barford, D. The structure of the cyld usp domain explains its specificity for lys63-linked polyubiquitin and reveals a b box module. Mol. Cell 2008, 29, 451–464. [Google Scholar] [CrossRef] [PubMed]
- Kristariyanto, Y.A.; Abdul Rehman, S.A.; Weidlich, S.; Knebel, A.; Kulathu, Y. A single miu motif of mindy-1 recognizes k48-linked polyubiquitin chains. EMBO Rep. 2017, 18, 392–402. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Okatsu, K.; Saeki, Y.; Yamano, K.; Matsuda, N.; Kaiho, A.; Yamagata, A.; Goto-Ito, S.; Ishikawa, M.; Hashimoto, Y.; et al. Structural basis for specific cleavage of lys6-linked polyubiquitin chains by usp30. Nat. Struct. Mol. Biol. 2017, 24, 911–919. [Google Scholar] [CrossRef] [PubMed]
- Peth, A.; Uchiki, T.; Goldberg, A.L. Atp-dependent steps in the binding of ubiquitin conjugates to the 26s proteasome that commit to degradation. Mol. Cell 2010, 40, 671–681. [Google Scholar] [CrossRef] [PubMed]
- Harrigan, J.A.; Jacq, X.; Martin, N.M.; Jackson, S.P. Deubiquitylating enzymes and drug discovery: Emerging opportunities. Nat. Rev. Drug Discov. 2018, 17, 57–77. [Google Scholar] [CrossRef] [PubMed]
- Duex, J.E.; Sorkin, A. Rna interference screen identifies usp18 as a regulator of epidermal growth factor receptor synthesis. Mol. Biol. Cell 2009, 20, 1833–1844. [Google Scholar] [CrossRef] [PubMed]
- Duex, J.E.; Comeau, L.; Sorkin, A.; Purow, B.; Kefas, B. Usp18 regulates epidermal growth factor (egf) receptor expression and cancer cell survival via microrna-7. J. Biol. Chem. 2011, 286, 25377–25386. [Google Scholar] [CrossRef] [PubMed]
- McCullough, J.; Clague, M.J.; Urbe, S. Amsh is an endosome-associated ubiquitin isopeptidase. J. Cell Biol. 2004, 166, 487–492. [Google Scholar] [CrossRef] [PubMed]
- McCullough, J.; Row, P.E.; Lorenzo, O.; Doherty, M.; Beynon, R.; Clague, M.J.; Urbe, S. Activation of the endosome-associated ubiquitin isopeptidase amsh by stam, a component of the multivesicular body-sorting machinery. Curr. Biol. CB 2006, 16, 160–165. [Google Scholar] [CrossRef] [PubMed]
- Savio, M.G.; Wollscheid, N.; Cavallaro, E.; Algisi, V.; Di Fiore, P.P.; Sigismund, S.; Maspero, E.; Polo, S. Usp9x controls egfr fate by deubiquitinating the endocytic adaptor eps15. Curr. Biol. CB 2016, 26, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.Y.; Lu, Y.; Zheng, Y.; Chen, X.R.; Dong, J.L.; Yuan, R.R.; Huang, S.H.; Yu, H.; Wang, Y.; Chen, Z.Y.; et al. Ubiquitin c-terminal hydrolase l1 (uch-l1) promotes hippocampus-dependent memory via its deubiquitinating effect on trkb. J. Neurosci. 2017, 37, 5978–5995. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, E.; Kobayashi, K.; Yamamoto, A.; Kitamura, N.; Komada, M. A deubiquitinating enzyme ubpy regulates the level of protein ubiquitination on endosomes. Traffic 2006, 7, 1017–1031. [Google Scholar] [CrossRef] [PubMed]
- Niendorf, S.; Oksche, A.; Kisser, A.; Lohler, J.; Prinz, M.; Schorle, H.; Feller, S.; Lewitzky, M.; Horak, I.; Knobeloch, K.P. Essential role of ubiquitin-specific protease 8 for receptor tyrosine kinase stability and endocytic trafficking in vivo. Mol. Cell. Biol. 2007, 27, 5029–5039. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.M.; Lee, S.B.; Choi, J.; Suh, H.Y.; Shim, S.; Song, Y.J.; Kim, B.; Lee, J.M.; Oh, S.J.; Jeong, Y.; et al. Usp8 modulates ubiquitination of lrig1 for met degradation. Sci. Rep. 2014, 4, 4980. [Google Scholar] [CrossRef] [PubMed]
- Berlin, I.; Schwartz, H.; Nash, P.D. Regulation of epidermal growth factor receptor ubiquitination and trafficking by the usp8.Stam complex. J. Biol. Chem. 2010, 285, 34909–34921. [Google Scholar] [CrossRef] [PubMed]
- Reincke, M.; Sbiera, S.; Hayakawa, A.; Theodoropoulou, M.; Osswald, A.; Beuschlein, F.; Meitinger, T.; Mizuno-Yamasaki, E.; Kawaguchi, K.; Saeki, Y.; et al. Mutations in the deubiquitinase gene usp8 cause cushing's disease. Nat. Genet. 2015, 47, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.A.; Fearnley, G.W.; Abdul-Zani, I.; Wheatcroft, S.B.; Tomlinson, D.C.; Harrison, M.A.; Ponnambalam, S. Vegfr2 trafficking, signaling and proteolysis is regulated by the ubiquitin isopeptidase usp8. Traffic 2016, 17, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Jian, F.; Cao, Y.; Bian, L.; Sun, Q. Usp8: A novel therapeutic target for cushing's disease. Endocrine 2015, 50, 292–296. [Google Scholar] [CrossRef] [PubMed]
- Byun, S.; Lee, S.Y.; Lee, J.; Jeong, C.H.; Farrand, L.; Lim, S.; Reddy, K.; Kim, J.Y.; Lee, M.H.; Lee, H.J.; et al. Usp8 is a novel target for overcoming gefitinib resistance in lung cancer. Clin. Cancer Res. 2013, 19, 3894–3904. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Critchley, W.R.; Pellet-Many, C.; Ringham-Terry, B.; Harrison, M.A.; Zachary, I.C.; Ponnambalam, S. Receptor Tyrosine Kinase Ubiquitination and De-Ubiquitination in Signal Transduction and Receptor Trafficking. Cells 2018, 7, 22. https://doi.org/10.3390/cells7030022
Critchley WR, Pellet-Many C, Ringham-Terry B, Harrison MA, Zachary IC, Ponnambalam S. Receptor Tyrosine Kinase Ubiquitination and De-Ubiquitination in Signal Transduction and Receptor Trafficking. Cells. 2018; 7(3):22. https://doi.org/10.3390/cells7030022
Chicago/Turabian StyleCritchley, William R., Caroline Pellet-Many, Benjamin Ringham-Terry, Michael A. Harrison, Ian C. Zachary, and Sreenivasan Ponnambalam. 2018. "Receptor Tyrosine Kinase Ubiquitination and De-Ubiquitination in Signal Transduction and Receptor Trafficking" Cells 7, no. 3: 22. https://doi.org/10.3390/cells7030022
APA StyleCritchley, W. R., Pellet-Many, C., Ringham-Terry, B., Harrison, M. A., Zachary, I. C., & Ponnambalam, S. (2018). Receptor Tyrosine Kinase Ubiquitination and De-Ubiquitination in Signal Transduction and Receptor Trafficking. Cells, 7(3), 22. https://doi.org/10.3390/cells7030022





