Control of DNA Replication Initiation by Ubiquitin
Abstract
1. Introduction
2. Ubiquitin Modification
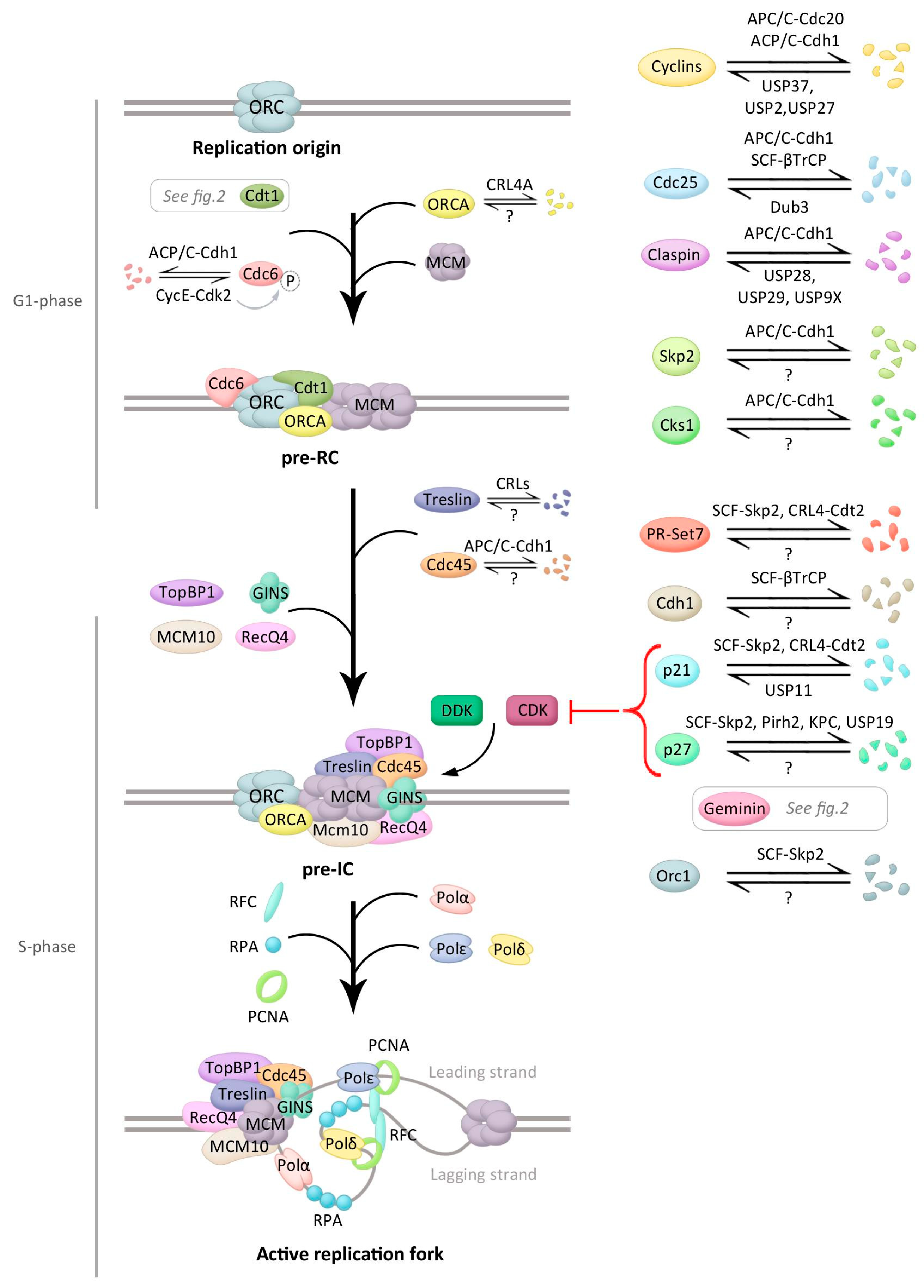
3. Initiation of DNA Replication
4. Ubiquitination Control during Initiation of DNA Replication
5. Cdt1 and Geminin Control to Avoid Re-Replication
6. Pathological Consequences of Dysregulation of DNA Replication
Author Contributions
Funding
Conflicts of Interest
References
- DePamphilis, M.L.; Blow, J.J.; Ghosh, S.; Saha, T.; Noguchi, K.; Vassilev, A. Regulating the licensing of DNA replication origins in metazoa. Curr. Opin. Cell Biol. 2006, 18, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef] [PubMed]
- Komander, D. The emerging complexity of protein ubiquitination. Biochem. Soc. Trans. 2009, 37, 937–953. [Google Scholar] [CrossRef] [PubMed]
- Komander, D.; Rape, M. The ubiquitin code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef] [PubMed]
- Deshaies, R.J.; Joazeiro, C.A.P. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009, 78, 399–434. [Google Scholar] [CrossRef] [PubMed]
- Michelle, C.; Vourc’h, P.; Mignon, L.; Andres, C.R. What was the set of ubiquitin and ubiquitin-like conjugating enzymes in the eukaryote common ancestor? J. Mol. Evol. 2009, 68, 616–628. [Google Scholar] [CrossRef] [PubMed]
- Rizzardi, L.F.; Cook, J.G. Flipping the switch from g1 to s phase with e3 ubiquitin ligases. Genes Cancer 2012, 3, 634–648. [Google Scholar] [CrossRef] [PubMed]
- Gilberto, S.; Peter, M. Dynamic ubiquitin signaling in cell cycle regulation. J. Cell Biol. 2017, 216, 2259–2271. [Google Scholar] [CrossRef] [PubMed]
- Yu, H. Cdc20: A WD40 activator for a cell cycle degradation machine. Mol. Cell 2007, 27, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Skaar, J.R.; Pagano, M. Cdh1: A master G0/G1 regulator. Nat. Cell Biol. 2008, 10, 755–757. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Turcu, F.E.; Ventii, K.H.; Wilkinson, K.D. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 2009, 78, 363–397. [Google Scholar] [CrossRef] [PubMed]
- Smits, V.A.J.; Freire, R. USP7/HAUSP: A SUMO deubiquitinase at the heart of DNA replication. Bioessays 2016, 38, 863–868. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Chen, D.; Shiloh, A.; Luo, J.; Nikolaev, A.Y.; Qin, J.; Gu, W. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature 2002, 416, 648–653. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Gu, L.; Li, M.; Jeffrey, P.D.; Gu, W.; Shi, Y. Structural basis of competitive recognition of p53 and MDM2 by HAUSP/USP7: Implications for the regulation of the p53-MDM2 pathway. PLoS Biol. 2006, 4, 228–239. [Google Scholar] [CrossRef] [PubMed]
- Lecona, E.; Rodriguez-Acebes, S.; Specks, J.; Lopez-Contreras, A.J.; Ruppen, I.; Murga, M.; Muñoz, J.; Méndez, J.; Fernandez-Capetillo, O. USP7 is a SUMO deubiquitinase essential for DNA replication. Nat. Struct. Mol. Biol. 2016, 23, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Méchali, M. Eukaryotic DNA replication origins: Many choices for appropriate answers. Nat. Rev. Mol. Cell Biol. 2010, 11, 728–738. [Google Scholar] [CrossRef] [PubMed]
- Cayrou, C.; Coulombe, P.; Vigneron, A.; Stanojcic, S.; Ganier, O.; Peiffer, I.; Rivals, E.; Puy, A.; Laurent-Chabalier, S.; Desprat, R.; et al. Genome-scale analysis of metazoan replication origins reveals their organization in specific but flexible sites defined by conserved features. Genome Res. 2011, 21, 1438–1449. [Google Scholar] [CrossRef] [PubMed]
- Fragkos, M.; Ganier, O.; Coulombe, P.; Méchali, M. DNA replication origin activation in space and time. Nat. Rev. Mol. Cell Biol. 2015, 16, 360–374. [Google Scholar] [CrossRef] [PubMed]
- DePamphilis, M.L. Cell cycle dependent regulation of the origin recognition complex. Cell Cycle 2005, 4, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Sathyan, K.M.; Geng, Y.; Zheng, R.; Chakraborty, A.; Freeman, B.; Wang, F.; Prasanth, K.V.; Prasanth, S.G. A WD-repeat protein stabilizes ORC binding to chromatin. Mol. Cell 2010, 40, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Giri, S.; Prasanth, S.G. Association of ORCA/LRWD1 with repressive histone methyl transferases mediates heterochromatin organization. Nucleus 2015, 6, 435–441. [Google Scholar] [CrossRef] [PubMed]
- Labib, K. How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev. 2010, 24, 1208–1219. [Google Scholar] [CrossRef] [PubMed]
- Rainey, M.D.; Harhen, B.; Wang, G.-N.; Murphy, P.V.; Santocanale, C. Cdc7-dependent and -independent phosphorylation of Claspin in the induction of the DNA replication checkpoint. Cell Cycle 2013, 12, 1560–1568. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.-C.; Suzuki, M.; Yamakawa, S.; Uno, S.; Ishii, A.; Yamazaki, S.; Fukatsu, R.; Fujisawa, R.; Sakimura, K.; Tsurimoto, T.; et al. Claspin recruits Cdc7 kinase for initiation of DNA replication in human cells. Nat. Commun. 2016, 7, 12135. [Google Scholar] [CrossRef] [PubMed]
- Kliszczak, M.; Sedlackova, H.; Pitchai, G.P.; Streicher, W.W.; Krejci, L.; Hickson, I.D. Interaction of RECQ4 and MCM10 is important for efficient DNA replication origin firing in human cells. Oncotarget 2015, 6, 40464–40479. [Google Scholar] [CrossRef] [PubMed]
- Thu, Y.M.; Bielinsky, A.-K. MCM10: One tool for all-Integrity, maintenance and damage control. Semin. Cell Dev. Biol. 2014, 30, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Majka, J.; Burgers, P.M.J. The PCNA-RFC families of DNA clamps and clamp loaders. Prog. Nucleic Acid Res. Mol. Biol. 2004, 78, 227–260. [Google Scholar] [PubMed]
- Moldovan, G.-L.; Pfander, B.; Jentsch, S. PCNA, the maestro of the replication fork. Cell 2007, 129, 665–679. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, K.; DeGregori, J.; Leone, G.; Herendeen, D.R.; Kelly, T.J.; Nevins, J.R. Expression of the HsOrc1 gene, a human ORC1 homolog, is regulated by cell proliferation via the E2F transcription factor. Mol. Cell. Biol. 1996, 16, 6977–6984. [Google Scholar] [CrossRef] [PubMed]
- Kara, N.; Hossain, M.; Prasanth, S.G.; Stillman, B. Orc1 Binding to Mitotic Chromosomes Precedes Spatial Patterning during G1 Phase and Assembly of the Origin Recognition Complex in Human Cells. J. Biol. Chem. 2015, 290, 12355–12369. [Google Scholar] [CrossRef] [PubMed]
- Méndez, J.; Zou-Yang, X.H.; Kim, S.Y.; Hidaka, M.; Tansey, W.P.; Stillman, B. Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication. Mol. Cell 2002, 9, 481–491. [Google Scholar] [CrossRef]
- Li, C.-J.; DePamphilis, M.L. Mammalian Orc1 protein is selectively released from chromatin and ubiquitinated during the S-to-M transition in the cell division cycle. Mol. Cell. Biol. 2002, 22, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; DeGregori, J.; Shohet, R.; Leone, G.; Stillman, B.; Nevins, J.R.; Williams, R.S. Cdc6 is regulated by E2F and is essential for DNA replication in mammalian cells. Proc. Natl. Acad. Sci. USA 1998, 95, 3603–3608. [Google Scholar] [CrossRef] [PubMed]
- Petersen, B.O.; Lukas, J.; Sørensen, C.S.; Bartek, J.; Helin, K. Phosphorylation of mammalian CDC6 by cyclin A/CDK2 regulates its subcellular localization. EMBO J. 1999, 18, 396–410. [Google Scholar] [CrossRef] [PubMed]
- Petersen, B.O.; Wagener, C.; Marinoni, F.; Kramer, E.R.; Melixetian, M.; Lazzerini Denchi, E.; Gieffers, C.; Matteucci, C.; Peters, J.M.; Helin, K. Cell cycle- and cell growth-regulated proteolysis of mammalian CDC6 is dependent on APC-CDH1. Genes Dev. 2000, 14, 2330–2343. [Google Scholar] [CrossRef] [PubMed]
- Donzelli, M.; Squatrito, M.; Ganoth, D.; Hershko, A.; Pagano, M.; Draetta, G.F. Dual mode of degradation of Cdc25 A phosphatase. EMBO J. 2002, 21, 4875–4884. [Google Scholar] [CrossRef] [PubMed]
- Bashir, T.; Dorrello, N.V.; Amador, V.; Guardavaccaro, D.; Pagano, M. Control of the SCF(Skp2-Cks1) ubiquitin ligase by the APC/C(Cdh1) ubiquitin ligase. Nature 2004, 428, 190–193. [Google Scholar] [CrossRef] [PubMed]
- Saha, P.; Chen, J.; Thome, K.C.; Lawlis, S.J.; Hou, Z.H.; Hendricks, M.; Parvin, J.D.; Dutta, A. Human CDC6/Cdc18 associates with Orc1 and cyclin-cdk and is selectively eliminated from the nucleus at the onset of S phase. Mol. Cell. Biol. 1998, 18, 2758–2767. [Google Scholar] [CrossRef] [PubMed]
- Walter, D.; Hoffmann, S.; Komseli, E.-S.; Rappsilber, J.; Gorgoulis, V.; Sørensen, C.S. SCF(Cyclin F)-dependent degradation of CDC6 suppresses DNA re-replication. Nat. Commun. 2016, 7, 10530. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Prasanth, S.G. Orc2 protects ORCA from ubiquitin-mediated degradation. Cell Cycle 2012, 11, 3578–3589. [Google Scholar] [CrossRef] [PubMed]
- Pereg, Y.; Liu, B.Y.; O’Rourke, K.M.; Sagolla, M.; Dey, A.; Komuves, L.; French, D.M.; Dixit, V.M. Ubiquitin hydrolase Dub3 promotes oncogenic transformation by stabilizing Cdc25A. Nat. Cell Biol. 2010, 12, 400–406. [Google Scholar] [CrossRef] [PubMed]
- Weinreich, M.; Stillman, B. Cdc7p-Dbf4p kinase binds to chromatin during S phase and is regulated by both the APC and the RAD53 checkpoint pathway. EMBO J. 1999, 18, 5334–5346. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.F.; Santocanale, C.; Drury, L.S.; Diffley, J.F. Dbf4p, an essential S phase-promoting factor, is targeted for degradation by the anaphase-promoting complex. Mol. Cell. Biol. 2000, 20, 242–248. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Hsiao, J.Y.; Davey, N.E.; Van Voorhis, V.A.; Foster, S.A.; Tang, C.; Morgan, D.O. Multiple mechanisms determine the order of APC/C substrate degradation in mitosis. J. Cell Biol. 2014, 207, 23–39. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Collyer, T.; Hardy, C.F. Cell cycle regulation of DNA replication initiator factor Dbf4p. Mol. Cell. Biol. 1999, 19, 4270–4278. [Google Scholar] [CrossRef] [PubMed]
- Yamada, M.; Watanabe, K.; Mistrik, M.; Vesela, E.; Protivankova, I.; Mailand, N.; Lee, M.; Masai, H.; Lukas, J.; Bartek, J. ATR-Chk1-APC/CCdh1-dependent stabilization of Cdc7-ASK (Dbf4) kinase is required for DNA lesion bypass under replication stress. Genes Dev. 2013, 27, 2459–2472. [Google Scholar] [CrossRef] [PubMed]
- Bennett, L.N.; Clarke, P.R. Regulation of Claspin degradation by the ubiquitin-proteosome pathway during the cell cycle and in response to ATR-dependent checkpoint activation. FEBS Lett. 2006, 580, 4176–4181. [Google Scholar] [CrossRef] [PubMed]
- Mamely, I.; van Vugt, M.A.; Smits, V.A.J.; Semple, J.I.; Lemmens, B.; Perrakis, A.; Medema, R.H.; Freire, R. Polo-like kinase-1 controls proteasome-dependent degradation of Claspin during checkpoint recovery. Curr. Biol. 2006, 16, 1950–1955. [Google Scholar] [CrossRef] [PubMed]
- Mailand, N.; Bekker-Jensen, S.; Bartek, J.; Lukas, J. Destruction of Claspin by SCFbetaTrCP restrains Chk1 activation and facilitates recovery from genotoxic stress. Mol. Cell 2006, 23, 307–318. [Google Scholar] [CrossRef] [PubMed]
- Peschiaroli, A.; Dorrello, N.V.; Guardavaccaro, D.; Venere, M.; Halazonetis, T.; Sherman, N.E.; Pagano, M. SCFbetaTrCP-mediated degradation of Claspin regulates recovery from the DNA replication checkpoint response. Mol. Cell 2006, 23, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Bassermann, F.; Frescas, D.; Guardavaccaro, D.; Busino, L.; Peschiaroli, A.; Pagano, M. The Cdc14B-Cdh1-Plk1 axis controls the G2 DNA-damage-response checkpoint. Cell 2008, 134, 256–267. [Google Scholar] [CrossRef] [PubMed]
- Martín, Y.; Cabrera, E.; Amoedo, H.; Hernández-Pérez, S.; Domínguez-Kelly, R.; Freire, R. USP29 controls the stability of checkpoint adaptor Claspin by deubiquitination. Oncogene 2015, 34, 1058–1063. [Google Scholar] [CrossRef] [PubMed]
- McGarry, E.; Gaboriau, D.; Rainey, M.; Restuccia, U.; Bachi, A.; Santocanale, C. The deubiquitinase USP9X maintains DNA replication fork stability and DNA damage checkpoint responses by regulating CLASPIN during S-phase. Cancer Res. 2016, 76, 2384–2393. [Google Scholar] [CrossRef] [PubMed]
- Charrasse, S.; Gharbi-Ayachi, A.; Burgess, A.; Vera, J.; Hached, K.; Raynaud, P.; Schwob, E.; Lorca, T.; Castro, A. Ensa controls S-phase length by modulating Treslin levels. Nat. Commun. 2017, 8, 206. [Google Scholar] [CrossRef] [PubMed]
- Mochida, S.; Maslen, S.L.; Skehel, M.; Hunt, T. Greatwall phosphorylates an inhibitor of protein phosphatase 2A that is essential for mitosis. Science 2010, 330, 1670–1673. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Shamanna, R.A.; de Freitas, J.K.; Okur, M.; Khadka, P.; Kulikowicz, T.; Holland, P.P.; Tian, J.; Croteau, D.L.; Davis, A.J.; et al. Cell cycle-dependent phosphorylation regulates RECQL4 pathway choice and ubiquitination in DNA double-strand break repair. Nat. Commun. 2017, 8, 2039. [Google Scholar] [CrossRef] [PubMed]
- Tardat, M.; Brustel, J.; Kirsh, O.; Lefevbre, C.; Callanan, M.; Sardet, C.; Julien, E. The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat. Cell Biol. 2010, 12, 1086–1093. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Yu, V.C.; Zhu, G.; Chang, D.C. SET8 plays a role in controlling G1/S transition by blocking lysine acetylation in histone through binding to H4 N-terminal tail. Cell Cycle 2008, 7, 1423–1432. [Google Scholar] [CrossRef] [PubMed]
- Das-Bradoo, S.; Ricke, R.M.; Bielinsky, A.-K. Interaction between PCNA and diubiquitinated Mcm10 is essential for cell growth in budding yeast. Mol. Cell. Biol. 2006, 26, 4806–4817. [Google Scholar] [CrossRef] [PubMed]
- Okabe, H.; Lee, S.-H.; Phuchareon, J.; Albertson, D.G.; McCormick, F.; Tetsu, O. A critical role for FBXW8 and MAPK in cyclin D1 degradation and cancer cell proliferation. PLoS ONE 2006, 1, e128. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Yang, K.; Harwalkar, J.; Nye, J.M.; Mason, D.R.; Garrett, M.D.; Hitomi, M.; Stacey, D.W. Phosphorylation of cyclin D1 at Thr 286 during S phase leads to its proteasomal degradation and allows efficient DNA synthesis. Oncogene 2005, 24, 2599–2612. [Google Scholar] [CrossRef] [PubMed]
- Alt, J.R.; Cleveland, J.L.; Hannink, M.; Diehl, J.A. Phosphorylation-dependent regulation of cyclin D1 nuclear export and cyclin D1-dependent cellular transformation. Genes Dev. 2000, 14, 3102–3114. [Google Scholar] [CrossRef] [PubMed]
- Shan, J.; Zhao, W.; Gu, W. Suppression of cancer cell growth by promoting cyclin D1 degradation. Mol. Cell 2009, 36, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Singer, J.D.; Gurian-West, M.; Clurman, B.; Roberts, J.M. Cullin-3 targets cyclin E for ubiquitination and controls S phase in mammalian cells. Genes Dev. 1999, 13, 2375–2387. [Google Scholar] [CrossRef] [PubMed]
- Koepp, D.M.; Schaefer, L.K.; Ye, X.; Keyomarsi, K.; Chu, C.; Harper, J.W.; Elledge, S.J. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science 2001, 294, 173–177. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Yu, L.; Bai, C.; Liu, L.; Long, H.; Shi, L.; Lin, Z. USP27-mediated Cyclin E stabilization drives cell cycle progression and hepatocellular tumorigenesis. Oncogene 2018, 37, 2702–2713. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Summers, M.K.; Pham, V.; Lill, J.R.; Liu, J.; Lee, G.; Kirkpatrick, D.S.; Jackson, P.K.; Fang, G.; Dixit, V.M. Deubiquitinase USP37 is activated by CDK2 to antagonize APC(CDH1) and promote S phase entry. Mol. Cell 2011, 42, 511–523. [Google Scholar] [CrossRef] [PubMed]
- Hattori, T.; Isobe, T.; Abe, K.; Kikuchi, H.; Kitagawa, K.; Oda, T.; Uchida, C.; Kitagawa, M. Pirh2 promotes ubiquitin-dependent degradation of the cyclin-dependent kinase inhibitor p27Kip1. Cancer Res. 2007, 67, 10789–10795. [Google Scholar] [CrossRef] [PubMed]
- Kamura, T.; Hara, T.; Matsumoto, M.; Ishida, N.; Okumura, F.; Hatakeyama, S.; Yoshida, M.; Nakayama, K.; Nakayama, K.I. Cytoplasmic ubiquitin ligase KPC regulates proteolysis of p27(Kip1) at G1 phase. Nat. Cell Biol. 2004, 6, 1229–1235. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Adegoke, O.A.J.; Nepveu, A.; Nakayama, K.I.; Bedard, N.; Cheng, D.; Peng, J.; Wing, S.S. USP19 deubiquitinating enzyme supports cell proliferation by stabilizing KPC1, a ubiquitin ligase for p27Kip1. Mol. Cell. Biol. 2009, 29, 547–558. [Google Scholar] [CrossRef] [PubMed]
- Oakes, V.; Wang, W.; Harrington, B.; Lee, W.J.; Beamish, H.; Chia, K.M.; Pinder, A.; Goto, H.; Inagaki, M.; Pavey, S.; et al. Cyclin A/Cdk2 regulates Cdh1 and claspin during late S/G2 phase of the cell cycle. Cell Cycle 2014, 13, 3302–3311. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, H.; Ogura, K.; Wan, L.; Lu, Y.; Li, V.; Gao, D.; Liu, P.; Lau, A.W.; Wu, T.; Kirschner, M.W.; et al. SCF-mediated Cdh1 degradation defines a negative feedback system that coordinates cell-cycle progression. Cell Rep. 2013, 4, 803–816. [Google Scholar] [CrossRef] [PubMed]
- Rape, M.; Kirschner, M.W. Autonomous regulation of the anaphase-promoting complex couples mitosis to S-phase entry. Nature 2004, 432, 588–595. [Google Scholar] [CrossRef] [PubMed]
- Hsu, J.Y.; Reimann, J.D.R.; Sørensen, C.S.; Lukas, J.; Jackson, P.K. E2F-dependent accumulation of hEmi1 regulates S phase entry by inhibiting APC(Cdh1). Nat. Cell Biol. 2002, 4, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Cappell, S.D.; Mark, K.G.; Garbett, D.; Pack, L.R.; Rape, M.; Meyer, T. EMI1 switches from being a substrate to an inhibitor of APC/CCDH1 to start the cell cycle. Nature 2018, 558, 313–317. [Google Scholar] [CrossRef] [PubMed]
- Margottin-Goguet, F.; Hsu, J.Y.; Loktev, A.; Hsieh, H.M.; Reimann, J.D.R.; Jackson, P.K. Prophase destruction of Emi1 by the SCF (betaTrCP/Slimb) ubiquitin ligase activates the anaphase promoting complex to allow progression beyond prometaphase. Dev. Cell 2003, 4, 813–826. [Google Scholar] [CrossRef]
- Moshe, Y.; Boulaire, J.; Pagano, M.; Hershko, A. Role of Polo-like kinase in the degradation of early mitotic inhibitor 1, a regulator of the anaphase promoting complex/cyclosome. Proc. Natl. Acad. Sci. USA 2004, 101, 7937–7942. [Google Scholar] [CrossRef] [PubMed]
- Schmid, J.A.; Berti, M.; Walser, F.; Raso, M.C.; Schmid, F.; Krietsch, J.; Stoy, H.; Zwicky, K.; Ursich, S.; Freire, R.; et al. Histone ubiquitination by the DNA damage response is required for efficient DNA replication in unperturbed S Phase. Mol. Cell 2018. [Google Scholar] [CrossRef] [PubMed]
- Moreno, S.P.; Bailey, R.; Campion, N.; Herron, S.; Gambus, A. Polyubiquitylation drives replisome disassembly at the termination of DNA replication. Science 2014, 346, 477–481. [Google Scholar] [CrossRef] [PubMed]
- Maric, M.; Maculins, T.; De Piccoli, G.; Labib, K. Cdc48 and a ubiquitin ligase drive disassembly of the CMG helicase at the end of DNA replication. Science 2014, 346. [Google Scholar] [CrossRef] [PubMed]
- Dewar, J.M.; Low, E.; Mann, M.; Räschle, M.; Walter, J.C. CRL2Lrr1 promotes unloading of the vertebrate replisome from chromatin during replication termination. Genes Dev. 2017, 31, 275–290. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, J.F.; Beach, D. cdt1 is an essential target of the Cdc10/Sct1 transcription factor: Requirement for DNA replication and inhibition of mitosis. EMBO J. 1994, 13, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Hong, B.; Choi, J.M.; Kim, Y.; Watanabe, S.; Ishimi, Y.; Enomoto, T.; Tada, S.; Kim, Y.; Cho, Y. Structural basis for inhibition of the replication licensing factor Cdt1 by geminin. Nature 2004, 430, 913–917. [Google Scholar] [CrossRef] [PubMed]
- Khayrutdinov, B.I.; Bae, W.J.; Yun, Y.M.; Lee, J.H.; Tsuyama, T.; Kim, J.J.; Hwang, E.; Ryu, K.-S.; Cheong, H.-K.; Cheong, C.; et al. Structure of the Cdt1 C-terminal domain: Conservation of the winged helix fold in replication licensing factors. Protein Sci. 2009, 18, 2252–2264. [Google Scholar] [CrossRef] [PubMed]
- Liu, E.; Li, X.; Yan, F.; Zhao, Q.; Wu, X. Cyclin-dependent kinases phosphorylate human Cdt1 and induce its degradation. J. Biol. Chem. 2004, 279, 17283–17288. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, N.; Tatsumi, Y.; Tsurumi, T.; Matsukage, A.; Kiyono, T.; Nishitani, H.; Fujita, M. Cdt1 phosphorylation by cyclin A-dependent kinases negatively regulates its function without affecting geminin binding. J. Biol. Chem. 2004, 279, 19691–19697. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhao, Q.; Liao, R.; Sun, P.; Wu, X. The SCF(Skp2) ubiquitin ligase complex interacts with the human replication licensing factor Cdt1 and regulates Cdt1 degradation. J. Biol. Chem. 2003, 278, 30854–30858. [Google Scholar] [CrossRef] [PubMed]
- Nishitani, H.; Sugimoto, N.; Roukos, V.; Nakanishi, Y.; Saijo, M.; Obuse, C.; Tsurimoto, T.; Nakayama, K.I.; Nakayama, K.; Fujita, M.; et al. Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis. EMBO J. 2006, 25, 1126–1136. [Google Scholar] [CrossRef] [PubMed]
- Havens, C.G.; Walter, J.C. Docking of a specialized PIP Box onto chromatin-bound PCNA creates a degron for the ubiquitin ligase CRL4Cdt2. Mol. Cell 2009, 35, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Havens, C.G.; Walter, J.C. Mechanism of CRL4(Cdt2), a PCNA-dependent E3 ubiquitin ligase. Genes Dev. 2011, 25, 1568–1582. [Google Scholar] [CrossRef] [PubMed]
- Johansson, P.; Jeffery, J.; Al-Ejeh, F.; Schulz, R.B.; Callen, D.F.; Kumar, R.; Khanna, K.K. SCF-FBXO31 E3 ligase targets DNA replication factor Cdt1 for proteolysis in the G2 phase of cell cycle to prevent re-replication. J. Biol. Chem. 2014, 289, 18514–18525. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, N.; Kitabayashi, I.; Osano, S.; Tatsumi, Y.; Yugawa, T.; Narisawa-Saito, M.; Matsukage, A.; Kiyono, T.; Fujita, M. Identification of novel human Cdt1-binding proteins by a proteomics approach: Proteolytic regulation by APC/CCdh1. Mol. Biol. Cell 2008, 19, 1007–1021. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Pérez, S.; Cabrera, E.; Amoedo, H.; Rodriguez-Acebes, S.; Koundrioukoff, S.; Debatisse, M.; Méndez, J.; Freire, R. USP37 deubiquitinates Cdt1 and contributes to regulate DNA replication. Mol. Oncol. 2016, 10, 1196–1206. [Google Scholar] [CrossRef] [PubMed]
- Franz, A.; Orth, M.; Pirson, P.A.; Sonneville, R.; Blow, J.J.; Gartner, A.; Stemmann, O.; Hoppe, T. CDC-48/p97 coordinates CDT-1 degradation with GINS chromatin dissociation to ensure faithful DNA replication. Mol. Cell 2011, 44, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Wong, P.G.; Glozak, M.A.; Cao, T.V.; Vaziri, C.; Seto, E.; Alexandrow, M. Chromatin unfolding by Cdt1 regulates MCM loading via opposing functions of HBO1 and HDAC11-geminin. Cell Cycle 2010, 9, 4351–4363. [Google Scholar] [CrossRef] [PubMed]
- Ballabeni, A.; Zamponi, R.; Caprara, G.; Melixetian, M.; Bossi, S.; Masiero, L.; Helin, K. Human CDT1 associates with CDC7 and recruits CDC45 to chromatin during S phase. J. Biol. Chem. 2009, 284, 3028–3036. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Liu, C.; Wu, X.; Xu, N.; Zhou, B.; Liang, C.; Zhu, G. Characterization and structure determination of the Cdt1 binding domain of human minichromosome maintenance (Mcm) 6. J. Biol. Chem. 2010, 285, 12469–12473. [Google Scholar] [CrossRef] [PubMed]
- McGarry, T.J.; Kirschner, M.W. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell 1998, 93, 1043–1053. [Google Scholar] [CrossRef]
- Wohlschlegel, J.A.; Dwyer, B.T.; Dhar, S.K.; Cvetic, C.; Walter, J.C.; Dutta, A. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science 2000, 290, 2309–2312. [Google Scholar] [CrossRef] [PubMed]
- Xouri, G.; Squire, A.; Dimaki, M.; Geverts, B.; Verveer, P.J.; Taraviras, S.; Nishitani, H.; Houtsmuller, A.B.; Bastiaens, P.I.H.; Lygerou, Z. Cdt1 associates dynamically with chromatin throughout G1 and recruits Geminin onto chromatin. EMBO J. 2007, 26, 1303–1314. [Google Scholar] [CrossRef] [PubMed]
- Miotto, B.; Struhl, K. HBO1 histone acetylase is a coactivator of the replication licensing factor Cdt1. Genes Dev. 2008, 22, 2633–2638. [Google Scholar] [CrossRef] [PubMed]
- Okorokov, A.L.; Orlova, E.V.; Kingsbury, S.R.; Bagneris, C.; Gohlke, U.; Williams, G.H.; Stoeber, K. Molecular structure of human geminin. Nat. Struct. Mol. Biol. 2004, 11, 1021–1022. [Google Scholar] [CrossRef] [PubMed]
- Caillat, C.; Pefani, D.-E.; Gillespie, P.J.; Taraviras, S.; Blow, J.J.; Lygerou, Z.; Perrakis, A. The Geminin and Idas coiled coils preferentially form a heterodimer that inhibits Geminin function in DNA replication licensing. J. Biol. Chem. 2013, 288, 31624–31634. [Google Scholar] [CrossRef] [PubMed]
- Benjamin, J.M.; Torke, S.J.; Demeler, B.; McGarry, T.J. Geminin has dimerization, Cdt1-binding, and destruction domains that are required for biological activity. J. Biol. Chem. 2004, 279, 45957–45968. [Google Scholar] [CrossRef] [PubMed]
- Machida, Y.J.; Dutta, A. The APC/C inhibitor, Emi1, is essential for prevention of rereplication. Genes Dev. 2007, 21, 184–194. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Y.; Li, T.T.; Yang, H.L.; Hu, T.; Nie, L.; Wang, F.; Alcala, M.; Zang, H.C. The spindle checkpoint, APC/C(Cdc20), and APC/C(Cdh1) play distinct roles in connecting mitosis to S phase. J. Innov. Opt. Health Sci. 2013, 201, 1013–1026. [Google Scholar]
- Boos, A.; Lee, A.; Thompson, D.M.; Kroll, K.L. Subcellular translocation signals regulate Geminin activity during embryonic development. Biol. Cell 2006, 98, 363–375. [Google Scholar] [CrossRef] [PubMed]
- Sakaue-Sawano, A.; Ohtawa, K.; Hama, H.; Kawano, M.; Ogawa, M.; Miyawaki, A. Tracing the silhouette of individual cells in S/G2/M phases with fluorescence. Chem. Biol. 2008, 15, 1243–1248. [Google Scholar] [CrossRef] [PubMed]
- De Marco, V.; Gillespie, P.J.; Li, A.; Karantzelis, N.; Christodoulou, E.; Klompmaker, R.; van Gerwen, S.; Fish, A.; Petoukhov, M.V.; Iliou, M.S.; et al. Quaternary structure of the human Cdt1-Geminin complex regulates DNA replication licensing. Proc. Natl. Acad. Sci. USA 2009, 106, 19807–19812. [Google Scholar] [CrossRef] [PubMed]
- Ballabeni, A.; Melixetian, M.; Zamponi, R.; Masiero, L.; Marinoni, F.; Helin, K. Human geminin promotes pre-RC formation and DNA replication by stabilizing CDT1 in mitosis. EMBO J. 2004, 23, 3122–3132. [Google Scholar] [CrossRef] [PubMed]
- Tsunematsu, T.; Takihara, Y.; Ishimaru, N.; Pagano, M.; Takata, T.; Kudo, Y. Aurora-A controls pre-replicative complex assembly and DNA replication by stabilizing geminin in mitosis. Nat. Commun. 2013, 4, 1885. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Pérez, S.; Cabrera, E.; Salido, E.; Lim, M.; Reid, L.; Lakhani, S.R.; Khanna, K.K.; Saunus, J.M.; Freire, R. DUB3 and USP7 de-ubiquitinating enzymes control replication inhibitor Geminin: Molecular characterization and associations with breast cancer. Oncogene 2017, 36, 4802–4809. [Google Scholar] [CrossRef] [PubMed]
- Tada, S.; Li, A.; Maiorano, D.; Méchali, M.; Blow, J.J. Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat. Cell Biol. 2001, 3, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Macheret, M.; Halazonetis, T.D. DNA replication stress as a hallmark of cancer. Annu. Rev. Pathol. 2015, 10, 425–448. [Google Scholar] [CrossRef] [PubMed]
- Monnat, R.J. Human RECQ helicases: Roles in DNA metabolism, mutagenesis and cancer biology. Semin. Cancer Biol. 2010, 20, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Bongers, E.M.; Opitz, J.M.; Fryer, A.; Sarda, P.; Hennekam, R.C.; Hall, B.D.; Superneau, D.W.; Harbison, M.; Poss, A.; van Bokhoven, H.; et al. Meier-Gorlin syndrome: Report of eight additional cases and review. Am. J. Med. Genet. 2001, 102, 115–124. [Google Scholar] [CrossRef] [PubMed]
- De Munnik, S.A.; Hoefsloot, E.H.; Roukema, J.; Schoots, J.; Knoers, N.V.A.M.; Brunner, H.G.; Jackson, A.P.; Bongers, E.M.H.F. Meier-Gorlin syndrome. Orphanet. J. Rare Dis. 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.; Stillman, B. Meier-Gorlin syndrome mutations disrupt an Orc1 CDK inhibitory domain and cause centrosome reduplication. Genes Dev. 2012, 26, 1797–1810. [Google Scholar] [CrossRef] [PubMed]
- Kuo, A.J.; Song, J.; Cheung, P.; Ishibe-Murakami, S.; Yamazoe, S.; Chen, J.K.; Patel, D.J.; Gozani, O. The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome. Nature 2012, 484, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Bleichert, F.; Balasov, M.; Chesnokov, I.; Nogales, E.; Botchan, M.R.; Berger, J.M. A Meier-Gorlin syndrome mutation in a conserved C-terminal helix of Orc6 impedes origin recognition complex formation. eLife 2013, 2, e00882. [Google Scholar] [CrossRef] [PubMed]
- Guernsey, D.L.; Matsuoka, M.; Jiang, H.; Evans, S.; Macgillivray, C.; Nightingale, M.; Perry, S.; Ferguson, M.; LeBlanc, M.; Paquette, J.; et al. Mutations in origin recognition complex gene ORC4 cause Meier-Gorlin syndrome. Nat. Genet. 2011, 43, 360–364. [Google Scholar] [CrossRef] [PubMed]
- Bicknell, L.S.; Bongers, E.M.H.F.; Leitch, A.; Brown, S.; Schoots, J.; Harley, M.E.; Aftimos, S.; Al-Aama, J.Y.; Bober, M.; Brown, P.A.J.; et al. Mutations in the pre-replication complex cause Meier-Gorlin syndrome. Nat. Genet. 2011, 43, 356–359. [Google Scholar] [CrossRef] [PubMed]
- Fenwick, A.L.; Kliszczak, M.; Cooper, F.; Murray, J.; Sanchez-Pulido, L.; Twigg, S.R.F.; Goriely, A.; McGowan, S.J.; Miller, K.A.; Taylor, I.B.; et al. Mutations in CDC45, Encoding an Essential Component of the Pre-initiation Complex, Cause Meier-Gorlin Syndrome and Craniosynostosis. Am. J. Hum. Genet. 2016, 99, 125–138. [Google Scholar] [CrossRef] [PubMed]
- Burrage, L.C.; Charng, W.-L.; Eldomery, M.K.; Willer, J.R.; Davis, E.E.; Lugtenberg, D.; Zhu, W.; Leduc, M.S.; Akdemir, Z.C.; Azamian, M.; et al. De Novo GMNN Mutations Cause Autosomal-Dominant Primordial Dwarfism Associated with Meier-Gorlin Syndrome. Am. J. Hum. Genet. 2015, 97, 904–913. [Google Scholar] [CrossRef] [PubMed]
- Shima, N.; Alcaraz, A.; Liachko, I.; Buske, T.R.; Andrews, C.A.; Munroe, R.J.; Hartford, S.A.; Tye, B.K.; Schimenti, J.C. A viable allele of Mcm4 causes chromosome instability and mammary adenocarcinomas in mice. Nat. Genet. 2007, 39, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Vetro, A.; Savasta, S.; Russo Raucci, A.; Cerqua, C.; Sartori, G.; Limongelli, I.; Forlino, A.; Maruelli, S.; Perucca, P.; Vergani, D.; et al. MCM5: A new actor in the link between DNA replication and Meier-Gorlin syndrome. Eur. J. Hum. Genet. 2017, 25, 646–650. [Google Scholar] [CrossRef] [PubMed]
- Whitfield, M.L.; George, L.K.; Grant, G.D.; Perou, C.M. Common markers of proliferation. Nat. Rev. Cancer 2006, 6, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Stoeber, K.; Tlsty, T.D.; Happerfield, L.; Thomas, G.A.; Romanov, S.; Bobrow, L.; Williams, E.D.; Williams, G.H. DNA replication licensing and human cell proliferation. J. Cell. Sci. 2001, 114, 2027–2041. [Google Scholar] [PubMed]
- Harada, H.; Nakagawa, H.; Takaoka, M.; Lee, J.; Herlyn, M.; Diehl, J.A.; Rustgi, A.K. Cleavage of MCM2 licensing protein fosters senescence in human keratinocytes. Cell Cycle 2008, 7, 3534–3538. [Google Scholar] [CrossRef] [PubMed]
- Tachibana, K.-E.K.; Gonzalez, M.A.; Coleman, N. Cell-cycle-dependent regulation of DNA replication and its relevance to cancer pathology. J. Pathol. 2005, 205, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Williams, G.H.; Romanowski, P.; Morris, L.; Madine, M.; Mills, A.D.; Stoeber, K.; Marr, J.; Laskey, R.A.; Coleman, N. Improved cervical smear assessment using antibodies against proteins that regulate DNA replication. Proc. Natl. Acad. Sci. USA 1998, 95, 14932–14937. [Google Scholar] [CrossRef] [PubMed]
- Davies, R.J.; Freeman, A.; Morris, L.S.; Bingham, S.; Dilworth, S.; Scott, I.; Laskey, R.A.; Miller, R.; Coleman, N. Analysis of minichromosome maintenance proteins as a novel method for detection of colorectal cancer in stool. Lancet 2002, 359, 1917–1919. [Google Scholar] [CrossRef]
- Gonzalez, M.A.; Pinder, S.E.; Callagy, G.; Vowler, S.L.; Morris, L.S.; Bird, K.; Bell, J.A.; Laskey, R.A.; Coleman, N. Minichromosome maintenance protein 2 is a strong independent prognostic marker in breast cancer. J. Clin. Oncol. 2003, 21, 4306–4313. [Google Scholar] [CrossRef] [PubMed]
- Meng, M.V.; Grossfeld, G.D.; Williams, G.H.; Dilworth, S.; Stoeber, K.; Mulley, T.W.; Weinberg, V.; Carroll, P.R.; Tlsty, T.D. Minichromosome maintenance protein 2 expression in prostate: Characterization and association with outcome after therapy for cancer. Clin. Cancer Res. 2001, 7, 2712–2718. [Google Scholar] [PubMed]
- Ramnath, N.; Hernandez, F.J.; Tan, D.F.; Huberman, J.A.; Natarajan, N.; Beck, A.F.; Hyland, A.; Todorov, I.T.; Brooks, J.S.; Bepler, G. MCM2 is an independent predictor of survival in patients with non-small-cell lung cancer. J. Clin. Oncol. 2001, 19, 4259–4266. [Google Scholar] [CrossRef] [PubMed]
- Jackson, A.P.; Laskey, R.A.; Coleman, N. Replication proteins and human disease. Cold Spring Harb. Perspect. Biol. 2014, 6, a013060. [Google Scholar] [CrossRef] [PubMed]
- Petropoulou, C.; Kotantaki, P.; Karamitros, D.; Taraviras, S. Cdt1 and Geminin in cancer: Markers or triggers of malignant transformation? Front. Biosci. 2008, 13, 4485–4494. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, M.A.; Tachibana, K.-E.K.; Chin, S.-F.; Callagy, G.; Madine, M.A.; Vowler, S.L.; Pinder, S.E.; Laskey, R.A.; Coleman, N. Geminin predicts adverse clinical outcome in breast cancer by reflecting cell-cycle progression. J. Pathol. 2004, 204, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Sundara Rajan, S.; Hanby, A.M.; Horgan, K.; Thygesen, H.H.; Speirs, V. The potential utility of geminin as a predictive biomarker in breast cancer. Breast Cancer Res. Treat. 2014, 143, 91–98. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernández-Carralero, E.; Cabrera, E.; Alonso-de Vega, I.; Hernández-Pérez, S.; Smits, V.A.J.; Freire, R. Control of DNA Replication Initiation by Ubiquitin. Cells 2018, 7, 146. https://doi.org/10.3390/cells7100146
Hernández-Carralero E, Cabrera E, Alonso-de Vega I, Hernández-Pérez S, Smits VAJ, Freire R. Control of DNA Replication Initiation by Ubiquitin. Cells. 2018; 7(10):146. https://doi.org/10.3390/cells7100146
Chicago/Turabian StyleHernández-Carralero, Esperanza, Elisa Cabrera, Ignacio Alonso-de Vega, Santiago Hernández-Pérez, Veronique A. J. Smits, and Raimundo Freire. 2018. "Control of DNA Replication Initiation by Ubiquitin" Cells 7, no. 10: 146. https://doi.org/10.3390/cells7100146
APA StyleHernández-Carralero, E., Cabrera, E., Alonso-de Vega, I., Hernández-Pérez, S., Smits, V. A. J., & Freire, R. (2018). Control of DNA Replication Initiation by Ubiquitin. Cells, 7(10), 146. https://doi.org/10.3390/cells7100146



