CRISPR Disruption of scaRNA1 Reduces Pseudouridylation in Spliceosomal RNA U2 at U89 and Perturbs the Transcriptome in HEK293T Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Guide RNA Design
2.2. HEK293T Cell Culture and CRISPR-Cas9 Transfection
2.3. DNA Isolation, Mutation Screening (Sanger Sequencing), and Validation by TIDE Analysis
2.4. RNA Extraction and cDNA Synthesis
2.5. qPCR for RNA Expression Analysis
2.6. CMC Reverse Transcription Stop Assay for Pseudouridylation Detection
2.7. RNA-Seq Library Preparation and Analysis
3. Results
3.1. Edit of the Genomic Sequence of scaRNA1 Using CRISPR-Cas9
3.2. TIDE Analysis Reveals Distinct Clonal Editing Profiles
3.3. Disruption of scaRNA1 Results in Altered Expression
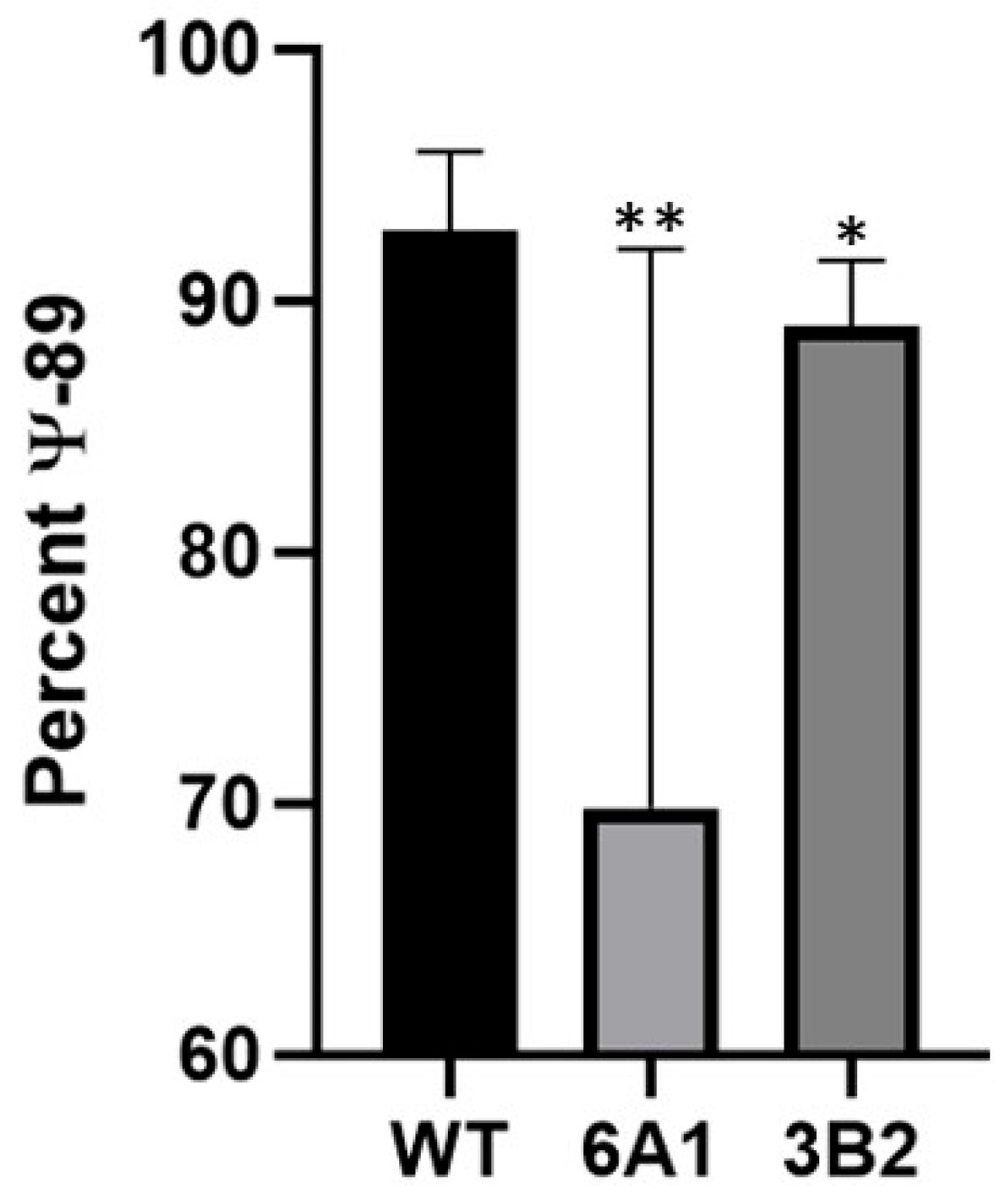
3.4. Pseudouridylation Is Reduced Following CRISPR-Cas9 Editing of scaRNA1
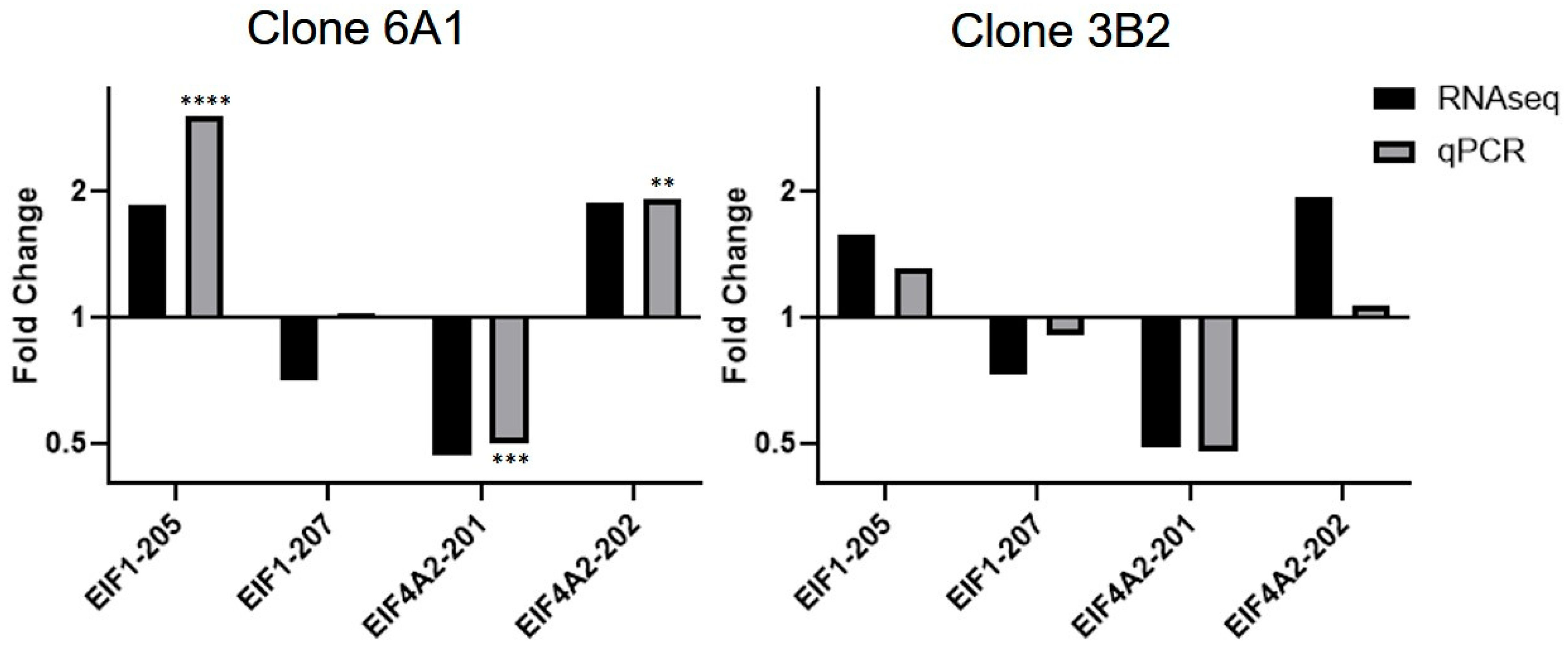
3.5. RNA-Seq
4. Discussion
4.1. Functional Disruption of scaRNA1 Impairs Expression and Pseudouridylation Activity
4.2. scaRNA1 Disruption Alters Alternative Splicing of RNA-Binding Genes
5. Limitations and Future Directions
6. Biological and Clinical Significance
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Adachi, H.; Yu, Y.-T. Insight into the mechanisms and functions of spliceosomal snRNA pseudouridylation. World J. Biol. Chem. 2014, 5, 398. [Google Scholar] [CrossRef]
- Antonicka, H.; Choquet, K.; Lin, Z.; Gingras, A.; Kleinman, C.L.; Shoubridge, E.A. A pseudouridine synthase module is essential for mitochondrial protein synthesis and cell viability. EMBO Rep. 2017, 18, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Baralle, F.E.; Giudice, J. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 2017, 18, 437. [Google Scholar] [CrossRef]
- Beneventi, G.; Munita, R.; Ngoc, P.C.T.; Madej, M.; Cieśla, M.; Muthukumar, S.; Krogh, N.; Nielsen, H.; Swaminathan, V.; Bellodi, C. The small Cajal body-specific RNA 15 (SCARNA15) directs p53 and redox homeostasis via selective splicing in cancer cells. NAR Cancer 2021, 3, zcab026. [Google Scholar] [CrossRef]
- Black, D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003, 72, 291–336. [Google Scholar] [CrossRef]
- Bohnsack, M.T.; Sloan, K.E. Modifications in small nuclear RNAs and their roles in spliceosome assembly and function. Biol. Chem. 2018, 399, 1265–1276. [Google Scholar] [CrossRef]
- Brinkman, E.K.; van Steensel, B. Rapid Quantitative Evaluation of CRISPR Genome Editing by TIDE and TIDER. Methods Mol. Biol. 2019, 1961, 29–44. [Google Scholar] [CrossRef]
- Concordet, J.P.; Haeussler, M. CRISPOR: Intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018, 46, W242–W245. [Google Scholar] [CrossRef]
- Ge, J.; Yu, Y.T. RNA pseudouridylation: New insights into an old modification. Trends Biochem. Sci. 2013, 38, 210. [Google Scholar] [CrossRef]
- Gelb, B.D.; Chung, W.K. Complex genetics and the etiology of human congenital heart disease. Cold Spring Harb. Perspect. Med. 2014, 4, a013953. [Google Scholar] [CrossRef]
- Harrison, P.W.; Amode, M.R.; Austine-Orimoloye, O.; Azov, A.G.; Barba, M.; Barnes, I.; Becker, A.; Bennett, R.; Berry, A.; Bhai, J.; et al. Ensembl 2024. Nucleic Acids Res. 2024, 52, D891–D899. [Google Scholar] [CrossRef] [PubMed]
- Karijolich, J.; Yu, Y.T. Spliceosomal snRNA modifications and their function. RNA Biol. 2010, 7, 192–204. [Google Scholar] [CrossRef] [PubMed]
- Kiss, A.M.; Jády, B.E.; Bertrand, E.; Kiss, T. Human box H/ACA pseudouridylation guide RNA machinery. Mol. Cell. Biol. 2004, 24, 5797–5807. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhao, X.; Yu, Y.T. Pseudouridylation (Ψ) of U2 snRNA in S. cerevisiae is catalyzed by an RNA-independent mechanism. EMBO J. 2003, 22, 1889–1897. [Google Scholar] [CrossRef]
- Nagasawa, C.K.; Kibiryeva, N.; Marshall, J.; O’Brien, J.E., Jr.; Bittel, D.C. scaRNA1 Levels Alter Pseudouridylation in Spliceosomal RNA U2 Affecting Alternative mRNA Splicing and Embryonic Development. Pediatr. Cardiol. 2020, 41, 341–349. [Google Scholar] [CrossRef]
- Patil, P.; Kibiryeva, N.; Uechi, T.; Marshall, J.; O’Brien, J.E.; Artman, M.; Kenmochi, N.; Bittel, D.C. scaRNAs regulate splicing and vertebrate heart development. Biochim. Biophys. Acta-Mol. Basis Dis. 2015, 1852, 1619–1629. [Google Scholar] [CrossRef]
- Patton, J.R. Ribonucleoprotein particle assembly and modification of U2 small nuclear RNA containing 5-fluorouridine. Biochemistry 1993, 32, 8939–8944. [Google Scholar] [CrossRef]
- Kelemen, O.; Convertini, P.; Zhang, Z.; Wen, Y.; Shen, M.; Falaleeva, M.; Stamm, S. Function of alternative splicing. Gene 2005, 514, 1–30. [Google Scholar] [CrossRef]
- O’Brien, J.E.; Kibiryeva, N.; Zhou, X.G.; Marshall, J.A.; Lofland, G.K.; Artman, M.; Chen, J.; Bittel, D.C. Noncoding RNA expression in myocardium from infants with tetralogy of fallot. Circ. Cardiovasc. Genet. 2012, 5, 279–286. [Google Scholar] [CrossRef]
- Wang, C.; Ulry, N.; Herzel, L.; Pythoud, N.; Kleiber, N.; Guérineau, V.; Jactel, V.; Moritz, C.; Bohnsack, M.T.; Carapito, C.; et al. N 2-methylguanosine modifications on human tRNAs and snRNA U6 are important for cell proliferation, protein translation and pre-mRNA splicing. Nucleic Acids Res. 2023, 51, 7496–7519. [Google Scholar] [CrossRef]
- Yu, Y.T.; Shu, M.D.; Steitz, J.A. Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J. 1998, 17, 5783. [Google Scholar] [CrossRef]





| gRNA | Sequence | Strand | PAM | Cut Site in scaRNA1 (bp) |
|---|---|---|---|---|
| gRNA1 | AGTGTATCTTTACACACTGGGCT | Antisense | TGG | 115 |
| gRNA2 | CACTCTGAGTTATAACTCAG | Sense | AGG | 162 |
| Primer Name | Sequence (5′–3′) | Direction | Amplicon Size, bp |
|---|---|---|---|
| Forward Primer | GACAGGGTTGCCTAGTCTTAAT | Sense | 990 |
| Reverse Primer | CATACTAGAGATTTAGGGATTTCTTTGAGAGG | Antisense |
| Primer Name | Sequence (5′–3′) | Direction | Amplicon Size, bp |
|---|---|---|---|
| RN7SL Forward | ATCGGGTGTCCGCACTAAGTT | Sense | 125 |
| RN7SL Reverse | CAGCACGGGAGTTTTGACCT | Antisense | |
| scaRNA1 Forward | GCACTTGATACTAACCGAGCTG | Sense | 138 |
| scaRNA1 Reverse | AGTGTATCTTTACACACTGGGCT | Antisense | |
| PPP1R8 Forward | CCCTCCCGGTTTACATCT | Sense | 202 |
| PPP1R8 Reverse | ACTGTTGAGATCTATCAGGAAA | Antisense | |
| EIF1 205 Forward | CTGCTGGCACTGAGGATTAT | Sense | |
| EIF1 205 Probe | CAATACATACAACACAGCTGGCACCC | Antisense | |
| EIF1 205 Reverse | ACGAATTAGGCCTAGCAAGAG | Antisense | 253 |
| EIF1 207 Forward | CTGAAGGTTCATGGGTTTAAAAACCTC | Sense | |
| EIF1 207 Probe | ACTTGGACTAGTGTAACTCCTTCATGCA | Sense | |
| EIF1 207 Reverse | CAAGACTAGACAGCATGGCTCTT | Antisense | 126 |
| EIF4A2 201 Forward | GTGCAACAAGTGTCTTTGGTTAT | Sense | |
| EIF4A2 201 Probe | CTATATTCACAGAATTGGCAGAGG | Sense | |
| EIF1 201 Reverse | CCACACCTTTCCTCCCAAAT | Antisense | 103 |
| EIF4A2 202 Forward | AACTATATTCACAGGAGTCGATAGC | Sense | |
| EIF4A2 202 Probe | CAGTTGGTGACGAGATGGCACTCA | Sense | |
| EIF1 202 Reverse | CAGTGTGCTGTTTCGCTTATG | Antisense | 105 |
| Primer Name | Primer Sequence | Description |
|---|---|---|
| HuU2-Ψ89UpF-CN-CMC | CTGATACGTCCTCTATCCGA | Upstream Forward |
| HuU2-Ψ89DnF-CN-CMC | TGGAGCAGGGAGATGGAATAGG | Downstream Forward |
| HuU2-Ψ89shR-CN-CMC | TACTGCAATACCAGGTCGATGCGT | Shared Reverse |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gardner-Kay, A.; Le, L.; Filla, M.; Kibiryeva, N.; O’Brien, J.E., Jr.; Bittel, D.C. CRISPR Disruption of scaRNA1 Reduces Pseudouridylation in Spliceosomal RNA U2 at U89 and Perturbs the Transcriptome in HEK293T Cells. Cells 2025, 14, 1882. https://doi.org/10.3390/cells14231882
Gardner-Kay A, Le L, Filla M, Kibiryeva N, O’Brien JE Jr., Bittel DC. CRISPR Disruption of scaRNA1 Reduces Pseudouridylation in Spliceosomal RNA U2 at U89 and Perturbs the Transcriptome in HEK293T Cells. Cells. 2025; 14(23):1882. https://doi.org/10.3390/cells14231882
Chicago/Turabian StyleGardner-Kay, Amanda, Lynndy Le, Michael Filla, Nataliya Kibiryeva, James E. O’Brien, Jr., and Douglas C. Bittel. 2025. "CRISPR Disruption of scaRNA1 Reduces Pseudouridylation in Spliceosomal RNA U2 at U89 and Perturbs the Transcriptome in HEK293T Cells" Cells 14, no. 23: 1882. https://doi.org/10.3390/cells14231882
APA StyleGardner-Kay, A., Le, L., Filla, M., Kibiryeva, N., O’Brien, J. E., Jr., & Bittel, D. C. (2025). CRISPR Disruption of scaRNA1 Reduces Pseudouridylation in Spliceosomal RNA U2 at U89 and Perturbs the Transcriptome in HEK293T Cells. Cells, 14(23), 1882. https://doi.org/10.3390/cells14231882







