Differential Responses of Human iPSC-Derived Microglia to Stimulation with Diverse Inflammogens
Abstract
1. Introduction
2. Materials and Methods
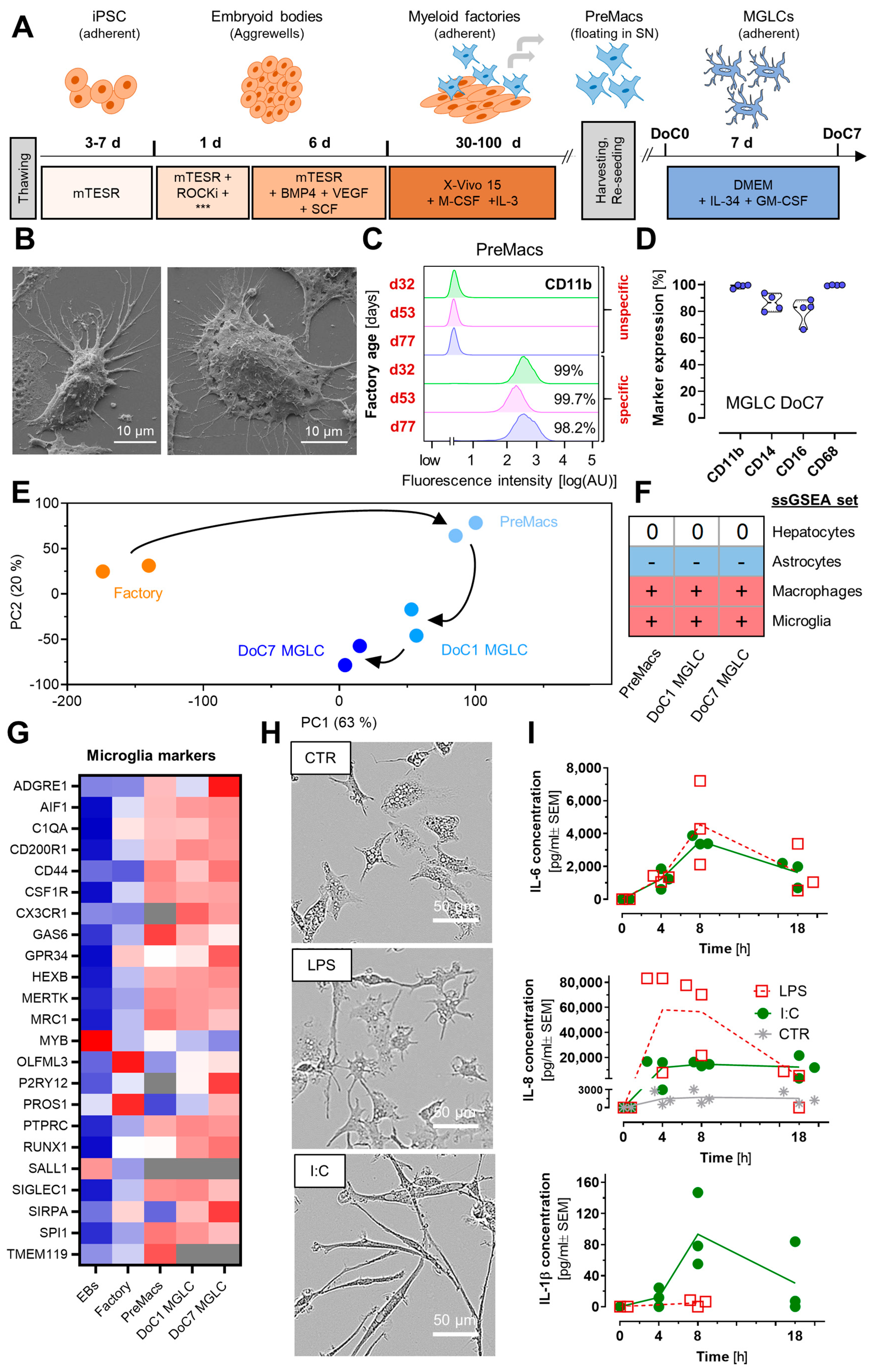
2.1. Stem Cell Culture
2.2. Factory Differentiation and PreMacs Generation
2.3. Harvest of Macrophage Precursor Cells (PreMacs)
2.4. Maturation to Microglia-like Cells (MGLCs)
2.5. Differentiation and Culture of iPSC-Derived Astrocytes
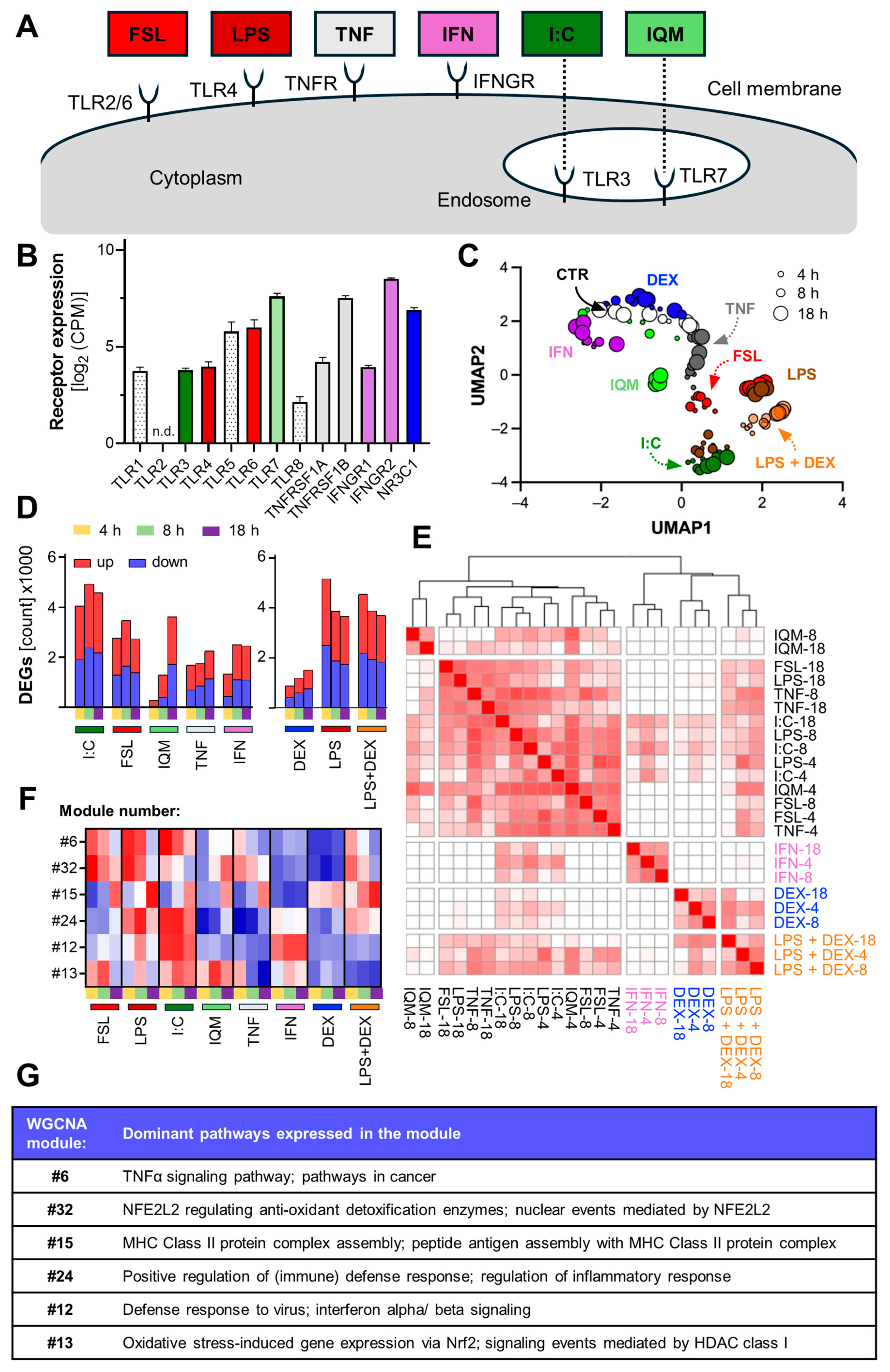
2.6. Inflammogens and Treatments
2.7. Flow Cytometry
2.8. Scanning Electron Microscopy
2.9. Bright Field Imaging
2.10. Treatment of MGLCs for Transcriptome and Cytokine Sample Generation
2.11. Cytokine Analyses by Multiplexed Microsphere-Based Sandwich Immunoassays
2.12. Transcriptome Data Generation and Analysis
2.13. Immunofluorescence Staining
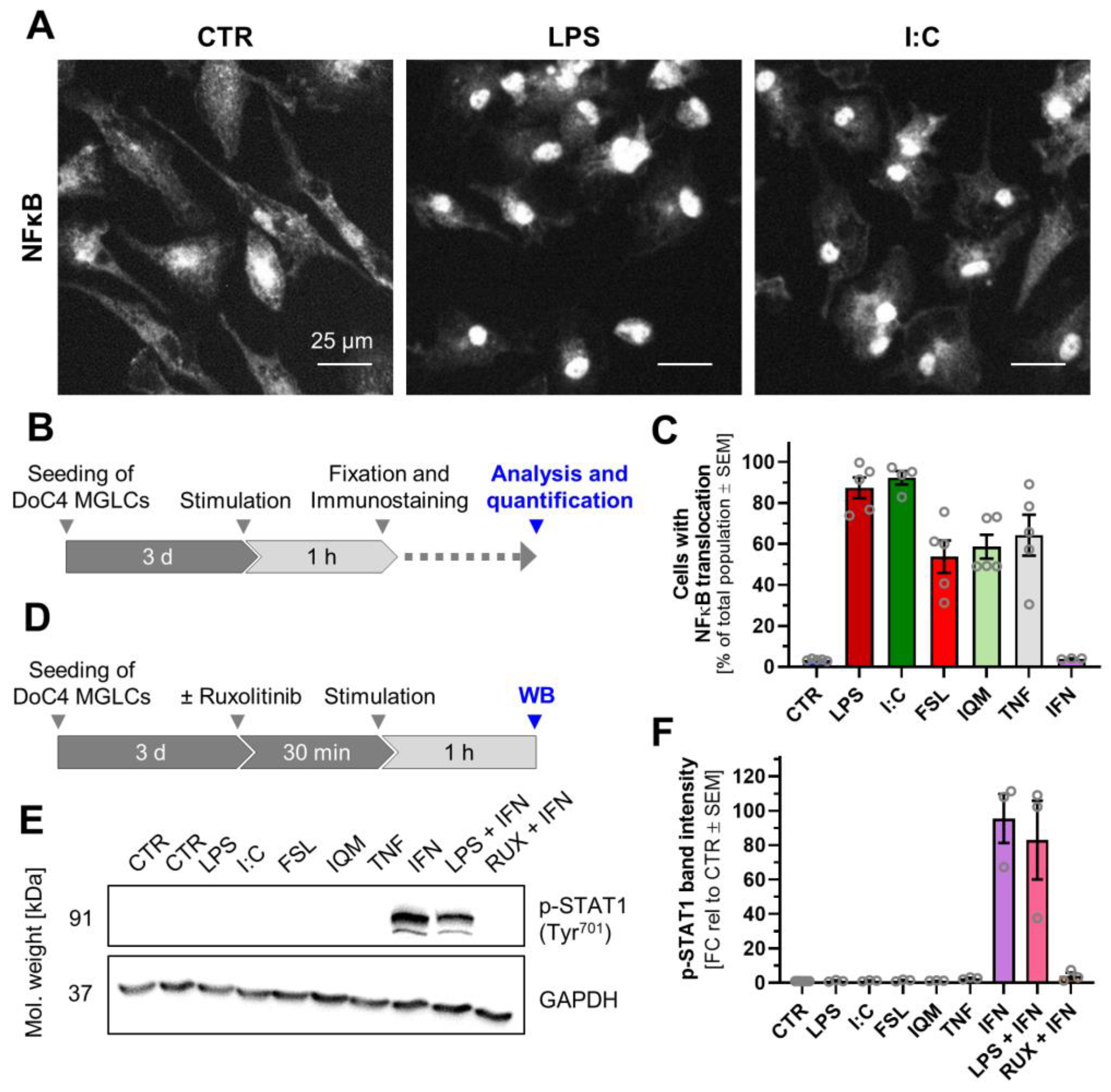
2.14. NFκB Translocation
2.15. Supernatant Transfer and TNFα Neutralization
2.16. Western Blot
2.17. Statistical Analysis
3. Results and Discussion
3.1. Characterization of Human Microglia-like Cells and Their Responses
3.2. Inflammation-Induced Transcriptome Changes in Microglia-like Cells
3.3. Divergent Signal Transduction in MGLCs After Exposure to Inflammatory Stimuli
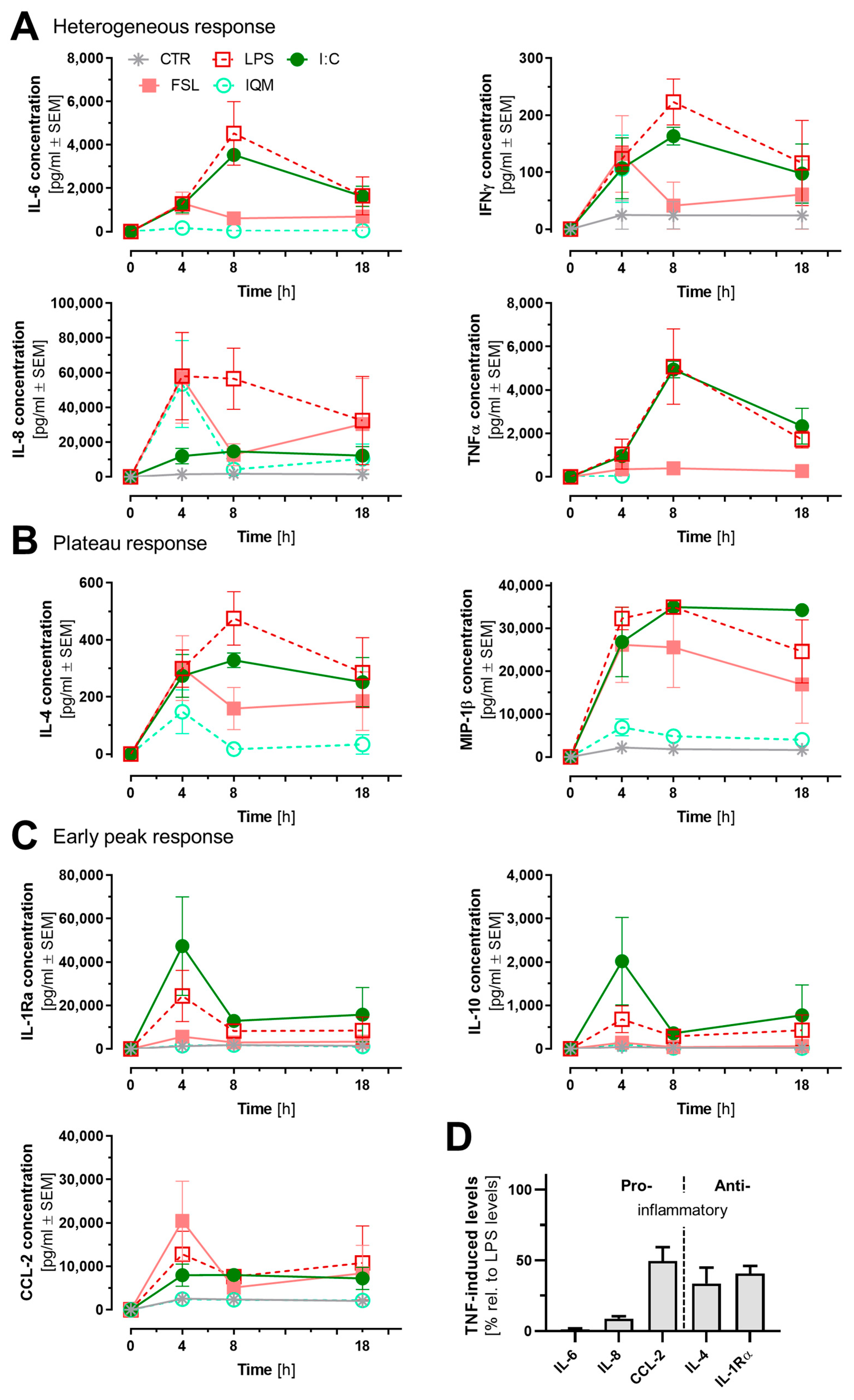
3.4. Release of Inflammatory Mediators from Microglia-like Cells
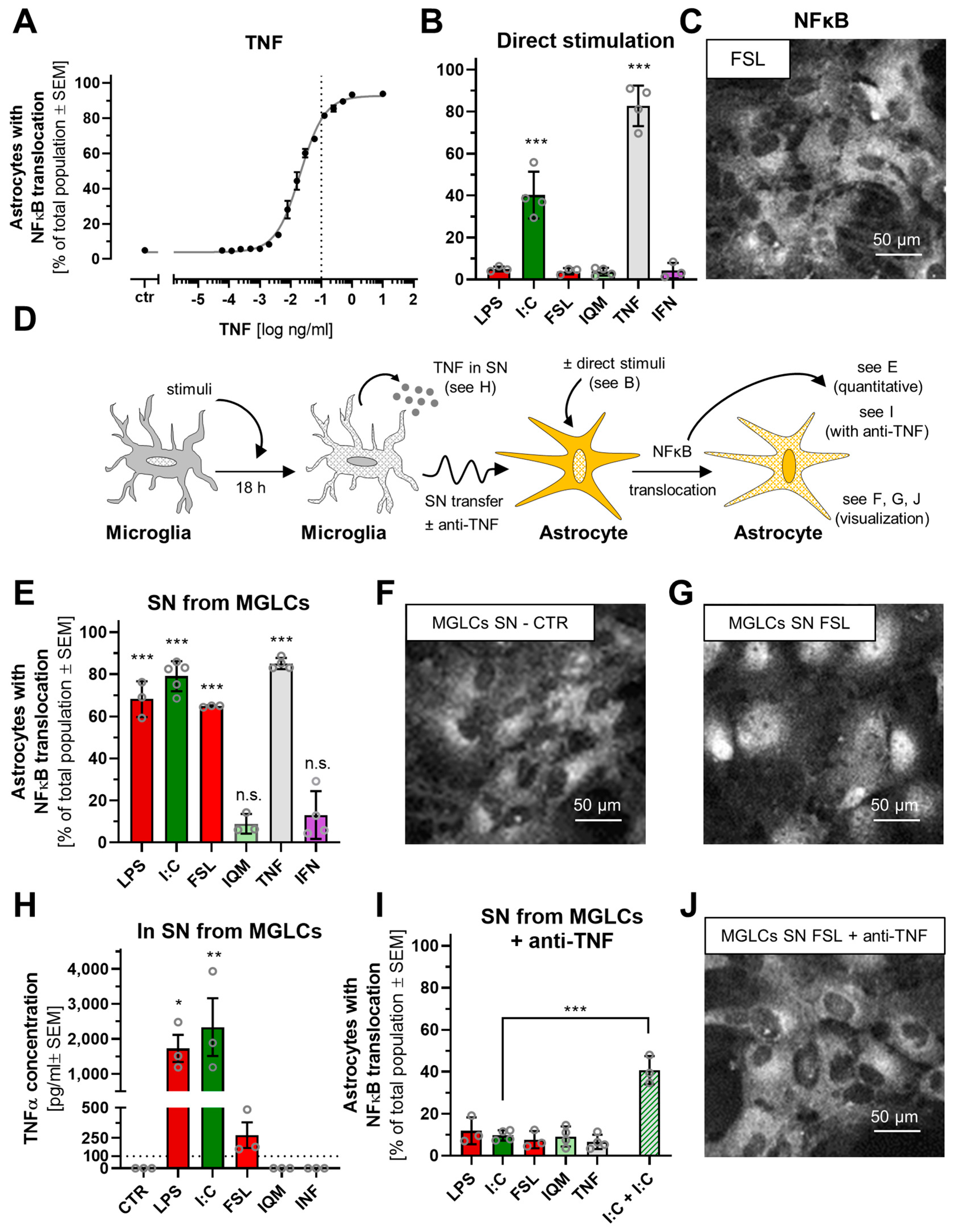
3.5. Differential Astrocyte Activation by Microglia-like Cells Exposed to Different Inflammogens
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CNS | Central nervous system |
| DEG | Differentially expressed gene |
| DEX | Dexamethasone |
| DoC | Day of culture |
| EB | Embryoid body |
| FSL | FSL-1 |
| I:C | Poly(I:C) |
| IFN | Interferon-γ |
| iPSC | Induced pluripotent stem cell |
| IQM | Imiquimod |
| LLOQ | Lower limit of quantification |
| LPS | Lipopolysaccharide |
| MGLC | Microglia-like cell |
| NFκB | Nuclear factor kappa-light-chain-enhancer of activated B cell |
| PCA | Principical component analysis |
| PreMacs | Macrophage precursor cells |
| SN | Supernatant |
| TF | Transcription factor |
| TLR | Toll-like receptor |
| TNF | Tumor necrosis factor-α |
References
- Block, M.L.; Zecca, L.; Hong, J.S. Microglia-mediated neurotoxicity: Uncovering the molecular mechanisms. Nat. Rev. Neurosci. 2007, 8, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Fagiani, F.; Pedrini, E.; Taverna, S.; Brambilla, E.; Murtaj, V.; Podini, P.; Ruffini, F.; Butti, E.; Braccia, C.; Andolfo, A.; et al. A glia-enriched stem cell 3D model of the human brain mimics the glial-immune neurodegenerative phenotypes of multiple sclerosis. Cell Rep. Med. 2024, 5, 101680. [Google Scholar] [CrossRef]
- Fritsche, E.; Tigges, J.; Hartmann, J.; Kapr, J.; Serafini, M.M.; Viviani, B. Neural In Vitro Models for Studying Substances Acting on the Central Nervous System. Handb. Exp. Pharmacol. 2021, 265, 111–141. [Google Scholar] [CrossRef]
- Woodburn, S.C.; Bollinger, J.L.; Wohleb, E.S. The semantics of microglia activation: Neuroinflammation, homeostasis, and stress. J. Neuroinflamm. 2021, 18, 258. [Google Scholar] [CrossRef]
- Yuan, N.Y.; Richards, W.D.; Parham, K.T.; Clark, S.G.; Greuel, K.; Polzin, B.; Smith, S.W.; Lebakken, C.S. Neural organoids incorporating microglia to assess neuroinflammation and toxicities induced by known developmental neurotoxins. Curr. Res. Toxicol. 2025, 9, 100252. [Google Scholar] [CrossRef]
- Zong, C.; Hasegawa, R.; Urushitani, M.; Zhang, L.; Nagashima, D.; Sakurai, T.; Ichihara, S.; Ohsako, S.; Ichihara, G. Role of microglial activation and neuroinflammation in neurotoxicity of acrylamide in vivo and in vitro. Arch. Toxicol. 2019, 93, 2007–2019. [Google Scholar] [CrossRef]
- Tan, Y.L.; Yuan, Y.; Tian, L. Microglial regional heterogeneity and its role in the brain. Mol. Psychiatry 2020, 25, 351–367. [Google Scholar] [CrossRef]
- Mittelbronn, M.; Dietz, K.; Schluesener, H.J.; Meyermann, R. Local distribution of microglia in the normal adult human central nervous system differs by up to one order of magnitude. Acta Neuropathol. 2001, 101, 249–255. [Google Scholar] [CrossRef]
- Perry, V.H.; Gordon, S. Macrophages and microglia in the nervous system. Trends Neurosci. 1988, 11, 273–277. [Google Scholar] [CrossRef] [PubMed]
- Nimmerjahn, A.; Kirchhoff, F.; Helmchen, F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 2005, 308, 1314–1318. [Google Scholar] [CrossRef] [PubMed]
- Sierra, A.; de Castro, F.; Del Río-Hortega, J.; Rafael Iglesias-Rozas, J.; Garrosa, M.; Kettenmann, H. The “Big-Bang” for modern glial biology: Translation and comments on Pío del Río-Hortega 1919 series of papers on microglia. Glia 2016, 64, 1801–1840. [Google Scholar] [CrossRef]
- Ransohoff, R.M.; Cardona, A.E. The myeloid cells of the central nervous system parenchyma. Nature 2010, 468, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Parakalan, R.; Jiang, B.; Nimmi, B.; Janani, M.; Jayapal, M.; Lu, J.; Tay, S.S.; Ling, E.A.; Dheen, S.T. Transcriptome analysis of amoeboid and ramified microglia isolated from the corpus callosum of rat brain. BMC Neurosci. 2012, 13, 64. [Google Scholar] [CrossRef]
- Cherry, J.D.; Olschowka, J.A.; O’Banion, M.K. Neuroinflammation and M2 microglia: The good, the bad, and the inflamed. J. Neuroinflamm. 2014, 11, 98. [Google Scholar] [CrossRef] [PubMed]
- Paolicelli, R.C.; Sierra, A.; Stevens, B.; Tremblay, M.E.; Aguzzi, A.; Ajami, B.; Amit, I.; Audinat, E.; Bechmann, I.; Bennett, M.; et al. Microglia states and nomenclature: A field at its crossroads. Neuron 2022, 110, 3458–3483. [Google Scholar] [CrossRef]
- Murray, P.J.; Allen, J.E.; Biswas, S.K.; Fisher, E.A.; Gilroy, D.W.; Goerdt, S.; Gordon, S.; Hamilton, J.A.; Ivashkiv, L.B.; Lawrence, T.; et al. Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity 2014, 41, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Spreng, A.S.; Brüll, M.; Leisner, H.; Suciu, I.; Leist, M. Distinct and Dynamic Transcriptome Adaptations of iPSC-Generated Astrocytes after Cytokine Stimulation. Cells 2022, 11, 2644. [Google Scholar] [CrossRef]
- Escartin, C.; Galea, E.; Lakatos, A.; O’Callaghan, J.P.; Petzold, G.C.; Serrano-Pozo, A.; Steinhäuser, C.; Volterra, A.; Carmignoto, G.; Agarwal, A.; et al. Reactive astrocyte nomenclature, definitions, and future directions. Nat. Neurosci. 2021, 24, 312–325. [Google Scholar] [CrossRef]
- Edison, P. Astroglial activation: Current concepts and future directions. Alzheimers Dement. 2024, 20, 3034–3053. [Google Scholar] [CrossRef]
- Hanke, M.L.; Kielian, T. Toll-like receptors in health and disease in the brain: Mechanisms and therapeutic potential. Clin. Sci. 2011, 121, 367–387. [Google Scholar] [CrossRef]
- Kettenmann, H.; Hanisch, U.K.; Noda, M.; Verkhratsky, A. Physiology of microglia. Physiol. Rev. 2011, 91, 461–553. [Google Scholar] [CrossRef]
- Fitzgerald, K.A.; Kagan, J.C. Toll-like Receptors and the Control of Immunity. Cell 2020, 180, 1044–1066. [Google Scholar] [CrossRef]
- Dantzer, R.; O’Connor, J.C.; Freund, G.G.; Johnson, R.W.; Kelley, K.W. From inflammation to sickness and depression: When the immune system subjugates the brain. Nat. Rev. Neurosci. 2008, 9, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Mendiola, A.S.; Yan, Z.; Dixit, K.; Johnson, J.R.; Bouhaddou, M.; Meyer-Franke, A.; Shin, M.G.; Yong, Y.; Agrawal, A.; MacDonald, E.; et al. Defining blood-induced microglia functions in neurodegeneration through multiomic profiling. Nat. Immunol. 2023, 24, 1173–1187. [Google Scholar] [CrossRef] [PubMed]
- Bsibsi, M.; Ravid, R.; Gveric, D.; van Noort, J.M. Broad expression of Toll-like receptors in the human central nervous system. J. Neuropathol. Exp. Neurol. 2002, 61, 1013–1021. [Google Scholar] [CrossRef]
- Olson, J.K.; Miller, S.D. Microglia initiate central nervous system innate and adaptive immune responses through multiple TLRs. J. Immunol. 2004, 173, 3916–3924. [Google Scholar] [CrossRef]
- Jung, D.Y.; Lee, H.; Jung, B.Y.; Ock, J.; Lee, M.S.; Lee, W.H.; Suk, K. TLR4, but not TLR2, signals autoregulatory apoptosis of cultured microglia: A critical role of IFN-beta as a decision maker. J. Immunol. 2005, 174, 6467–6476. [Google Scholar] [CrossRef]
- Stöberl, N.; Maguire, E.; Salis, E.; Shaw, B.; Hall-Roberts, H. Human iPSC-derived glia models for the study of neuroinflammation. J. Neuroinflamm. 2023, 20, 231. [Google Scholar] [CrossRef] [PubMed]
- Town, T.; Jeng, D.; Alexopoulou, L.; Tan, J.; Flavell, R.A. Microglia recognize double-stranded RNA via TLR3. J. Immunol. 2006, 176, 3804–3812. [Google Scholar] [CrossRef]
- Akira, S.; Takeda, K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004, 4, 499–511. [Google Scholar] [CrossRef]
- Darnell, J.E., Jr.; Kerr, I.M.; Stark, G.R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 1994, 264, 1415–1421. [Google Scholar] [CrossRef]
- Horvath, C.M. The Jak-STAT pathway stimulated by interferon gamma. Sci. STKE 2004, 2004, tr8. [Google Scholar] [CrossRef] [PubMed]
- Platanias, L.C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005, 5, 375–386. [Google Scholar] [CrossRef]
- Halonen, S.K.; Woods, T.; McInnerney, K.; Weiss, L.M. Microarray analysis of IFN-gamma response genes in astrocytes. J. Neuroimmunol. 2006, 175, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Stark, G.R.; Darnell, J.E., Jr. The JAK-STAT pathway at twenty. Immunity 2012, 36, 503–514. [Google Scholar] [CrossRef]
- Garland, E.F.; Hartnell, I.J.; Boche, D. Microglia and Astrocyte Function and Communication: What Do We Know in Humans? Front. Neurosci. 2022, 16, 824888. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Jiang, J.; Tan, Y.; Chen, S. Microglia in neurodegenerative diseases: Mechanism and potential therapeutic targets. Signal Transduct. Target. Ther. 2023, 8, 359. [Google Scholar] [CrossRef]
- Matejuk, A.; Ransohoff, R.M. Crosstalk Between Astrocytes and Microglia: An Overview. Front. Immunol. 2020, 11, 1416. [Google Scholar] [CrossRef]
- Liu, L.R.; Liu, J.C.; Bao, J.S.; Bai, Q.Q.; Wang, G.Q. Interaction of Microglia and Astrocytes in the Neurovascular Unit. Front. Immunol. 2020, 11, 1024. [Google Scholar] [CrossRef]
- Falsig, J.; van Beek, J.; Hermann, C.; Leist, M. Molecular basis for detection of invading pathogens in the brain. J. Neurosci. Res. 2008, 86, 1434–1447. [Google Scholar] [CrossRef]
- Liddelow, S.A.; Guttenplan, K.A.; Clarke, L.E.; Bennett, F.C.; Bohlen, C.J.; Schirmer, L.; Bennett, M.L.; Münch, A.E.; Chung, W.S.; Peterson, T.C.; et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature 2017, 541, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Hickman, S.E.; Kingery, N.D.; Ohsumi, T.K.; Borowsky, M.L.; Wang, L.C.; Means, T.K.; El Khoury, J. The microglial sensome revealed by direct RNA sequencing. Nat. Neurosci. 2013, 16, 1896–1905. [Google Scholar] [CrossRef] [PubMed]
- Martinez, F.O.; Gordon, S. The M1 and M2 paradigm of macrophage activation: Time for reassessment. F1000Prime Rep. 2014, 6, 13. [Google Scholar] [CrossRef]
- Bjornson-Hooper, Z.B.; Fragiadakis, G.K.; Spitzer, M.H.; Chen, H.; Madhireddy, D.; Hu, K.; Lundsten, K.; McIlwain, D.R.; Nolan, G.P. A Comprehensive Atlas of Immunological Differences Between Humans, Mice, and Non-Human Primates. Front. Immunol. 2022, 13, 867015. [Google Scholar] [CrossRef]
- Seok, J.; Warren, H.S.; Cuenca, A.G.; Mindrinos, M.N.; Baker, H.V.; Xu, W.; Richards, D.R.; McDonald-Smith, G.P.; Gao, H.; Hennessy, L.; et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl. Acad. Sci. USA 2013, 110, 3507–3512. [Google Scholar] [CrossRef]
- Leist, M.; Hartung, T. Reprint: Inflammatory findings on species extrapolations: Humans are definitely no 70-kg mice. Altex 2013, 30, 227–230. [Google Scholar] [CrossRef]
- Copeland, S.; Warren, H.S.; Lowry, S.F.; Calvano, S.E.; Remick, D. Acute inflammatory response to endotoxin in mice and humans. Clin. Diagn. Lab. Immunol. 2005, 12, 60–67. [Google Scholar] [CrossRef]
- Woolf, Z.; Stevenson, T.J.; Lee, K.; Highet, B.; Macapagal Foliaki, J.; Ratiu, R.; Rustenhoven, J.; Correia, J.; Schweder, P.; Heppner, P.; et al. In vitro models of microglia: A comparative study. Sci. Rep. 2025, 15, 15621. [Google Scholar] [CrossRef] [PubMed]
- Nutma, E.; Fancy, N.; Weinert, M.; Tsartsalis, S.; Marzin, M.C.; Muirhead, R.C.J.; Falk, I.; Breur, M.; de Bruin, J.; Hollaus, D.; et al. Translocator protein is a marker of activated microglia in rodent models but not human neurodegenerative diseases. Nat. Commun. 2023, 14, 5247. [Google Scholar] [CrossRef]
- Sabogal-Guáqueta, A.M.; Marmolejo-Garza, A.; Trombetta-Lima, M.; Oun, A.; Hunneman, J.; Chen, T.; Koistinaho, J.; Lehtonen, S.; Kortholt, A.; Wolters, J.C.; et al. Species-specific metabolic reprogramming in human and mouse microglia during inflammatory pathway induction. Nat. Commun. 2023, 14, 6454. [Google Scholar] [CrossRef]
- Haenseler, W.; Sansom, S.N.; Buchrieser, J.; Newey, S.E.; Moore, C.S.; Nicholls, F.J.; Chintawar, S.; Schnell, C.; Antel, J.P.; Allen, N.D.; et al. A Highly Efficient Human Pluripotent Stem Cell Microglia Model Displays a Neuronal-Co-culture-Specific Expression Profile and Inflammatory Response. Stem Cell Rep. 2017, 8, 1727–1742. [Google Scholar] [CrossRef]
- Muffat, J.; Li, Y.; Yuan, B.; Mitalipova, M.; Omer, A.; Corcoran, S.; Bakiasi, G.; Tsai, L.H.; Aubourg, P.; Ransohoff, R.M.; et al. Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat. Med. 2016, 22, 1358–1367. [Google Scholar] [CrossRef]
- Abud, E.M.; Ramirez, R.N.; Martinez, E.S.; Healy, L.M.; Nguyen, C.H.H.; Newman, S.A.; Yeromin, A.V.; Scarfone, V.M.; Marsh, S.E.; Fimbres, C.; et al. iPSC-Derived Human Microglia-like Cells to Study Neurological Diseases. Neuron 2017, 94, 278–293.e279. [Google Scholar] [CrossRef]
- Speicher, A.M.; Wiendl, H.; Meuth, S.G.; Pawlowski, M. Generating microglia from human pluripotent stem cells: Novel in vitro models for the study of neurodegeneration. Mol. Neurodegener. 2019, 14, 46. [Google Scholar] [CrossRef]
- Reich, M.; Paris, I.; Ebeling, M.; Dahm, N.; Schweitzer, C.; Reinhardt, D.; Schmucki, R.; Prasad, M.; Köchl, F.; Leist, M.; et al. Alzheimer’s Risk Gene TREM2 Determines Functional Properties of New Type of Human iPSC-Derived Microglia. Front. Immunol. 2020, 11, 617860. [Google Scholar] [CrossRef]
- Washer, S.J.; Perez-Alcantara, M.; Chen, Y.; Steer, J.; James, W.S.; Trynka, G.; Bassett, A.R.; Cowley, S.A. Single-cell transcriptomics defines an improved, validated monoculture protocol for differentiation of human iPSC to microglia. Sci. Rep. 2022, 12, 19454. [Google Scholar] [CrossRef]
- Henn, A.; Kirner, S.; Leist, M. TLR2 hypersensitivity of astrocytes as functional consequence of previous inflammatory episodes. J. Immunol. 2011, 186, 3237–3247. [Google Scholar] [CrossRef]
- Gutbier, S.; Wanke, F.; Dahm, N.; Rümmelin, A.; Zimmermann, S.; Christensen, K.; Köchl, F.; Rautanen, A.; Hatje, K.; Geering, B.; et al. Large-Scale Production of Human iPSC-Derived Macrophages for Drug Screening. Int. J. Mol. Sci. 2020, 21, 4808. [Google Scholar] [CrossRef]
- Brüll, M.; Geese, N.; Celardo, I.; Laumann, M.; Leist, M. Preparation of Viable Human Neurites for Neurobiological and Neurodegeneration Studies. Cells 2024, 13, 242. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, M.; Hartmann, H.; Jakobi, M.; März, L.; Bichmann, L.; Freudenmann, L.K.; Mühlenbruch, L.; Segan, S.; Rammensee, H.G.; Schneiderhan-Marra, N.; et al. The Impact of Biomaterial Cell Contact on the Immunopeptidome. Front. Bioeng. Biotechnol. 2020, 8, 571294. [Google Scholar] [CrossRef] [PubMed]
- Carson, R.T.; Vignali, D.A. Simultaneous quantitation of 15 cytokines using a multiplexed flow cytometric assay. J. Immunol. Methods 1999, 227, 41–52. [Google Scholar] [CrossRef]
- Klima, S.; Brüll, M.; Spreng, A.S.; Suciu, I.; Falt, T.; Schwamborn, J.C.; Waldmann, T.; Karreman, C.; Leist, M. A human stem cell-derived test system for agents modifying neuronal N-methyl-D-aspartate-type glutamate receptor Ca(2+)-signalling. Arch. Toxicol. 2021, 95, 1703–1722. [Google Scholar] [CrossRef] [PubMed]
- Yeakley, J.M.; Shepard, P.J.; Goyena, D.E.; VanSteenhouse, H.C.; McComb, J.D.; Seligmann, B.E. A trichostatin A expression signature identified by TempO-Seq targeted whole transcriptome profiling. PLoS ONE 2017, 12, e0178302. [Google Scholar] [CrossRef]
- Barbie, D.A.; Tamayo, P.; Boehm, J.S.; Kim, S.Y.; Moody, S.E.; Dunn, I.F.; Schinzel, A.C.; Sandy, P.; Meylan, E.; Scholl, C.; et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 2009, 462, 108–112. [Google Scholar] [CrossRef]
- Verheijen, M.C.; Meier, M.J.; Asensio, J.O.; Gant, T.W.; Tong, W.; Yauk, C.L.; Caiment, F. R-ODAF: Omics data analysis framework for regulatory application. Regul. Toxicol. Pharmacol. 2022, 131, 105143. [Google Scholar] [CrossRef]
- Harrill, J.A.; Everett, L.J.; Haggard, D.E.; Sheffield, T.; Bundy, J.L.; Willis, C.M.; Thomas, R.S.; Shah, I.; Judson, R.S. High-Throughput Transcriptomics Platform for Screening Environmental Chemicals. Toxicol. Sci. 2021, 181, 68–89. [Google Scholar] [CrossRef]
- Becht, E.; McInnes, L.; Healy, J.; Dutertre, C.A.; Kwok, I.W.H.; Ng, L.G.; Ginhoux, F.; Newell, E.W. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 2018, 37, 38–44. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Badia, I.M.P.; Vélez Santiago, J.; Braunger, J.; Geiss, C.; Dimitrov, D.; Müller-Dott, S.; Taus, P.; Dugourd, A.; Holland, C.H.; Ramirez Flores, R.O.; et al. decoupleR: Ensemble of computational methods to infer biological activities from omics data. Bioinform. Adv. 2022, 2, vbac016. [Google Scholar] [CrossRef] [PubMed]
- Henn, A.; Lund, S.; Hedtjärn, M.; Schrattenholz, A.; Pörzgen, P.; Leist, M. The suitability of BV2 cells as alternative model system for primary microglia cultures or for animal experiments examining brain inflammation. Altex 2009, 26, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Wolfbeisz, C.; Leist, M. Differential Responses of Human iPSC-Derived Microglia to the Stimulation with Diverse Inflammogens—Supplementary Data. 2025. Available online: https://doi.org/10.5281/zenodo.17199105 (accessed on 25 September 2025).
- Jurga, A.M.; Paleczna, M.; Kuter, K.Z. Overview of General and Discriminating Markers of Differential Microglia Phenotypes. Front. Cell. Neurosci. 2020, 14, 198. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Taylor, N.; Yao, X.; Bhattacharya, A. Mouse primary microglia respond differently to LPS and poly(I:C) in vitro. Sci. Rep. 2021, 11, 10447. [Google Scholar] [CrossRef]
- Tiemeijer, B.M.; Heester, S.; Sturtewagen, A.Y.W.; Smits, A.; Tel, J. Single-cell analysis reveals TLR-induced macrophage heterogeneity and quorum sensing dictate population wide anti-inflammatory feedback in response to LPS. Front. Immunol. 2023, 14, 1135223. [Google Scholar] [CrossRef] [PubMed]
- McInnes, L.; Healy, J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv 2018. [Google Scholar] [CrossRef]
- Allaoui, M.; Kherfi, M.L.; Cheriet, A. Considerably Improving Clustering Algorithms Using UMAP Dimensionality Reduction Technique: A Comparative Study. Image Signal Process. 2020, 12119, 317–325. [Google Scholar]
- Yang, Y.; Sun, H.; Zhang, Y.; Zhang, T.; Gong, J.; Wei, Y.; Duan, Y.G.; Shu, M.; Yang, Y.; Wu, D.; et al. Dimensionality reduction by UMAP reinforces sample heterogeneity analysis in bulk transcriptomic data. Cell Rep. 2021, 36, 109442. [Google Scholar] [CrossRef]
- Das, A.; Chai, J.C.; Kim, S.H.; Lee, Y.S.; Park, K.S.; Jung, K.H.; Chai, Y.G. Transcriptome sequencing of microglial cells stimulated with TLR3 and TLR4 ligands. BMC Genom. 2015, 16, 517. [Google Scholar] [CrossRef]
- Bormann, D.; Copic, D.; Klas, K.; Direder, M.; Riedl, C.J.; Testa, G.; Kühtreiber, H.; Poreba, E.; Hametner, S.; Golabi, B.; et al. Exploring the heterogeneous transcriptional response of the CNS to systemic LPS and Poly(I:C). Neurobiol. Dis. 2023, 188, 106339. [Google Scholar] [CrossRef] [PubMed]
- Ros-Bernal, F.; Hunot, S.; Herrero, M.T.; Parnadeau, S.; Corvol, J.C.; Lu, L.; Alvarez-Fischer, D.; Carrillo-de Sauvage, M.A.; Saurini, F.; Coussieu, C.; et al. Microglial glucocorticoid receptors play a pivotal role in regulating dopaminergic neurodegeneration in parkinsonism. Proc. Natl. Acad. Sci. USA 2011, 108, 6632–6637. [Google Scholar] [CrossRef] [PubMed]
- Frank, M.G.; Thompson, B.M.; Watkins, L.R.; Maier, S.F. Glucocorticoids mediate stress-induced priming of microglial pro-inflammatory responses. Brain Behav. Immun. 2012, 26, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Tong, Z.B.; Braisted, J.; Chu, P.H.; Gerhold, D. The MT1G Gene in LUHMES Neurons Is a Sensitive Biomarker of Neurotoxicity. Neurotox. Res. 2020, 38, 967–978. [Google Scholar] [CrossRef]
- Tong, Z.B.; Sakamuru, S.; Travers, J.; Xu, T.; Yang, S.; Xia, M.; Simeonov, A.; Huang, R.; Gerhold, D. MT1G activation in dopaminergic neurons identifies chelators and their relationships to cytotoxicity. SLAS Discov. 2025, 35, 100244. [Google Scholar] [CrossRef]
- Yang, R.; Roshani, D.; Gao, B.; Li, P.; Shang, N. Metallothionein: A Comprehensive Review of Its Classification, Structure, Biological Functions, and Applications. Antioxidants 2024, 13, 825. [Google Scholar] [CrossRef]
- Guo, Q.; Jin, Y.; Chen, X.; Ye, X.; Shen, X.; Lin, M.; Zeng, C.; Zhou, T.; Zhang, J. NF-κB in biology and targeted therapy: New insights and translational implications. Signal Transduct. Target. Ther. 2024, 9, 53. [Google Scholar] [CrossRef]
- Patel, P.; Drayman, N.; Liu, P.; Bilgic, M.; Tay, S. Computer vision reveals hidden variables underlying NF-κB activation in single cells. Sci. Adv. 2021, 7, eabg4135. [Google Scholar] [CrossRef]
- Lund, S.; Porzgen, P.; Mortensen, A.L.; Hasseldam, H.; Bozyczko-Coyne, D.; Morath, S.; Hartung, T.; Bianchi, M.; Ghezzi, P.; Bsibsi, M.; et al. Inhibition of microglial inflammation by the MLK inhibitor CEP-1347. J. Neurochem. 2005, 92, 1439–1451. [Google Scholar] [CrossRef]
- Dueck, H.; Eberwine, J.; Kim, J. Variation is function: Are single cell differences functionally important?: Testing the hypothesis that single cell variation is required for aggregate function. Bioessays 2016, 38, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Biswas, K. Microglia mediated neuroinflammation in neurodegenerative diseases: A review on the cell signaling pathways involved in microglial activation. J. Neuroimmunol. 2023, 383, 578180. [Google Scholar] [CrossRef]
- Li, J.; Shui, X.; Sun, R.; Wan, L.; Zhang, B.; Xiao, B.; Luo, Z. Microglial Phenotypic Transition: Signaling Pathways and Influencing Modulators Involved in Regulation in Central Nervous System Diseases. Front. Cell Neurosci. 2021, 15, 736310. [Google Scholar] [CrossRef] [PubMed]
- Knappe, E.; Rudolph, F.; Klein, C.; Seibler, P. Cytokine Profiling in Human iPSC-Derived Dopaminergic Neuronal and Microglial Cultures. Cells 2023, 12, 2535. [Google Scholar] [CrossRef]
- Tamai, R.; Sugawara, S.; Takeuchi, O.; Akira, S.; Takada, H. Synergistic effects of lipopolysaccharide and interferon-gamma in inducing interleukin-8 production in human monocytic THP-1 cells is accompanied by up-regulation of CD14, Toll-like receptor 4, MD-2 and MyD88 expression. J. Endotoxin Res. 2003, 9, 145–153. [Google Scholar] [CrossRef]
- Kang, K.; Bachu, M.; Park, S.H.; Kang, K.; Bae, S.; Park-Min, K.H.; Ivashkiv, L.B. IFN-γ selectively suppresses a subset of TLR4-activated genes and enhancers to potentiate macrophage activation. Nat. Commun. 2019, 10, 3320. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Giannopoulou, E.G.; Chan, C.H.; Park, S.H.; Gong, S.; Chen, J.; Hu, X.; Elemento, O.; Ivashkiv, L.B. Synergistic activation of inflammatory cytokine genes by interferon-γ-induced chromatin remodeling and toll-like receptor signaling. Immunity 2013, 39, 454–469. [Google Scholar] [CrossRef]
- Su, X.; Yu, Y.; Zhong, Y.; Giannopoulou, E.G.; Hu, X.; Liu, H.; Cross, J.R.; Rätsch, G.; Rice, C.M.; Ivashkiv, L.B. Interferon-γ regulates cellular metabolism and mRNA translation to potentiate macrophage activation. Nat. Immunol. 2015, 16, 838–849. [Google Scholar] [CrossRef]
- Falsig, J.; Pörzgen, P.; Lotharius, J.; Leist, M. Specific modulation of astrocyte inflammation by inhibition of mixed lineage kinases with CEP-1347. J. Immunol. 2004, 173, 2762–2770. [Google Scholar] [CrossRef]
- Kuegler, P.B.; Baumann, B.A.; Zimmer, B.; Keller, S.; Marx, A.; Kadereit, S.; Leist, M. GFAP-independent inflammatory competence and trophic functions of astrocytes generated from murine embryonic stem cells. Glia 2012, 60, 218–228. [Google Scholar] [CrossRef] [PubMed]
- Kleiderman, S.; Sá, J.V.; Teixeira, A.P.; Brito, C.; Gutbier, S.; Evje, L.G.; Hadera, M.G.; Glaab, E.; Henry, M.; Sachinidis, A.; et al. Functional and phenotypic differences of pure populations of stem cell-derived astrocytes and neuronal precursor cells. Glia 2016, 64, 695–715. [Google Scholar] [CrossRef] [PubMed]
- Lehnardt, S.; Lachance, C.; Patrizi, S.; Lefebvre, S.; Follett, P.L.; Jensen, F.E.; Rosenberg, P.A.; Volpe, J.J.; Vartanian, T. The toll-like receptor TLR4 is necessary for lipopolysaccharide-induced oligodendrocyte injury in the CNS. J. Neurosci. 2002, 22, 2478–2486. [Google Scholar] [CrossRef]
- Tarassishin, L.; Suh, H.S.; Lee, S.C. LPS and IL-1 differentially activate mouse and human astrocytes: Role of CD14. Glia 2014, 62, 999–1013. [Google Scholar] [CrossRef]
- Batiuk, M.Y.; Martirosyan, A.; Wahis, J.; de Vin, F.; Marneffe, C.; Kusserow, C.; Koeppen, J.; Viana, J.F.; Oliveira, J.F.; Voet, T.; et al. Identification of region-specific astrocyte subtypes at single cell resolution. Nat. Commun. 2020, 11, 1220. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Acioglu, C.; Heary, R.F.; Elkabes, S. Role of astroglial toll-like receptors (TLRs) in central nervous system infections, injury and neurodegenerative diseases. Brain Behav. Immun. 2021, 91, 740–755. [Google Scholar] [CrossRef]
- Trudler, D.; Nazor, K.L.; Eisele, Y.S.; Grabauskas, T.; Dolatabadi, N.; Parker, J.; Sultan, A.; Zhong, Z.; Goodwin, M.S.; Levites, Y.; et al. Soluble α-synuclein-antibody complexes activate the NLRP3 inflammasome in hiPSC-derived microglia. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef]
- Wallach, T.; Raden, M.; Hinkelmann, L.; Brehm, M.; Rabsch, D.; Weidling, H.; Krüger, C.; Kettenmann, H.; Backofen, R.; Lehnardt, S. Distinct SARS-CoV-2 RNA fragments activate Toll-like receptors 7 and 8 and induce cytokine release from human macrophages and microglia. Front. Immunol. 2022, 13, 1066456. [Google Scholar] [CrossRef]
- Wang, P.; Liu, J.B.; Wang, X.; Meng, F.Z.; Xiao, Q.H.; Liu, L.; Zhu, J.; Hu, W.H.; Ho, W.Z. Activation of Toll-like receptor 3 inhibits HIV infection of human iPSC-derived microglia. J. Med. Virol. 2023, 95, e29217. [Google Scholar] [CrossRef]
- Kim, W.G.; Mohney, R.P.; Wilson, B.; Jeohn, G.H.; Liu, B.; Hong, J.S. Regional difference in susceptibility to lipopolysaccharide-induced neurotoxicity in the rat brain: Role of microglia. J. Neurosci. 2000, 20, 6309–6316. [Google Scholar] [CrossRef]
- Lee, J.W.; Lee, Y.K.; Yuk, D.Y.; Choi, D.Y.; Ban, S.B.; Oh, K.W.; Hong, J.T. Neuro-inflammation induced by lipopolysaccharide causes cognitive impairment through enhancement of beta-amyloid generation. J. Neuroinflamm. 2008, 5, 37. [Google Scholar] [CrossRef]
- Catorce, M.N.; Gevorkian, G. LPS-induced Murine Neuroinflammation Model: Main Features and Suitability for Pre-clinical Assessment of Nutraceuticals. Curr. Neuropharmacol. 2016, 14, 155–164. [Google Scholar] [CrossRef]
- Mingo, Y.B.; Gabele, L.; Lonnemann, N.; Brône, B.; Korte, M.; Hosseini, S. The effects of urolithin A on poly I:C-induced microglial activation. Front. Cell Neurosci. 2024, 18, 1343562. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Wang, H.; Yin, Y. Microglia Polarization from M1 to M2 in Neurodegenerative Diseases. Front. Aging Neurosci. 2022, 14, 815347. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wolfbeisz, C.; Suess, J.; Dreser, N.; Leisner, H.; Brüll, M.; Fandrich, M.; Schneiderhan-Marra, N.; Poetz, O.; Hartung, T.; Leist, M. Differential Responses of Human iPSC-Derived Microglia to Stimulation with Diverse Inflammogens. Cells 2025, 14, 1687. https://doi.org/10.3390/cells14211687
Wolfbeisz C, Suess J, Dreser N, Leisner H, Brüll M, Fandrich M, Schneiderhan-Marra N, Poetz O, Hartung T, Leist M. Differential Responses of Human iPSC-Derived Microglia to Stimulation with Diverse Inflammogens. Cells. 2025; 14(21):1687. https://doi.org/10.3390/cells14211687
Chicago/Turabian StyleWolfbeisz, Chiara, Julian Suess, Nadine Dreser, Heidrun Leisner, Markus Brüll, Madeleine Fandrich, Nicole Schneiderhan-Marra, Oliver Poetz, Thomas Hartung, and Marcel Leist. 2025. "Differential Responses of Human iPSC-Derived Microglia to Stimulation with Diverse Inflammogens" Cells 14, no. 21: 1687. https://doi.org/10.3390/cells14211687
APA StyleWolfbeisz, C., Suess, J., Dreser, N., Leisner, H., Brüll, M., Fandrich, M., Schneiderhan-Marra, N., Poetz, O., Hartung, T., & Leist, M. (2025). Differential Responses of Human iPSC-Derived Microglia to Stimulation with Diverse Inflammogens. Cells, 14(21), 1687. https://doi.org/10.3390/cells14211687