The Role of the Cell Surface Heparan Sulfate Proteoglycan Syndecan-3 in Breast Cancer Pathophysiology
Abstract
1. Introduction
2. Materials and Methods
2.1. The Human Protein Atlas, TNM Plot, and Kaplan–Meier Plot Analysis
2.2. Cell Culture
2.3. siRNA Transfection
2.4. Flow Cytometry
2.5. Immunofluorescence
2.6. MTT Cell Viability Assay
2.7. Cell Cycle Analysis
2.8. Hanging Drop Assay for 3D Sphere Formation
2.9. Cell Migration Assay
2.10. Wound-Healing Assay
2.11. Total RNA Extraction and Quantitative Real-Time PCR
2.12. Western Blotting
2.13. In-Silico Protein Interaction Network Analysis
2.14. Statistical Analysis
3. Results
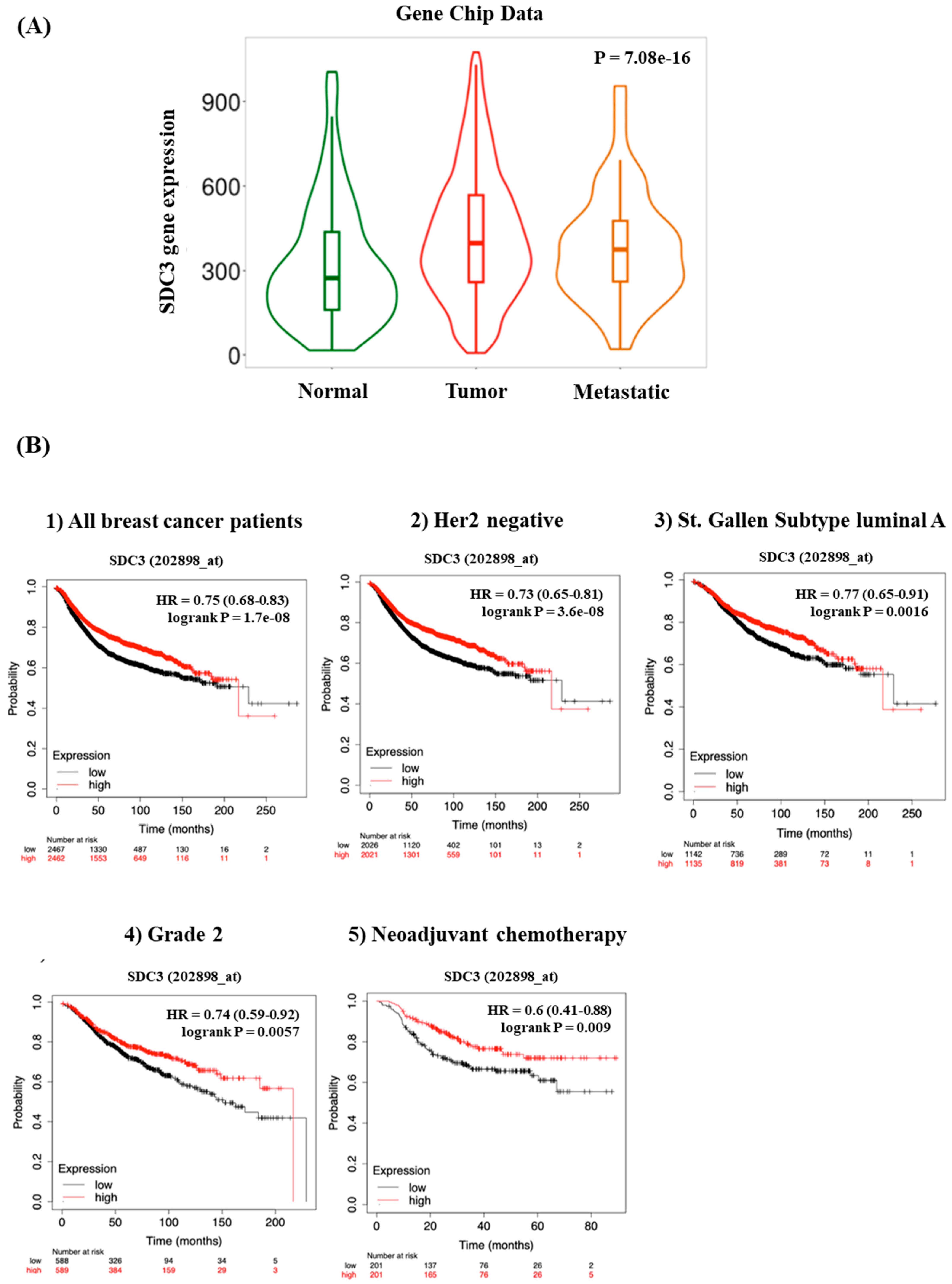
3.1. SDC3 Is Overexpressed in Breast Cancer
3.2. SDC3 Expression Affects the Prognosis and Survival of Breast Cancer Patients
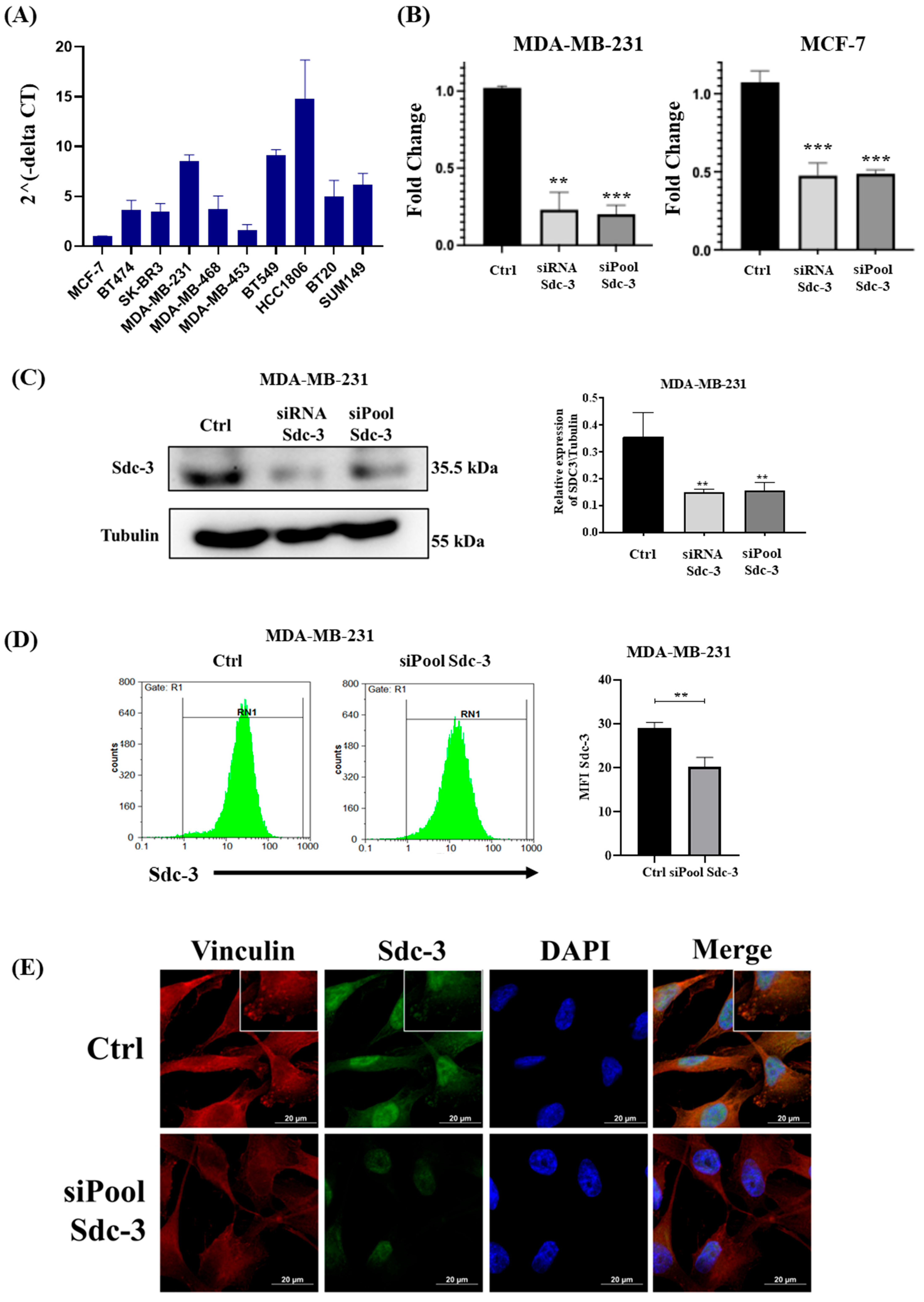
3.3. SDC3 RNA Expression Varies in Breast Cancer Cell Lines of Different Classification
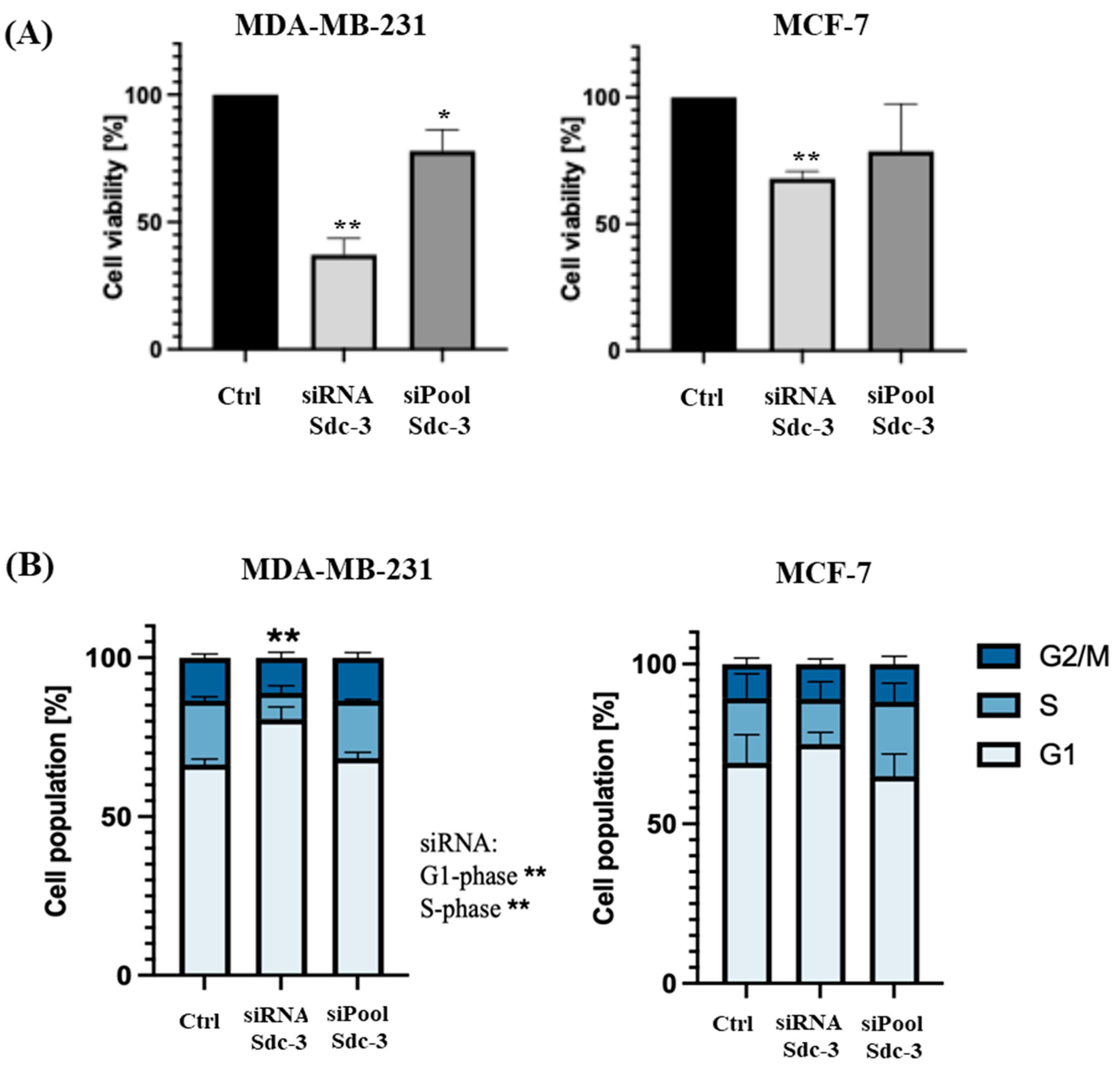
3.4. SDC3 Depletion Affects the Metabolic Activity and Cell Cycle of Human MDA-MB-231 and MCF-7 Breast Cancer Cells
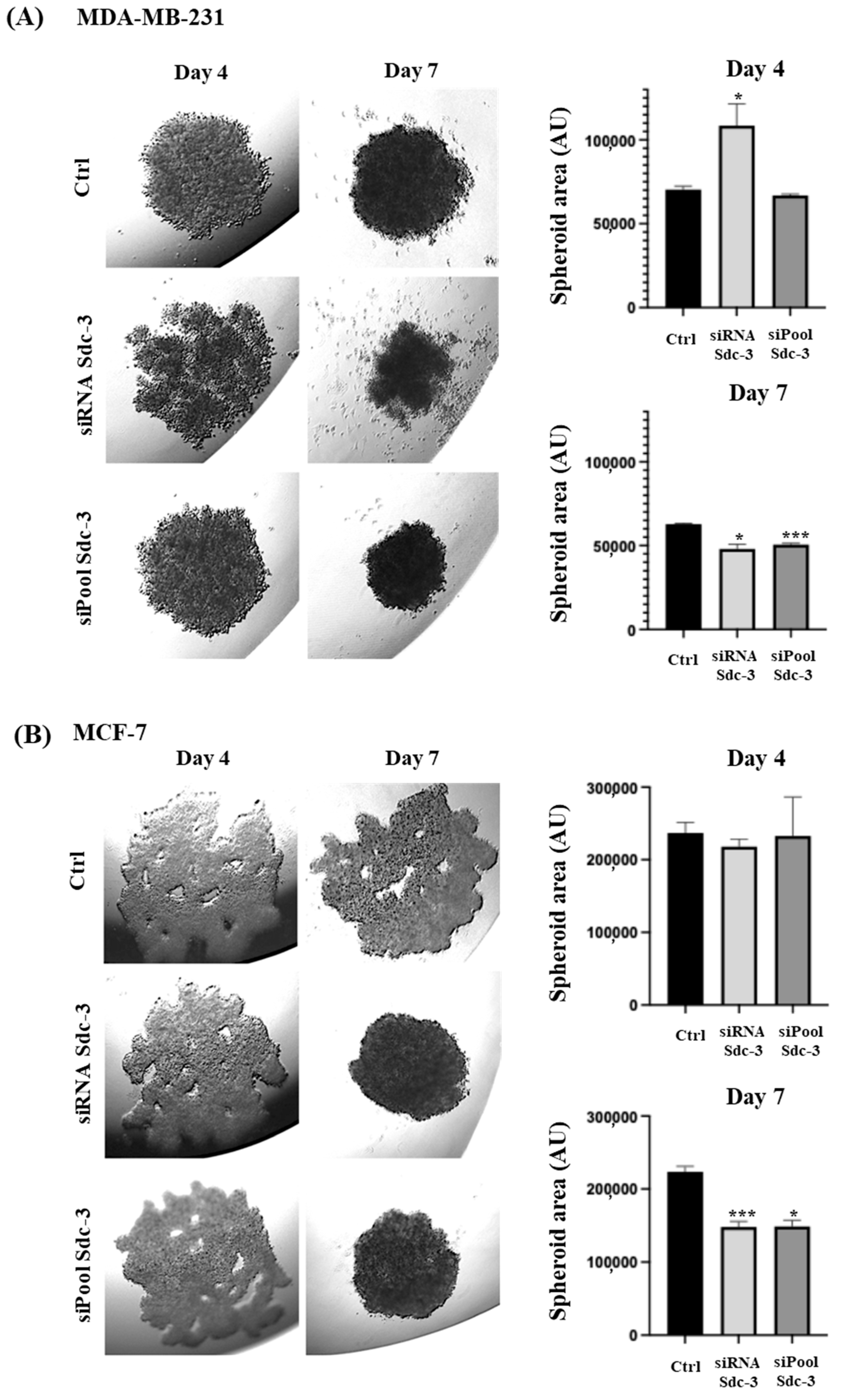
3.5. SDC3 Depletion Affects Three-Dimensional Spheroid Growth of Human MDA-MB-231 and MCF-7 Breast Cancer Cells
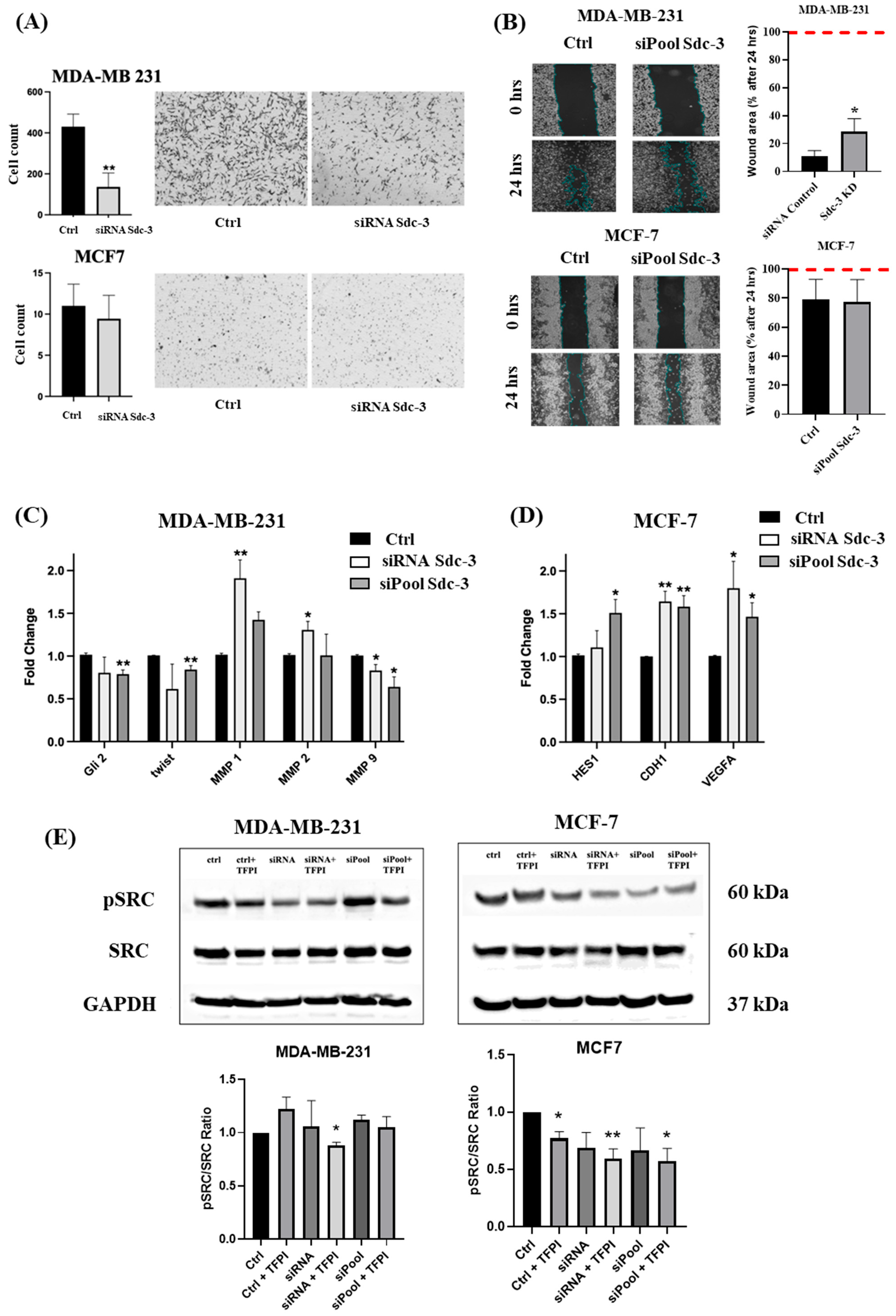
3.6. SDC3 Depletion Affects Cell Migration of Human MDA-MB-231 and MCF-7 Breast Cancer Cells
3.7. SDC3 Depletion Affects the RNA Expression of Target Genes Associated with Relevant Signaling Pathways in Breast Cancer
3.8. SDC3 Depletion and TFPI Treatment Synergistically Affect the Activation of SRC of Human MDA-MB-231 and MCF-7 Breast Cancer Cells
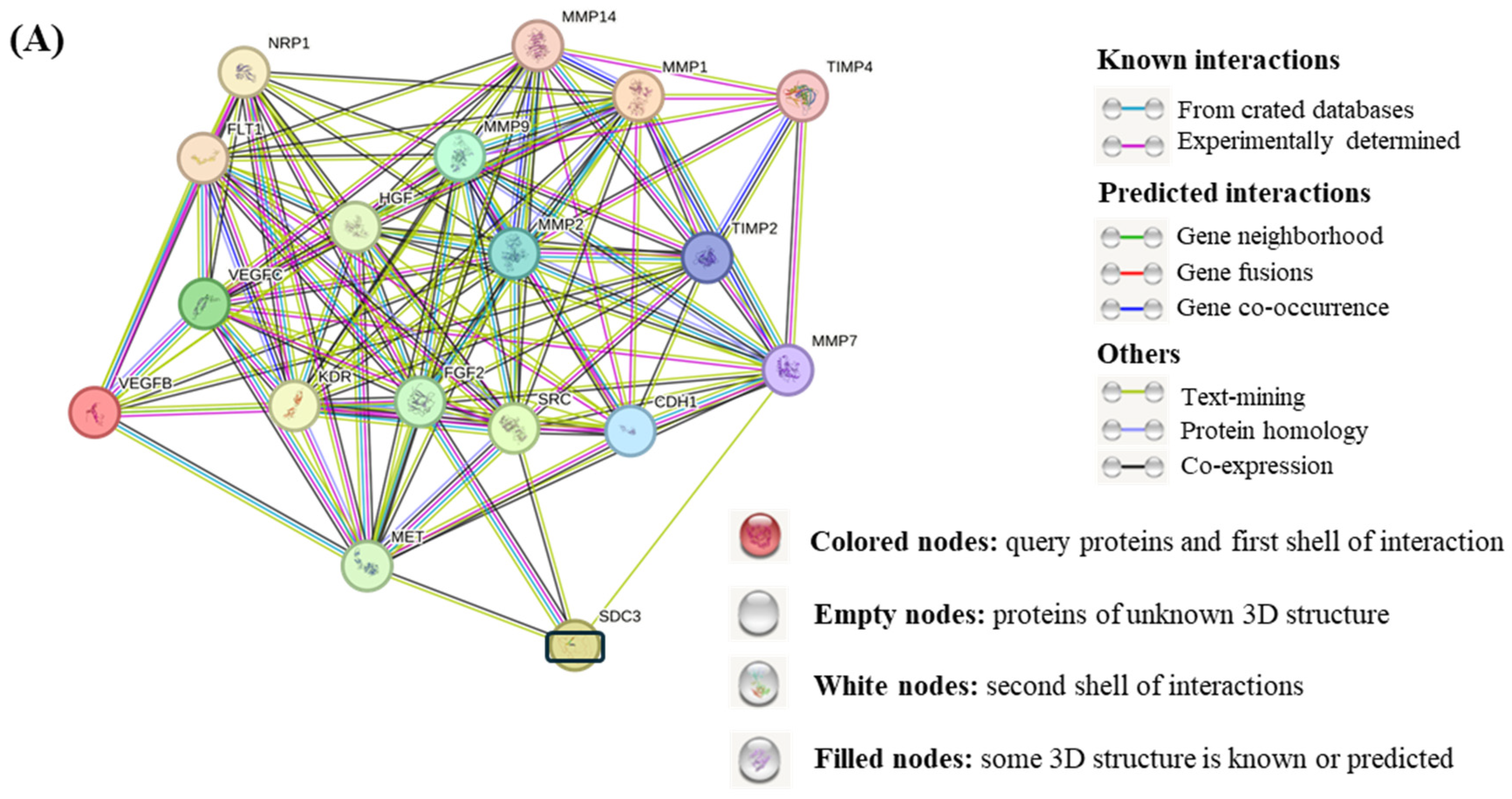
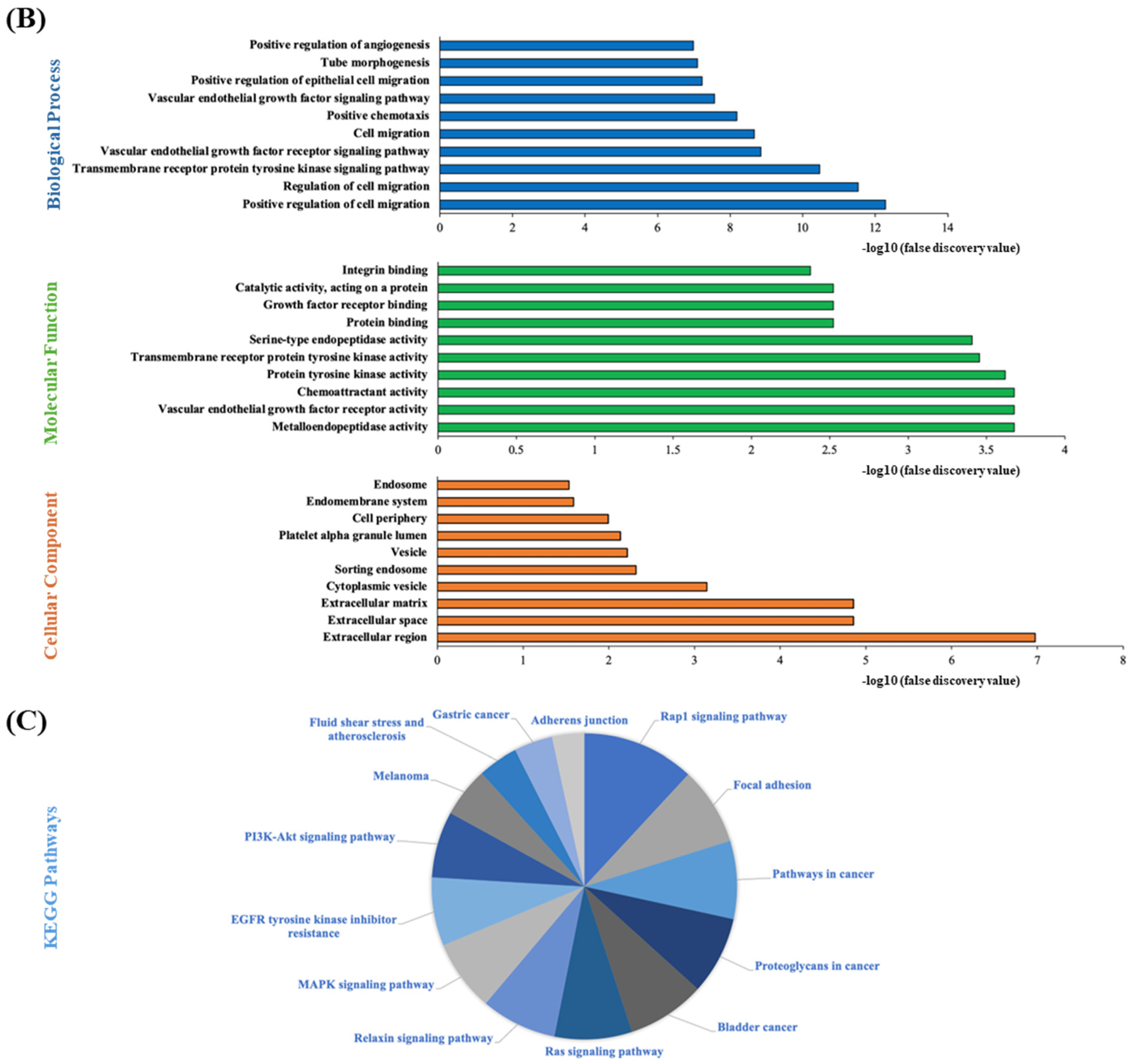
3.9. STRING Functional Enrichment Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Łukasiewicz, S.; Czeczelewski, M.; Forma, A.; Baj, J.; Sitarz, R.; Stanisławek, A. Breast Cancer—Epidemiology, Risk Factors, Classification, Prognostic Markers, and Current Treatment Strategies—An Updated Review. Cancers 2021, 13, 4287. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X. Molecular Classification of Breast Cancer Relevance and Challenges. Arch. Pathol. Lab. Med. 2023, 147, 46–51. [Google Scholar] [CrossRef]
- Smolarz, B.; Nowak, A.Z.; Romanowicz, H. Breast Cancer—Epidemiology, Classification, Pathogenesis and Treatment (Review of Literature). Cancers 2022, 14, 2569. [Google Scholar] [CrossRef]
- Piperigkou, Z.; Mangani, S.; Koletsis, N.E.; Koutsakis, C.; Mastronikolis, N.S.; Franchi, M.; Karamanos, N.K. Principal mechanisms of extracellular matrix-mediated cell–cell communication in physiological and tumor microenvironments. FEBS J. 2025. [Google Scholar] [CrossRef]
- Theocharis, A.D.; Skandalis, S.S.; Gialeli, C.; Karamanos, N.K. Extracellular matrix structure. Adv. Drug Deliv. Rev. 2016, 97, 4–27. [Google Scholar] [CrossRef] [PubMed]
- Pickup, M.W.; Mouw, J.K.; Weaver, V.M. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 2014, 15, 1243–1253. [Google Scholar] [CrossRef]
- Lu, P.; Weaver, V.M.; Werb, Z. The extracellular matrix: A dynamic niche in cancer progression. J. Cell Biol. 2012, 196, 395–406. [Google Scholar] [CrossRef]
- Schaefer, L.; Schaefer, R.M. Proteoglycans: From structural compounds to signaling molecules. Cell Tissue Res. 2010, 339, 237–246. [Google Scholar] [CrossRef]
- Wei, J.; Hu, M.; Huang, K.; Lin, S.; Du, H. Molecular Sciences Roles of Proteoglycans and Glycosaminoglycans in Cancer Development and Progression. Int. J. Mol. Sci. 2020, 21, 5983. [Google Scholar] [CrossRef]
- Mouw, J.K.; Ou, G.; Weaver, V.M. Extracellular matrix assembly: A multiscale deconstruction. Nat. Rev. Mol. Cell Biol. 2014, 15, 771–785. [Google Scholar] [CrossRef]
- Couchman, J.R. Transmembrane signaling proteoglycans. Annu. Rev. Cell Dev. Biol. 2010, 26, 89–114. [Google Scholar] [CrossRef] [PubMed]
- Hassan, N.; Greve, B.; Espinoza-Sánchez, N.A.; Götte, M. Cell-surface heparan sulfate proteoglycans as multifunctional integrators of signaling in cancer. Cell Signal. 2021, 77, 109822. [Google Scholar] [CrossRef]
- Kim, S.; Yang, H.; Cho, S.; Jang, Y.; Han, I.-O.; Oh, E.-S. Correlation of syndecan gene amplification with metastatic potential and clinical outcomes in carcinomas. Am. J. Physiol. Cell Physiol. 2024, 327, C380–C386. [Google Scholar] [CrossRef]
- Motta, J.M.; Hassan, H.; Ibrahim, S.A. Revisiting the Syndecans: Master Signaling Regulators with Prognostic and Targetable Therapeutic Values in Breast Carcinoma. Cancers 2023, 15, 1794. [Google Scholar] [CrossRef] [PubMed]
- Hillemeyer, L.; Espinoza-Sanchez, N.A.; Greve, B.; Hassan, N.; Chelariu-Raicu, A.; Kiesel, L.; Götte, M. The Cell Surface Heparan Sulfate Proteoglycan Syndecan-3 Promotes Ovarian Cancer Pathogenesis. Int. J. Mol. Sci. 2022, 23, 5793. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Zhang, L.L.; Huang, X.M.; Li, W.Y.; Gao, S.G. Pleiotrophin and N-syndecan promote perineural invasion and tumor progression in an orthotopic mouse model of pancreatic cancer. World J. Gastroenterol. 2017, 23, 3907–3914. [Google Scholar] [CrossRef]
- Yamada, Y.; Arai, T.; Kojima, S.; Sugawara, S.; Kato, M.; Okato, A.; Yamazaki, K.; Naya, Y.; Ichikawa, T.; Seki, N. Regulation of antitumor miR-144-5p targets oncogenes: Direct regulation of syndecan-3 and its clinical significance. Cancer Sci. 2018, 109, 2919–2936. [Google Scholar] [CrossRef]
- Digre, A.; Lindskog, C. The Human Protein Atlas—Spatial localization of the human proteome in health and disease. Protein Sci. 2021, 30, 218–233. [Google Scholar] [CrossRef]
- Bartha, Á.; Győrffy, B. TNMplot.com: A Web Tool for the Comparison of Gene Expression in Normal, Tumor and Metastatic Tissues. Int. J. Mol. Sci. 2021, 22, 2622. [Google Scholar] [CrossRef]
- Győrffy, B. Survival analysis across the entire transcriptome identifies biomarkers with the highest prognostic power in breast cancer. Comput. Struct. Biotechnol. J. 2021, 19, 4101–4109. [Google Scholar] [CrossRef]
- Stejskalová, A.; Fincke, V.; Nowak, M.; Schmidt, Y.; Borrmann, K.; von Wahlde, M.-K.; Schäfer, S.D.; Kiesel, L.; Greve, B.; Götte, M. Collagen I triggers directional migration, invasion and matrix remodeling of stroma cells in a 3D spheroid model of endometriosis. Sci. Rep. 2021, 11, 4115. [Google Scholar] [CrossRef]
- Rao, X.; Huang, X.; Zhou, Z.; Lin, X. An improvement of the 2ˆ(−delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinforma. Biomath. 2013, 3, 71–85. [Google Scholar]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, 638–646. [Google Scholar] [CrossRef]
- Györffy, B.; Lanczky, A.; Eklund, A.C.; Denkert, C.; Budczies, J.; Li, Q.; Szallasi, Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 2010, 123, 725–731. [Google Scholar] [CrossRef]
- Dai, X.; Cheng, H.; Bai, Z.; Li, J. Breast Cancer Cell Line Classification and Its Relevance with Breast Tumor Subtyping. J. Cancer 2017, 8, 3131–3141. [Google Scholar] [CrossRef]
- Holliday, D.L.; Speirs, V. Choosing the right cell line for breast cancer research. Breast Cancer Res. 2011, 13, 215. [Google Scholar] [CrossRef]
- Banerjee, M.; Bhonde, R.R. Application of hanging drop technique for stem cell differentiation and cytotoxicity studies. Cytotechnology 2006, 51, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Kurosawa, H. Methods for inducing embryoid body formation: In vitro differentiation system of embryonic stem cells. J. Biosci. Bioeng. 2007, 103, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Vitale, D.; Kumar Katakam, S.; Greve, B.; Jang, B.; Oh, E.; Alaniz, L.; Götte, M. Proteoglycans and glycosaminoglycans as regulators of cancer stem cell function and therapeutic resistance. FEBS J. 2019, 286, 2870–2882. [Google Scholar] [CrossRef] [PubMed]
- Corti, F.; Ristori, E.; Rivera-Molina, F.; Toomre, D.; Zhang, J.; Mihailovic, J.; Zhuang, Z.W.; Simons, M. Syndecan-2 selectively regulates VEGF-induced vascular permeability. Nat. Cardiovasc. Res. 2022, 1, 518–528. [Google Scholar] [CrossRef]
- Jang, B.; Kim, A.; Hwang, J.; Song, H.-K.; Kim, Y.; Oh, E.-S. Emerging Role of Syndecans in Extracellular Matrix Remodeling in Cancer. J. Histochem. Cytochem. 2022, 68, 863–870. [Google Scholar] [CrossRef]
- Huang, X.; Reye, G.; Momot, K.I.; Blick, T.; Lloyd, T.; Tilley, W.D.; Hickey, T.E.; Snell, C.E.; Okolicsanyi, R.K.; Haupt, L.M.; et al. Heparanase Promotes Syndecan-1 Expression to Mediate Fibrillar Collagen and Mammographic Density in Human Breast Tissue Cultured ex vivo. Front. Cell Dev. Biol. 2020, 8, 599. [Google Scholar] [CrossRef]
- Lacroix, M.; Leclercq, G. Relevance of breast cancer cell lines as models for breast tumours: An update. Breast Cancer Res. Treat. 2004, 83, 249–289. [Google Scholar] [CrossRef]
- Liang, H.; Xiao, J.; Zhou, Z.; Wu, J.; Ge, F.; Li, Z.; Zhang, H.; Sun, J.; Li, F.; Liu, R. Hypoxia induces miR-153 through the IRE1α-XBP1 pathway to fine tune the HIF1α/VEGFA axis in breast cancer angiogenesis. Oncogene 2018, 37, 1961–1975. [Google Scholar] [CrossRef] [PubMed]
- Kinnunen, T.; Kaksonen, M.; Saarinen, J.; Kalkkinen, N.; Peng, H.B.; Rauvala, H. Cortactin-Src kinase signaling pathway is involved in N-syndecan-dependent neurite outgrowth. J. Biol. Chem. 1998, 273, 10702–10708. [Google Scholar] [CrossRef] [PubMed]
- Siegbahn, A.; Johnell, M.; Nordin, A.; Åberg, M.; Velling, T. TF/FVIIa Transactivate PDGFRβ to Regulate PDGF-BB–Induced Chemotaxis in Different Cell Types: Involvement of Src And PLC. Arterioscler. Thromb. Vasc. Biol. 2008, 28, 135–141. [Google Scholar] [CrossRef]
- Luo, J.; Zou, H.; Guo, Y.; Tong, T.; Ye, L.; Zhu, C.; Deng, L.; Wang, B.; Pan, Y.; Li, P. SRC kinase-mediated signaling pathways and targeted therapies in breast cancer. Breast Cancer Res. 2022, 24, 99. [Google Scholar] [CrossRef]
- Jiang, B.-H.; Agani, F.; Passaniti, A.; Semenza, G.L. V-SRC induces expression of hypoxia-inducible factor 1 (HIF-1) and transcription of genes encoding vascular endothelial growth factor and enolase 1: Involvement of HIF-1 in tumor progression. Cancer Res. 1997, 57, 5328–5335. [Google Scholar]
- Hassan, N.; Bückreiß, N.; Efing, J.; Schulz-Fincke, M.; König, P.; Greve, B.; Bendas, G.; Götte, M. The heparan sulfate proteoglycan syndecan-1 triggers breast cancer cell-induced coagulability by induced expression of tissue factor. Cells 2023, 12, 910. [Google Scholar] [CrossRef] [PubMed]
- Tinholt, M.; Stavik, B.; Louch, W.; Carlson, C.R.; Sletten, M.; Ruf, W.; Skretting, G.; Sandset, P.M.; Iversen, N. Syndecan-3 and TFPI Colocalize on the Surface of Endothelial-, Smooth Muscle-, and Cancer Cells. PLoS ONE 2015, 10, e0117404. [Google Scholar] [CrossRef]
- Czarnowski, D. Syndecans in cancer: A review of function, expression, prognostic value, and therapeutic significance. Cancer Treat. Res. Commun. 2021, 27, 100312. [Google Scholar] [CrossRef]
- Prieto-Fernández, E.; Egia-Mendikute, L.; Bosch, A.; García del Río, A.; Jimenez-Lasheras, B.; Antoñana-Vildosola, A.; Lee, S.Y.; Palazon, A. Hypoxia Promotes Syndecan-3 Expression in the Tumor Microenvironment. Front. Immunol. 2020, 11, 586977. [Google Scholar] [CrossRef]
- Lee, S.Y.; Prieto-Fernández, E.; Egia-Mendikute, L.; Antoñana-Vildosola, A.; Velasco-Beltrán, P.; Bosch, A.; Jimenez-Lasheras, B.; de Blas, A.; de Durana, J.E.-D.; Valdaliso-Díez, E.; et al. Syndecan-3 positively regulates the pro-inflammatory function of macrophages. Cell Mol. Life Sci. 2025, 82, 145. [Google Scholar] [CrossRef]
- Wu, Z.-S.; Pandey, V.; Wu, W.-Y.; Ye, S.; Zhu, T.; Lobie, P.E. Prognostic significance of the expression of GFRα1, GFRα3 and Syndecan-3, proteins binding ARTEMIN, in mammary carcinoma. BMC Cancer 2013, 13, 34. [Google Scholar] [CrossRef]
- Jiang, F.; Wu, C.; Wang, M.; Wei, K.; Wang, J. Identification of novel cell glycolysis related gene signature predicting survival in patients with breast cancer. Sci. Rep. 2021, 11, 3986. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, H.; Fukuda, A.; Ikeda, N.; Sato, M.; Hashimoto, K.; Miyamoto, Y. Syndecan-3 regulates the time of transition from cell cycle exit to initial differentiation stage in mouse cerebellar granule cell precursors. Brain Res. 2023, 1807, 148317. [Google Scholar] [CrossRef]
- Pisconti, A.; Cornelison, D.D.W.; Olguín, H.C.; Antwine, T.L.; Olwin, B.B. Syndecan-3 and Notch cooperate in regulating adult myogenesis. J. Cell Biol. 2010, 190, 427–441. [Google Scholar] [CrossRef] [PubMed]
- Ordaz-Ramos, A.; Tellez-Jimenez, O.; Vazquez-Santillan, K. Signaling pathways governing the maintenance of breast cancer stem cells and their therapeutic implications. Front. Cell Dev. Biol. 2023, 11, 1221175. [Google Scholar] [CrossRef] [PubMed]
- Valla, S.; Hassan, N.; Vitale, D.L.; Madanes, D.; Spinelli, F.M.; Teixeira, F.C.; Greve, B.; Espinoza-Sánchez, N.A.; Cristina, C.; Alaniz, L. Syndecan-1 depletion has a differential impact on hyaluronic acid metabolism and tumor cell behavior in luminal and triple-negative breast cancer cells. Int. J. Mol. Sci. 2021, 22, 5874. [Google Scholar] [CrossRef]
- Jones, F.K.; Stefan, A.; Kay, A.G.; Hyland, M.; Morgan, R.; Forsyth, N.R.; Pisconti, A.; Kehoe, O. Syndecan-3 regulates MSC adhesion, ERK and AKT signalling in vitro and its deletion enhances MSC efficacy in a model of inflammatory arthritis in vivo. Sci. Rep. 2020, 10, 20487. [Google Scholar] [CrossRef]
- Inatani, M.; Haruta, M.; Honjo, M.; Oohira, A.; Kido, N.; Takahashi, M.; Honda, Y.; Tanihara, H. Upregulated expression of N-syndecan, a transmembrane heparan sulfate proteoglycan, in differentiated neural stem cells. Brain Res. 2001, 920, 217–221. [Google Scholar] [CrossRef]
- Edwards, A.; Brennan, K. Notch Signalling in Breast Development and Cancer. Front. Cell Dev. Biol. 2001, 9, 1–24. [Google Scholar] [CrossRef]
- Depau, L.; Brunetti, J.; Falciani, C.; Mandarini, E.; Zanchi, M.; Paolocci, M.F.; Garfì, M.; Pini, A.; Bracci, L. Targeting heparan sulfate proteoglycans as an effective strategy for inhibiting cancer cell migration and invasiveness compared to heparin. Front. Cell Dev. Biol. 2025, 12, 1505680. [Google Scholar] [CrossRef] [PubMed]
- Hienola, A.; Tumova, S.; Kulesskiy, E.; Rauvala, H. N-syndecan deficiency impairs neural migration in brain. J. Cell Biol. 2006, 174, 569–580. [Google Scholar] [CrossRef] [PubMed]
- Bespalov, M.M.; Sidorova, Y.A.; Tumova, S.; Ahonen-Bishopp, A.; Magalhães, A.C.; Kulesskiy, E.; Paveliev, M.; Rivera, C.; Rauvala, H.; Saarma, M. Heparan sulfate proteoglycan syndecan-3 is a novel receptor for GDNF, neurturin, and artemin. J. Cell Biol. 2011, 192, 153–169. [Google Scholar] [CrossRef] [PubMed]
- Kehoe, O.; Kalia, N.; King, S.; Eustace, A.; Boyes, C.; Reizes, O.; Williams, A.; Patterson, A.; Middleton, J. Syndecan-3 is selectively pro-inflammatory in the joint and contributes to antigen-induced arthritis in mice. Arthritis Res. Ther. 2014, 16, R148. [Google Scholar] [CrossRef]
- Eustace, A.D.; McNaughton, E.F.; King, S.; Kehoe, O.; Kungl, A.; Mattey, D.; Nobbs, A.H.; Williams, N.; Middleton, J. Soluble syndecan-3 binds chemokines, reduces leukocyte migration in vitro and ameliorates disease severity in models of rheumatoid arthritis. Arthritis Res. Ther. 2019, 21, 172. [Google Scholar] [CrossRef]
- Felipe Lima, J.; Nofech-Mozes, S.; Bayani, J.; Bartlett, J. EMT in Breast Carcinoma—A Review. J. Clin. Med. 2016, 5, 65. [Google Scholar] [CrossRef]
- Kessenbrock, K.; Plaks, V.; Werb, Z. Matrix Metalloproteinases: Regulators of the Tumor Microenvironment. Cell 2010, 141, 52–67. [Google Scholar] [CrossRef]
- Dufour, A.; Sampson, N.S.; Zucker, S.; Cao, J. Role of the hemopexin domain of matrix metalloproteinases in cell migration. J. Cell Physiol. 2008, 217, 643–651. [Google Scholar] [CrossRef]
- Asundi, V.K.; Erdman, R.; Stahl, R.C.; Carey, D.J. Matrix metalloproteinase-dependent shedding of syndecan-3, a transmembrane heparan sulfate proteoglycan, in Schwann cells. J. Neurosci. Res. 2003, 73, 593–602. [Google Scholar] [CrossRef]
- Arokiasamy, S.; Balderstone, M.J.M.; De Rossi, G.; Whiteford, J.R. Syndecan-3 in Inflammation and Angiogenesis. Front. Immunol. 2020, 10, 3031. [Google Scholar] [CrossRef]
- Bertrand, J.; Bollmann, M. Soluble syndecans: Biomarkers for diseases and therapeutic options. Br. J. Pharmacol. 2019, 176, 67–81. [Google Scholar] [CrossRef]
- Onyeisi, J.O.S.; El-Shorafa, H.M.; Greve, B.; Götte, M. Role of syndecan-4 in angiogenesis and vasculogenic mimicry in triple negative breast cancer cells. Matrix Biol. 2025, 136, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Whiteford, J.R. A novel role for syndecan-3 in angiogenesis. F1000Research 2013, 2, 270. [Google Scholar] [CrossRef]
- Jannaway, M.; Yang, X.; Meegan, J.E.; Coleman, D.C.; Yuan, S.Y. Thrombin-cleaved syndecan-3/-4 ectodomain fragments mediate endothelial barrier dysfunction. PLoS ONE 2019, 14, e0214737. [Google Scholar] [CrossRef]
- Johnson, D.; Agochiya, M.; Samejima, K.; Earnshaw, W.; Frame, M.; Wyke, J. Regulation of both apoptosis and cell survival by the v-Src oncoprotein. Cell Death Differ. 2000, 7, 685–696. [Google Scholar] [CrossRef]
- Zhang, S.; Yu, D. Targeting Src family kinases in anti-cancer therapies: Turning promise into triumph. Trends Pharmacol. Sci. 2012, 33, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Hanna, S.C.; Krishnan, B.; Bailey, S.T.; Moschos, S.J.; Kuan, P.-F.; Shimamura, T.; Osborne, L.D.; Siegel, M.B.; Duncan, L.M.; O’Brien, E.T., III; et al. HIF1α and HIF2α independently activate SRC to promote melanoma metastases. J. Clin. Investig. 2013, 123, 2078–2093. [Google Scholar] [CrossRef] [PubMed]
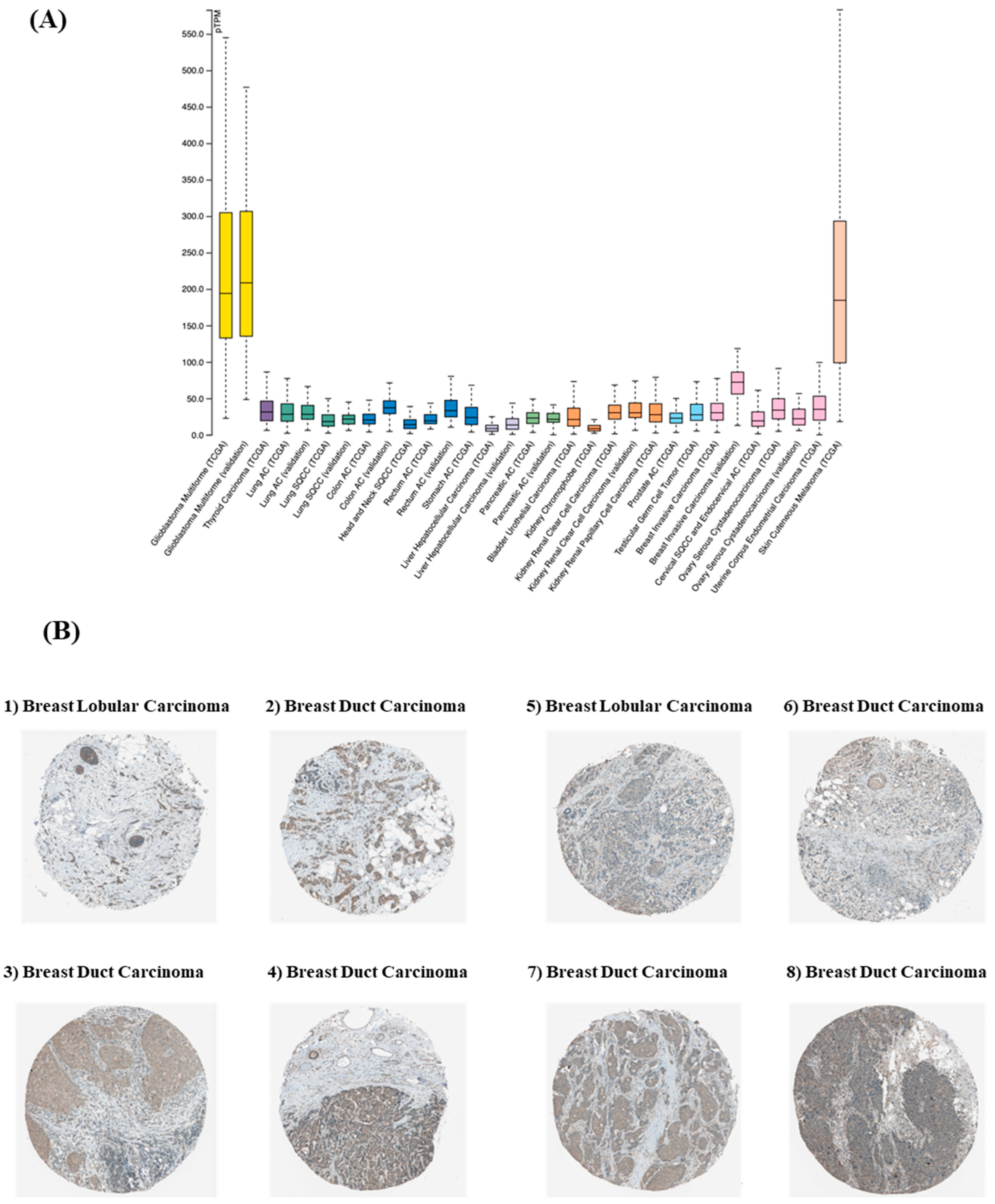
| Sample | Patient ID | Staining | Intensity | Quantity | Location |
|---|---|---|---|---|---|
| 1 | #2898 | medium | moderate | >75% | cytoplasmic, membranous |
| 2 | #3257 | medium | moderate | ||
| 3 | #2392 | medium | moderate | ||
| 4 | #1939 | medium | moderate | ||
| 5 | #2805 | low | weak | ||
| 6 | #1874 | low | weak | ||
| 7 | #2428 | low | weak | ||
| 8 | #2174 | low | weak |
| Classification | Status | Cases | Hazard Ratio | p Value |
|---|---|---|---|---|
| All breast cancer patients | 4929 | 0.75 (0.68–0.83) | Log-rank p = 1.7 × 10−8 | |
| Estrogen receptor (ER) | Positive | 2561 | 0.87 (0.75–1.02) | Log-rank p = 0.079 |
| Negative | 796 | 0.81 (0.64–1.03) | Log-rank p =0.086 | |
| Progesterone receptor (PR) | Positive | 926 | 0.95 (0.71–1.26) | Log-rank p = 0.72 |
| Negative | 925 | 0.91 (0.72–1.14) | Log-rank p = 0.41 | |
| Her2 | Positive | 882 | 0.85 (0.68–1.06) | Log-rank p = 0.15 |
| Negative | 4047 | 0.73 (0.65–0.81) | Log-rank p = 3.6 × 10−8 | |
| ER, PR, Her2 | Negative | 392 | 0.9 (0.63–1.29) | Log-rank p = 0.56 |
| St. Gallen subtype | Luminal A | 2277 | 0.77 (0.65–0.91) | Log-rank p = 0.0016 |
| Luminal B | 1419 | 0.71 (0.59–0.85) | Log-rank p = 0.00015 | |
| Her2 positive | 315 | 0.9 (0.64–1.29) | Log-rank p = 0.6 | |
| Basal | 846 | 0.72 (0.58–0.9) | Log-rank p = 0.004 | |
| PAM50 subtype | Luminal A | 1809 | 0.88 (0.72–1.08) | Log-rank p = 0.23 |
| Luminal B | 1353 | 0.83 (0.7–0.99) | Log-rank p = 0.038 | |
| Her2 positive | 695 | 0.86 (0.68–1.1) | Log-rank p = 0.23 | |
| Basal | 953 | 0.69 (0.56–0.86) | Log-rank p = 7 × 10−4 | |
| Lymph node | Positive | 1656 | 0.97 (0.82–1.15) | Log-rank p = 0.75 |
| Negative | 2368 | 0.74 (0.63–0.87) | Log-rank p = 0.00027 | |
| Grade | 1 | 397 | 1.12 (0.73–2) | Log-rank p = 0.47 |
| 2 | 1177 | 0.74 (0.59–0.92) | Log-rank p = 0.0057 | |
| 3 | 1300 | 0.88 (0.73–1.06) | Log-rank p = 0.16 | |
| Treatment with chemotherapy | Neoadjuvant | 402 | 0.6 (0.41–0.88) | Log-rank p = 0.009 |
| Adjuvant | 458 | 0.93 (0.67–1.3) | Log-rank p = 0.68 | |
| Treatment with endocrine therapy | 867 | 0.86 (0.66–1.12) | Log-rank p = 0.25 | |
| Treatment with chemotherapy and endocrine therapy | 510 | 1.22 (0.81–1.83) | Log-rank p = 0.34 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Habenicht, L.; Hassan, N.; Espinoza-Sànchez, N.A.; Onyeisi, J.O.S.; Győrffy, B.; Hanker, L.; Greve, B.; Götte, M. The Role of the Cell Surface Heparan Sulfate Proteoglycan Syndecan-3 in Breast Cancer Pathophysiology. Cells 2025, 14, 1612. https://doi.org/10.3390/cells14201612
Habenicht L, Hassan N, Espinoza-Sànchez NA, Onyeisi JOS, Győrffy B, Hanker L, Greve B, Götte M. The Role of the Cell Surface Heparan Sulfate Proteoglycan Syndecan-3 in Breast Cancer Pathophysiology. Cells. 2025; 14(20):1612. https://doi.org/10.3390/cells14201612
Chicago/Turabian StyleHabenicht, Lena, Nourhan Hassan, Nancy A. Espinoza-Sànchez, Jessica Oyie Sousa Onyeisi, Balázs Győrffy, Lars Hanker, Burkhard Greve, and Martin Götte. 2025. "The Role of the Cell Surface Heparan Sulfate Proteoglycan Syndecan-3 in Breast Cancer Pathophysiology" Cells 14, no. 20: 1612. https://doi.org/10.3390/cells14201612
APA StyleHabenicht, L., Hassan, N., Espinoza-Sànchez, N. A., Onyeisi, J. O. S., Győrffy, B., Hanker, L., Greve, B., & Götte, M. (2025). The Role of the Cell Surface Heparan Sulfate Proteoglycan Syndecan-3 in Breast Cancer Pathophysiology. Cells, 14(20), 1612. https://doi.org/10.3390/cells14201612








