Galectin-3 Mediated Endocytosis of the Orphan G-Protein-Coupled Receptor GPRC5A
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Purification of Recombinant Galectin-3
2.3. HepG2 Transient Transfection by GFP-Gal-3 Plasmid
2.4. Cell Lysis and Protein Extraction
2.5. Co-Immunoprecipitation Experiments
2.6. Immunoprecipitation Experiments for Interactomic Study
2.7. Tunicamycin Treatment
2.8. Western Blot Analyses
2.9. Mass Spectrometry Analysis
2.10. Database Search and Mass Spectrometry Data Post-Processing
2.11. Gal-3 Dependent Endocytosis Assays
3. Results
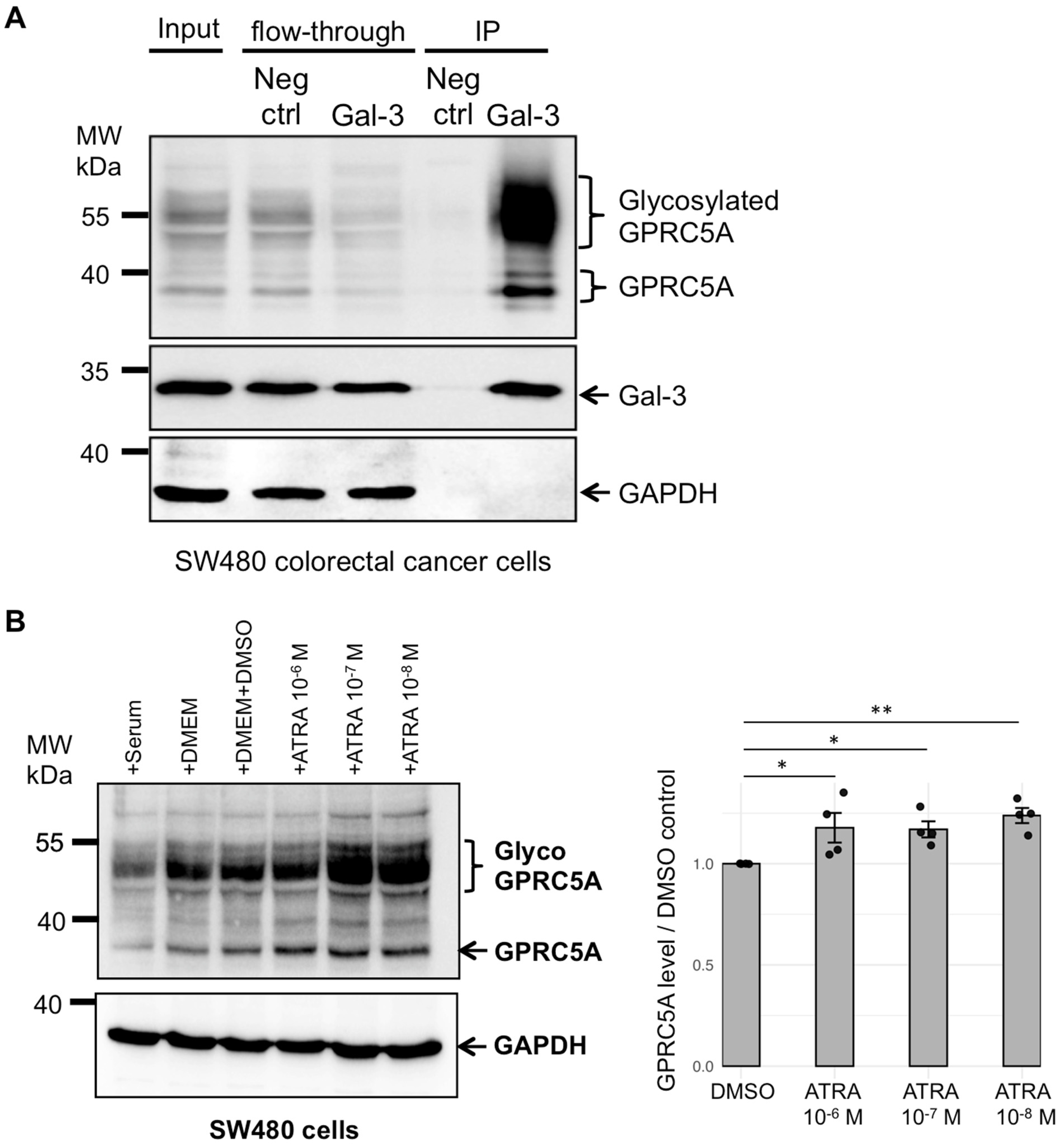
3.1. Identification of GPRC5A as a Gal-3 Binding Partner
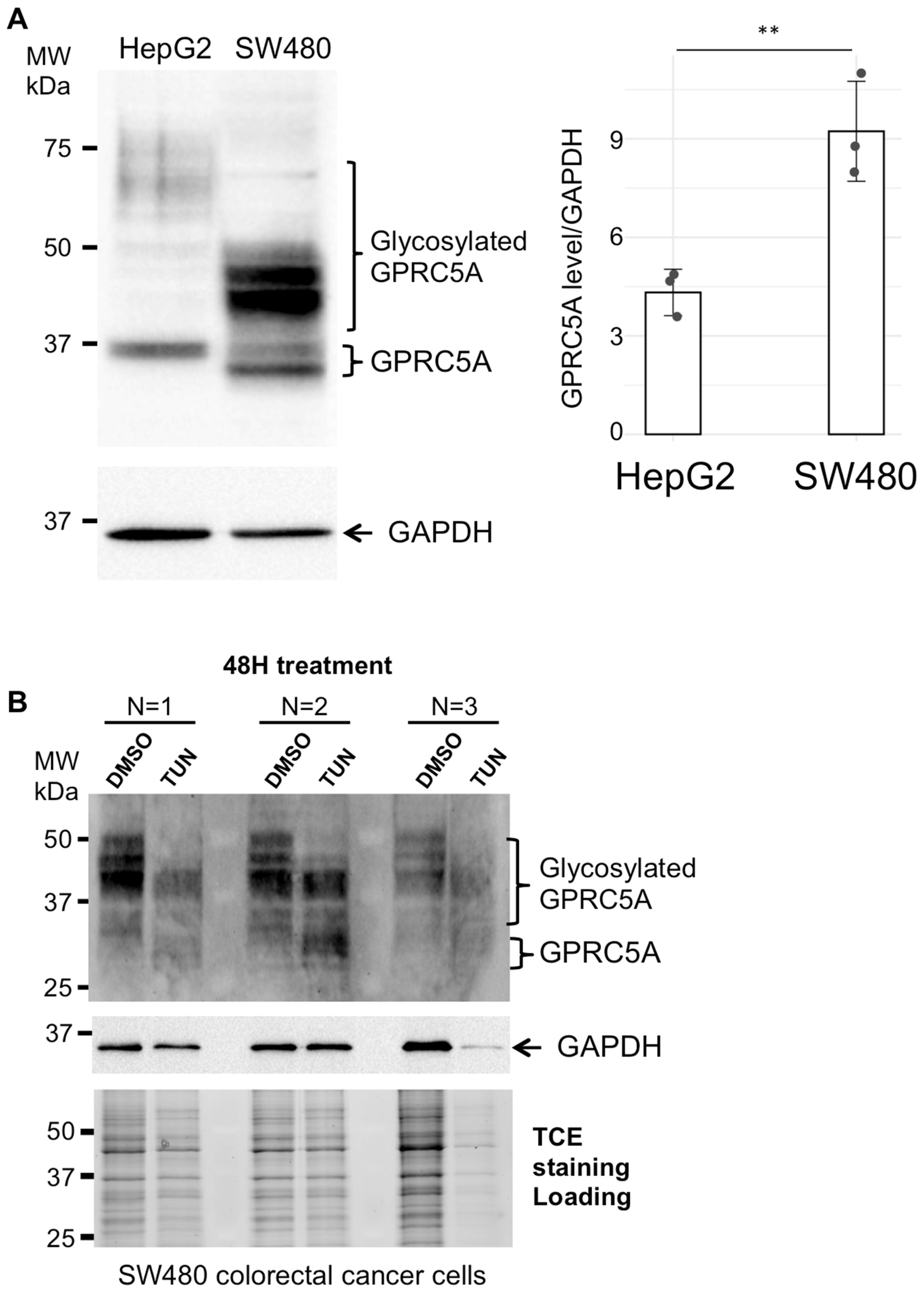
3.2. GPRC5A Glycosylation in SW480 Colon Cancer Cells
3.3. Addition of Retinoic Acid Leads to Increased Levels of GPRC5A Proteins
3.4. GPRC5A Endocytosis Is Mediated by Extracellular Gal-3 Addition
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thiemann, S.; Baum, L.G. Galectins and immune responses-just how do they do those things they do? Annu. Rev. Immunol. 2016, 34, 243–264. [Google Scholar] [CrossRef]
- Lin, Y.H.; Qiu, D.C.; Chang, W.H.; Yeh, Y.Q.; Jeng, U.S.; Liu, F.T.; Huang, J.R. The intrinsically disordered n-terminal domain of galectin-3 dynamically mediates multisite self-association of the protein through fuzzy interactions. J. Biol. Chem. 2017, 292, 17845–17856. [Google Scholar] [CrossRef]
- Bouffette, S.; Botez, I.; De Ceuninck, F. Targeting galectin-3 in inflammatory and fibrotic diseases. Trends Pharmacol. Sci. 2023, 44, 519–531. [Google Scholar] [CrossRef]
- Boutas, I.; Potiris, A.; Brenner, W.; Lebrecht, A.; Hasenburg, A.; Kalantaridou, S.; Schmidt, M. The expression of galectin-3 in breast cancer and its association with chemoresistance: A systematic review of the literature. Arch. Gynecol. Obs. 2019, 300, 1113–1120. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.N.; Dang, Y.F.; Gong, F.L.; Guo, X.L. Role and regulation mechanism of gal-3 in non-small cell lung cancer and its potential clinical therapeutic significance. Chem. Biol. Interact. 2019, 309, 108724. [Google Scholar] [CrossRef] [PubMed]
- Portacci, A.; Diaferia, F.; Santomasi, C.; Dragonieri, S.; Boniello, E.; Di Serio, F.; Carpagnano, G.E. Galectin-3 as prognostic biomarker in patients with covid-19 acute respiratory failure. Respir. Med. 2021, 187, 106556. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, J.; Xiao, W.; Zhang, Y.; Lv, Y.; Yu, X.; Zheng, J. The therapeutic potential of galectin-3 in the treatment of intrahepatic cholangiocarcinoma patients and those compromised with covid-19. Front. Mol. Biosci. 2021, 8, 666054. [Google Scholar] [CrossRef]
- Gilson, R.C.; Gunasinghe, S.D.; Johannes, L.; Gaus, K. Galectin-3 modulation of t-cell activation: Mechanisms of membrane remodelling. Prog. Lipid Res. 2019, 76, 101010. [Google Scholar] [CrossRef]
- Johannes, L.; Wunder, C.; Shafaq-Zadah, M. Glycolipids and lectins in endocytic uptake processes. J. Mol. Biol. 2016, 428, 4792–4818. [Google Scholar] [CrossRef]
- Lakshminarayan, R.; Wunder, C.; Becken, U.; Howes, M.T.; Benzing, C.; Arumugam, S.; Sales, S.; Ariotti, N.; Chambon, V.; Lamaze, C.; et al. Galectin-3 drives glycosphingolipid-dependent biogenesis of clathrin-independent carriers. Nat. Cell Biol. 2014, 16, 595–606. [Google Scholar] [CrossRef]
- Ivashenka, A.; Wunder, C.; Chambon, V.; Sandhoff, R.; Jennemann, R.; Dransart, E.; Podsypanina, K.; Lombard, B.; Loew, D.; Lamaze, C.; et al. Glycolipid-dependent and lectin-driven transcytosis in mouse enterocytes. Commun. Biol. 2021, 4, 173. [Google Scholar] [CrossRef]
- Zhang, C.; Shafaq-Zadah, M.; Pawling, J.; Hesketh, G.G.; Dransart, E.; Pacholczyk, K.; Longo, J.; Gingras, A.C.; Penn, L.Z.; Johannes, L.; et al. Slc3a2 n-glycosylation and golgi remodeling regulate slc7a amino acid exchangers and stress mitigation. J. Biol. Chem. 2023, 299, 105416. [Google Scholar] [CrossRef]
- MacDonald, E.; Forrester, A.; Valades-Cruz, C.A.; Madsen, T.D.; Hetmanski, J.H.R.; Dransart, E.; Ng, Y.; Godbole, R.; Shp, A.A.; Leconte, L.; et al. Growth factor-triggered de-sialylation controls glycolipid-lectin-driven endocytosis. Nat. Cell Biol. 2025, 27, 449–463. [Google Scholar] [CrossRef]
- Johannes, L.; Shafaq-Zadah, M.; Dransart, E.; Wunder, C.; Leffler, H. Endocytic roles of glycans on proteins and lipids. Cold Spring Harb. Perspect. Biol. 2024, 16, a041398. [Google Scholar] [CrossRef] [PubMed]
- Hauser, A.S.; Attwood, M.M.; Rask-Andersen, M.; Schioth, H.B.; Gloriam, D.E. Trends in gpcr drug discovery: New agents, targets and indications. Nat. reviews. Drug Discov. 2017, 16, 829–842. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Lotan, R. Molecular cloning and characterization of a novel retinoic acid-inducible gene that encodes a putative g protein-coupled receptor. J. Biol. Chem. 1998, 273, 35008–35015. [Google Scholar] [CrossRef] [PubMed]
- Robbins, M.J.; Michalovich, D.; Hill, J.; Calver, A.R.; Medhurst, A.D.; Gloger, I.; Sims, M.; Middlemiss, D.N.; Pangalos, M.N. Molecular cloning and characterization of two novel retinoic acid-inducible orphan g-protein-coupled receptors (gprc5b and gprc5c). Genomics 2000, 67, 8–18. [Google Scholar] [CrossRef]
- Zhao, X.; Stein, K.R.; Chen, V.; Griffin, M.E.; Lairson, L.L.; Hang, H.C. Chemoproteomics reveals microbiota-derived aromatic monoamine agonists for gprc5a. Nat. Chem. Biol. 2023, 19, 1205–1214. [Google Scholar] [CrossRef]
- Dai, L.; Jin, X.; Liu, Z. Prognostic and clinicopathological significance of gprc5a in various cancers: A systematic review and meta-analysis. PLoS ONE 2021, 16, e0249040. [Google Scholar] [CrossRef]
- Iglesias González, P.A.; Valdivieso, A.G.; Santa-Coloma, T.A. The g protein-coupled receptor gprc5a-a phorbol ester and retinoic acid-induced orphan receptor with roles in cancer, inflammation, and immunity. Biochem. Cell Biol. Biochim. Et Biol. Cell. 2023, 101, 465–480. [Google Scholar] [CrossRef]
- Maejima, I.; Takahashi, A.; Omori, H.; Kimura, T.; Takabatake, Y.; Saitoh, T.; Yamamoto, A.; Hamasaki, M.; Noda, T.; Isaka, Y.; et al. Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J. 2013, 32, 2336–2347. [Google Scholar] [CrossRef]
- Ladner, C.L.; Yang, J.; Turner, R.J.; Edwards, R.A. Visible fluorescent detection of proteins in polyacrylamide gels without staining. Anal. Biochem. 2004, 326, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Chicois, C.; Scheer, H.; Garcia, S.; Zuber, H.; Mutterer, J.; Chicher, J.; Hammann, P.; Gagliardi, D.; Garcia, D. The upf1 interactome reveals interaction networks between rna degradation and translation repression factors in arabidopsis. Plant J. 2018, 96, 119–132. [Google Scholar] [CrossRef] [PubMed]
- R-Core-Team. R: A Language and Environment for Statistical Computing. Available online: https://www.R-project.org/ (accessed on 22 July 2025).
- Posit-Team. Rstudio: Integrated Development Environment for R. Available online: http://www.posit.co/ (accessed on 22 July 2025).
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Ginestet, C. Ggplot2: Elegant graphics for data analysis. J. R. Stat. Soc. Ser. A Stat. Soc. 2011, 174, 245–246. [Google Scholar] [CrossRef]
- Wickham, H.; Bryan, J. Readxl: Read Excel Files. Available online: https://readxl.tidyverse.org (accessed on 22 July 2025).
- Schauberger, P.; Walker, A. Openxlsx: Read, Write and Edit Xlsx Files. Available online: https://ycphs.github.io/openxlsx/index.html (accessed on 22 July 2025).
- Slowikowski, K. Ggrepel: Automatically Position Non-Overlapping Text Labels with ‘ggplot2’. Available online: https://ggrepel.slowkow.com/ (accessed on 22 July 2025).
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; García-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The pride database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef]
- Debard, S.; Bader, G.; De Craene, J.O.; Enkler, L.; Bar, S.; Laporte, D.; Hammann, P.; Myslinski, E.; Senger, B.; Friant, S.; et al. Nonconventional localizations of cytosolic aminoacyl-trna synthetases in yeast and human cells. Methods 2017, 113, 91–104. [Google Scholar] [CrossRef]
- Stoetzel, C.; Bar, S.; De Craene, J.O.; Scheidecker, S.; Etard, C.; Chicher, J.; Reck, J.R.; Perrault, I.; Geoffroy, V.; Chennen, K.; et al. A mutation in vps15 (pik3r4) causes a ciliopathy and affects ift20 release from the cis-golgi. Nat. Commun. 2016, 7, 13586. [Google Scholar] [CrossRef]
- Lin, T.W.; Chang, H.T.; Chen, C.H.; Chen, C.H.; Lin, S.W.; Hsu, T.L.; Wong, C.H. Galectin-3 binding protein and galectin-1 interaction in breast cancer cell aggregation and metastasis. J. Am. Chem. Soc. 2015, 137, 9685–9693. [Google Scholar] [CrossRef]
- Mathew, M.P.; Donaldson, J.G. Distinct cargo-specific response landscapes underpin the complex and nuanced role of galectin-glycan interactions in clathrin-independent endocytosis. J. Biol. Chem. 2018, 293, 7222–7237. [Google Scholar] [CrossRef]
- Joeh, E.; O’Leary, T.; Li, W.; Hawkins, R.; Hung, J.R.; Parker, C.G.; Huang, M.L. Mapping glycan-mediated galectin-3 interactions by live cell proximity labeling. Proc. Natl. Acad. Sci. USA 2020, 117, 27329–27338. [Google Scholar] [CrossRef]
- Couves, E.C.; Gardner, S.; Voisin, T.B.; Bickel, J.K.; Stansfeld, P.J.; Tate, E.W.; Bubeck, D. Structural basis for membrane attack complex inhibition by cd59. Nat. Commun. 2023, 14, 890. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Edwards, C.K., 3rd; Zhou, L. The biological function and clinical utilization of cd147 in human diseases: A review of the current scientific literature. Int. J. Mol. Sci. 2014, 15, 17411–17441. [Google Scholar] [CrossRef] [PubMed]
- Capone, E.; Iacobelli, S.; Sala, G. Role of galectin 3 binding protein in cancer progression: A potential novel therapeutic target. J. Transl. Med. 2021, 19, 405. [Google Scholar] [CrossRef] [PubMed]
- Priglinger, C.S.; Szober, C.M.; Priglinger, S.G.; Merl, J.; Euler, K.N.; Kernt, M.; Gondi, G.; Behler, J.; Geerlof, A.; Kampik, A.; et al. Galectin-3 induces clustering of cd147 and integrin-β1 transmembrane glycoprotein receptors on the rpe cell surface. PLoS ONE 2013, 8, e70011. [Google Scholar] [CrossRef]
- Greenhough, A.; Bagley, C.; Heesom, K.J.; Gurevich, D.B.; Gay, D.; Bond, M.; Collard, T.J.; Paraskeva, C.; Martin, P.; Sansom, O.J.; et al. Cancer cell adaptation to hypoxia involves a hif-gprc5a-yap axis. EMBO Mol. Med. 2018, 10, e8699. [Google Scholar] [CrossRef]
- Chen, Y.; Deng, J.; Fujimoto, J.; Kadara, H.; Men, T.; Lotan, D.; Lotan, R. Gprc5a deletion enhances the transformed phenotype in normal and malignant lung epithelial cells by eliciting persistent stat3 signaling induced by autocrine leukemia inhibitory factor. Cancer Res. 2010, 70, 8917–8926. [Google Scholar] [CrossRef]
- Mattox, D.E.; Bailey-Kellogg, C. Comprehensive analysis of lectin-glycan interactions reveals determinants of lectin specificity. PLoS Comput. Biol. 2021, 17, e1009470. [Google Scholar] [CrossRef]
- Ghyselinck, N.B.; Duester, G. Retinoic acid signaling pathways. Development 2019, 146, dev167502. [Google Scholar] [CrossRef]
- Bräuner-Osborne, H.; Jensen, A.A.; Sheppard, P.O.; Brodin, B.; Krogsgaard-Larsen, P.; O’Hara, P. Cloning and characterization of a human orphan family c g-protein coupled receptor gprc5d. Biochim. Et. Biophys. Acta 2001, 1518, 237–248. [Google Scholar] [CrossRef]
- Lagerström, M.C.; Schiöth, H.B. Structural diversity of g protein-coupled receptors and significance for drug discovery. Nat. reviews. Drug Discov. 2008, 7, 339–357. [Google Scholar] [CrossRef]
- Zhang, L.; Li, L.; Gao, G.; Wei, G.; Zheng, Y.; Wang, C.; Gao, N.; Zhao, Y.; Deng, J.; Chen, H.; et al. Elevation of gprc5a expression in colorectal cancer promotes tumor progression through vnn-1 induced oxidative stress. Int. J. Cancer 2017, 140, 2734–2747. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, M.M.; Biesbrock, D.; Hanover, J.A. Galectin-3: Integrator of signaling via hexosamine flux. Biomolecules 2025, 15, 1028. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Xiao, K.; Tian, Z. Comparative glycomics study of cell-surface n-glycomes of hepg2 versus lo2 cell lines. J. Proteome Res. 2019, 18, 372–379. [Google Scholar] [CrossRef] [PubMed]
- Holst, S.; Deuss, A.J.; van Pelt, G.W.; van Vliet, S.J.; Garcia-Vallejo, J.J.; Koeleman, C.A.; Deelder, A.M.; Mesker, W.E.; Tollenaar, R.A.; Rombouts, Y.; et al. N-glycosylation profiling of colorectal cancer cell lines reveals association of fucosylation with differentiation and caudal type homebox 1 (cdx1)/villin mrna expression. Mol. Cell. Proteom. 2016, 15, 124–140. [Google Scholar] [CrossRef]
- Shafaq-Zadah, M.; Dransart, E.; Mani, S.K.; Sampaio, J.L.; Bouidghaghen, L.; Nilsson, U.J.; Leffler, H.; Johannes, L. Exploration into galectin-3 driven endocytosis and lattices. Biomolecules 2024, 14, 1169. [Google Scholar] [CrossRef]
- Thottacherry, J.J.; Sathe, M.; Prabhakara, C.; Mayor, S. Spoiled for choice: Diverse endocytic pathways function at the cell surface. Annu. Rev. Cell Dev. Biol. 2019, 35, 55–84. [Google Scholar] [CrossRef]
- Yang, W.; Di Vizio, D.; Kirchner, M.; Steen, H.; Freeman, M.R. Proteome scale characterization of human s-acylated proteins in lipid raft-enriched and non-raft membranes. Mol. Cell. Proteom. 2010, 9, 54–70. [Google Scholar] [CrossRef]
- Chalhoub, G.; McCormick, P.J. Palmitoylation and g-protein coupled receptors. Prog. Mol. Biol. Transl. Sci. 2022, 193, 195–211. [Google Scholar]
- Thankamony, S.P.; Knudson, W. Acylation of cd44 and its association with lipid rafts are required for receptor and hyaluronan endocytosis. J. Biol. Chem. 2006, 281, 34601–34609. [Google Scholar] [CrossRef]
- Kim, Y.J.; Greimel, P.; Hirabayashi, Y. Gprc5b-mediated sphingomyelin synthase 2 phosphorylation plays a critical role in insulin resistance. iScience 2018, 8, 250–266. [Google Scholar] [CrossRef] [PubMed]
- Rog, T.; Vattulainen, I. Cholesterol, sphingolipids, and glycolipids: What do we know about their role in raft-like membranes? Chem. Phys. Lipids 2014, 184, 82–104. [Google Scholar] [CrossRef] [PubMed]
- Hirabayashi, Y.; Kim, Y.J. Roles of gprc5 family proteins: Focusing on gprc5b and lipid-mediated signalling. J. Biochem. 2020, 167, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Im, E.J.; Moon, P.G.; Baek, M.C. Discovery of a diagnostic biomarker for colon cancer through proteomic profiling of small extracellular vesicles. BMC Cancer 2018, 18, 1058. [Google Scholar] [CrossRef]
- Bänfer, S.; Jacob, R. Galectins in intra- and extracellular vesicles. Biomolecules 2020, 10, 1232. [Google Scholar] [CrossRef]
- Zhang, D.X.; Dang, X.T.T.; Vu, L.T.; Lim, C.M.H.; Yeo, E.Y.M.; Lam, B.W.S.; Leong, S.M.; Omar, N.; Putti, T.C.; Yeh, Y.C.; et al. Avβ1 integrin is enriched in extracellular vesicles of metastatic breast cancer cells: A mechanism mediated by galectin-3. J. Extracell. Vesicles 2022, 11, e12234. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boucheham, A.; Franco, J.M.; Bär, S.; MacDonald, E.; Zuttion, S.; Blagec, L.; Rinaldi, B.; Chicher, J.; Kuhn, L.; Hammann, P.; et al. Galectin-3 Mediated Endocytosis of the Orphan G-Protein-Coupled Receptor GPRC5A. Cells 2025, 14, 1571. https://doi.org/10.3390/cells14191571
Boucheham A, Franco JM, Bär S, MacDonald E, Zuttion S, Blagec L, Rinaldi B, Chicher J, Kuhn L, Hammann P, et al. Galectin-3 Mediated Endocytosis of the Orphan G-Protein-Coupled Receptor GPRC5A. Cells. 2025; 14(19):1571. https://doi.org/10.3390/cells14191571
Chicago/Turabian StyleBoucheham, Abdeldjalil, Jorge Mallor Franco, Séverine Bär, Ewan MacDonald, Solène Zuttion, Lana Blagec, Bruno Rinaldi, Johana Chicher, Laurianne Kuhn, Philippe Hammann, and et al. 2025. "Galectin-3 Mediated Endocytosis of the Orphan G-Protein-Coupled Receptor GPRC5A" Cells 14, no. 19: 1571. https://doi.org/10.3390/cells14191571
APA StyleBoucheham, A., Franco, J. M., Bär, S., MacDonald, E., Zuttion, S., Blagec, L., Rinaldi, B., Chicher, J., Kuhn, L., Hammann, P., Wunder, C., Johannes, L., Rechreche, H., & Friant, S. (2025). Galectin-3 Mediated Endocytosis of the Orphan G-Protein-Coupled Receptor GPRC5A. Cells, 14(19), 1571. https://doi.org/10.3390/cells14191571






