Author Contributions
Conceptualization, M.S. and K.-i.N.; methodology, L.J., H.T., and H.I.; software, R.S.; validation, G.L., H.I. and H.T.; formal analysis, R.S., S.C., and M.S.; investigation, R.S., C.A., L.J.; resources, C.A., L.J., and G.L.; data curation, L.G., F.L.B., and H.U.; writing—original draft preparation, M.S. and K.-i.N.; writing—review and editing, S.C. and F.L.B.; visualization, S.C. and H.T.; supervision, H.U. and K.-i.N.; project administration, M.S., S.C., and K.-i.N.; funding acquisition, K.-i.N. and S.C. All authors have read and agreed to the published version of the manuscript.
Figure 1.
Post-mortem clinical and imaging findings. (A) Prenatal brain MRI at 23 weeks’ gestation. Sagittal and axial T2-weighted MRI scans reveal white matter thinning with corpus callosum agenesis, accompanied by secondary septal defects. The lateral ventricles are enlarged and abnormally shaped. There is hypoplasia of the cerebellar vermis, resulting in enlargement of the cisterna magna, along with dysgenesis of the brain characterized by an elongated pons. (B) Autoptic findings. The fetus exhibits microcephaly and distinct dysmorphic features, including a receding forehead, large mouth with thin lips, an ogival palate, a short lingual frenulum, and large, malformed ears. Additionally, organomegaly is present, affecting the lungs, heart, kidneys, adrenal glands, and spleen. (C) Hand abnormalities. The fetus presents with clenched fists and flexion contractures involving the fingers.
Figure 1.
Post-mortem clinical and imaging findings. (A) Prenatal brain MRI at 23 weeks’ gestation. Sagittal and axial T2-weighted MRI scans reveal white matter thinning with corpus callosum agenesis, accompanied by secondary septal defects. The lateral ventricles are enlarged and abnormally shaped. There is hypoplasia of the cerebellar vermis, resulting in enlargement of the cisterna magna, along with dysgenesis of the brain characterized by an elongated pons. (B) Autoptic findings. The fetus exhibits microcephaly and distinct dysmorphic features, including a receding forehead, large mouth with thin lips, an ogival palate, a short lingual frenulum, and large, malformed ears. Additionally, organomegaly is present, affecting the lungs, heart, kidneys, adrenal glands, and spleen. (C) Hand abnormalities. The fetus presents with clenched fists and flexion contractures involving the fingers.
Figure 2.
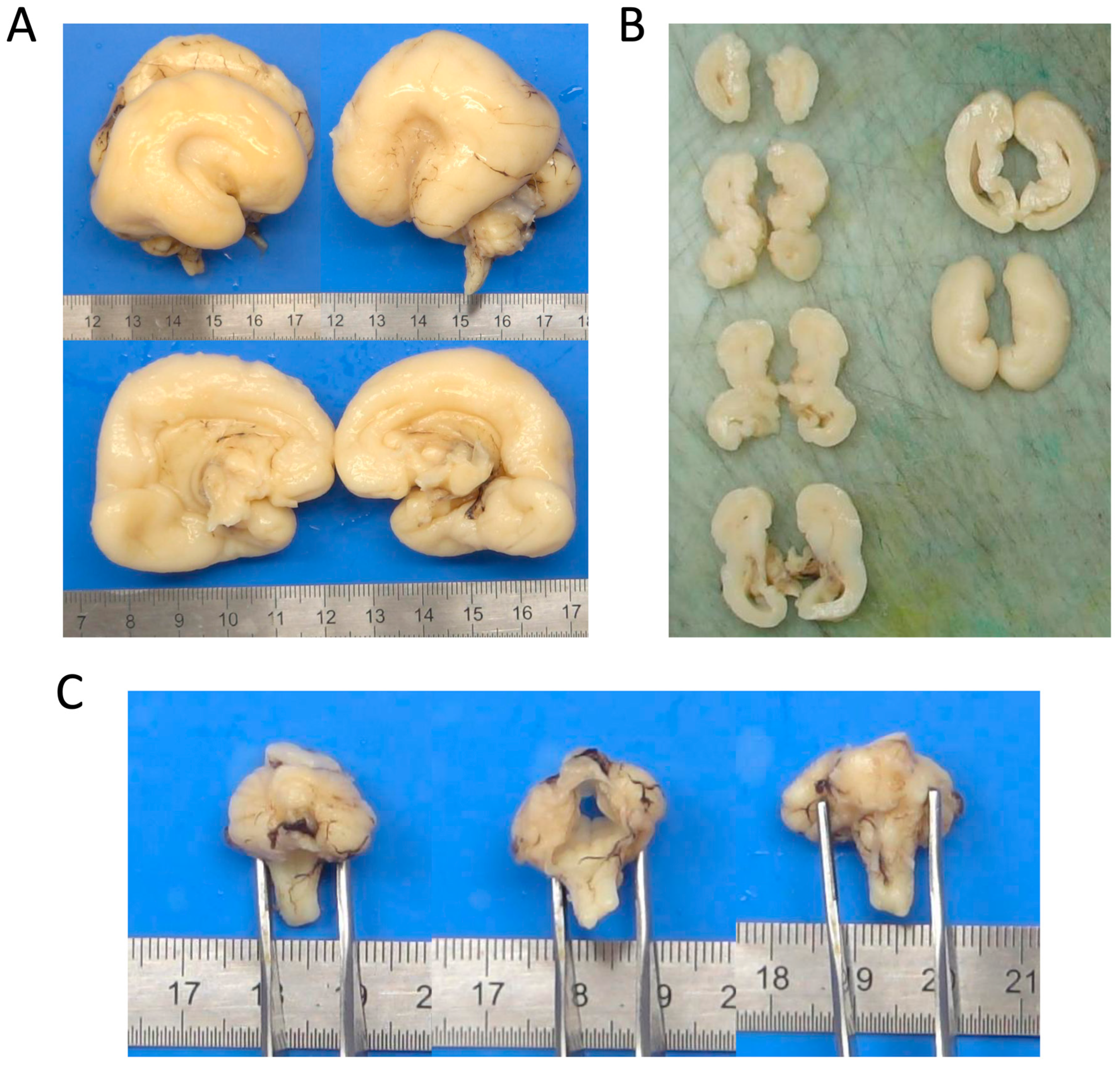
Post-mortem brain examination. (A) Sagittal section of the fetal brain. The brain is small, with complete agenesis of the corpus callosum, accompanied by a lack of opercularization of the sylvian fissures. (B) Axial section of the hemispheres. There is likely involvement of the white matter, with unusually sharp distinction between the cortical surface and the subcortical white matter, suggesting an underlying neuronal migration abnormality. (C) Detailed view of the brainstem. There is severe hypoplasia of the cerebellar vermis, accompanied by a small and dysmorphic pons.
Figure 2.
Post-mortem brain examination. (A) Sagittal section of the fetal brain. The brain is small, with complete agenesis of the corpus callosum, accompanied by a lack of opercularization of the sylvian fissures. (B) Axial section of the hemispheres. There is likely involvement of the white matter, with unusually sharp distinction between the cortical surface and the subcortical white matter, suggesting an underlying neuronal migration abnormality. (C) Detailed view of the brainstem. There is severe hypoplasia of the cerebellar vermis, accompanied by a small and dysmorphic pons.
Figure 3.
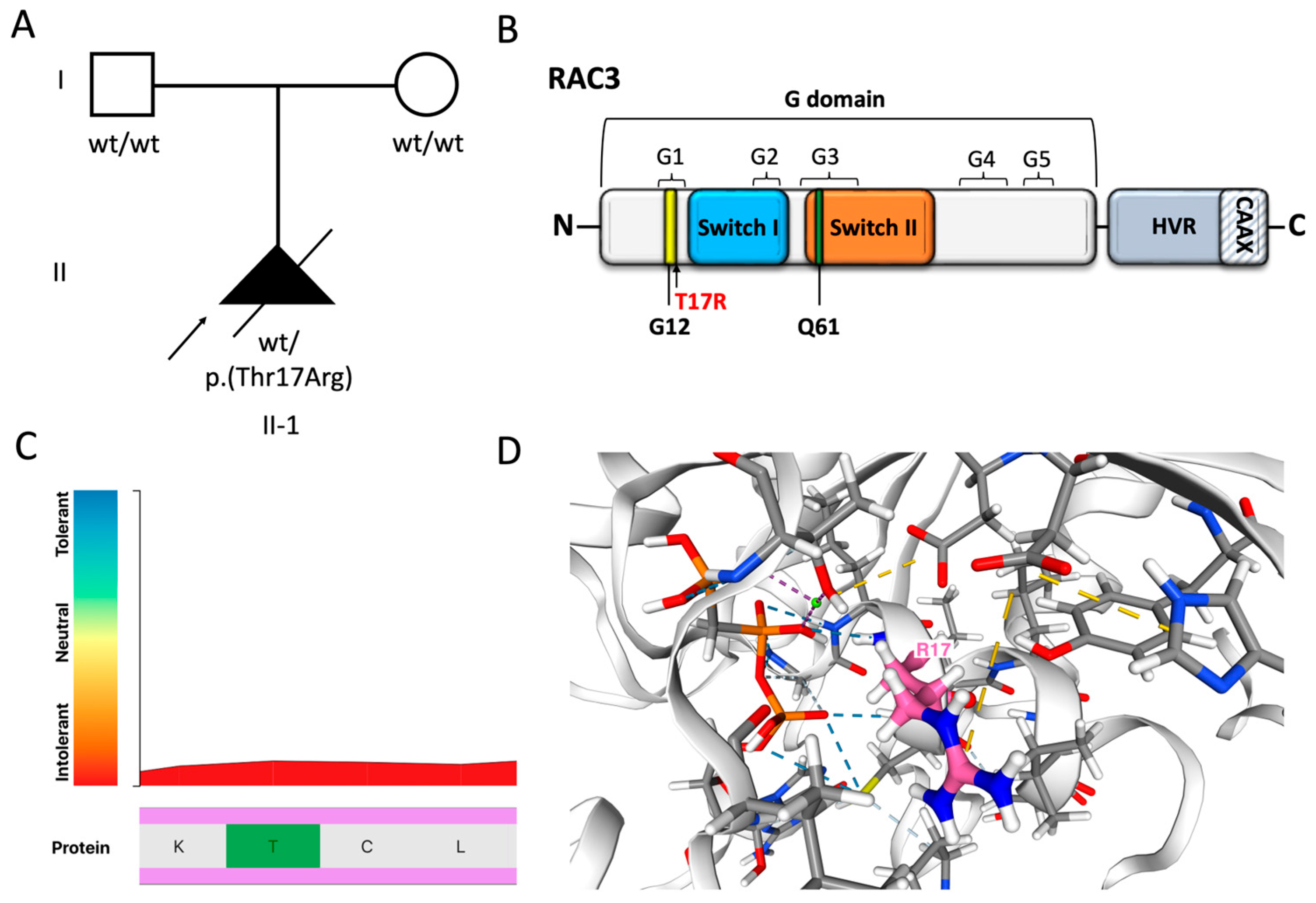
Genetic findings. (
A) Family pedigree. The proband was conceived by unrelated, healthy parents and was found to harbor a de novo variant in
RAC3, NM_005052.3: c.50C>G p.(T17R) (
WT = wild type;
Mut = mutation). (
B) Schematic illustration of RAC3 GTPase and mapping of the p.(T17R) variant. The G domain contains the Switch I and II regions and G1-G5 boxes, and includes highly conserved residues such as G12 and Q61. The C-terminal hypervariable region (HVR) contains the CAAX box, a consensus sequence necessary for posttranslational modifications that regulate subcellular localization of the protein. The p.(T17R) substitution affects the last residue of the G1 box and is located near the Switch I domain. (
C) Intolerance analysis according to Metadome (
https://stuart.radboudumc.nl/metadome/dashboard, accessed on 1 July 2025). Thr17 is highly intolerant to genetic variation (score = 0.15) and is located within a functionally constrained region of the protein, suggesting that alterations at this site are likely pathogenic. (
D) The p.(T17R) substitution is predicted to destabilize the protein structure, with an estimated ΔΔG of 3 kcal/mol. This residue is located 2.0 Å from the binding site for phosphomethylphosphonic acid-guanylate ester (GCP) and within the nucleotide phosphate-binding region for GTP, likely impacting these interactions. Furthermore, the substitution of Thr17 with Arg, which has a different shape and charge, is predicted to alter hydrogen bonding (blue and light blue) and ionic interactions (yellow) with neighboring residues. Predictions made according to Michelanglo (
https://michelanglo.sgc.ox.ac.uk/venus, accessed on 1 July 2025).
Figure 3.
Genetic findings. (
A) Family pedigree. The proband was conceived by unrelated, healthy parents and was found to harbor a de novo variant in
RAC3, NM_005052.3: c.50C>G p.(T17R) (
WT = wild type;
Mut = mutation). (
B) Schematic illustration of RAC3 GTPase and mapping of the p.(T17R) variant. The G domain contains the Switch I and II regions and G1-G5 boxes, and includes highly conserved residues such as G12 and Q61. The C-terminal hypervariable region (HVR) contains the CAAX box, a consensus sequence necessary for posttranslational modifications that regulate subcellular localization of the protein. The p.(T17R) substitution affects the last residue of the G1 box and is located near the Switch I domain. (
C) Intolerance analysis according to Metadome (
https://stuart.radboudumc.nl/metadome/dashboard, accessed on 1 July 2025). Thr17 is highly intolerant to genetic variation (score = 0.15) and is located within a functionally constrained region of the protein, suggesting that alterations at this site are likely pathogenic. (
D) The p.(T17R) substitution is predicted to destabilize the protein structure, with an estimated ΔΔG of 3 kcal/mol. This residue is located 2.0 Å from the binding site for phosphomethylphosphonic acid-guanylate ester (GCP) and within the nucleotide phosphate-binding region for GTP, likely impacting these interactions. Furthermore, the substitution of Thr17 with Arg, which has a different shape and charge, is predicted to alter hydrogen bonding (blue and light blue) and ionic interactions (yellow) with neighboring residues. Predictions made according to Michelanglo (
https://michelanglo.sgc.ox.ac.uk/venus, accessed on 1 July 2025).
![Cells 14 01499 g003 Cells 14 01499 g003]()
Figure 4.
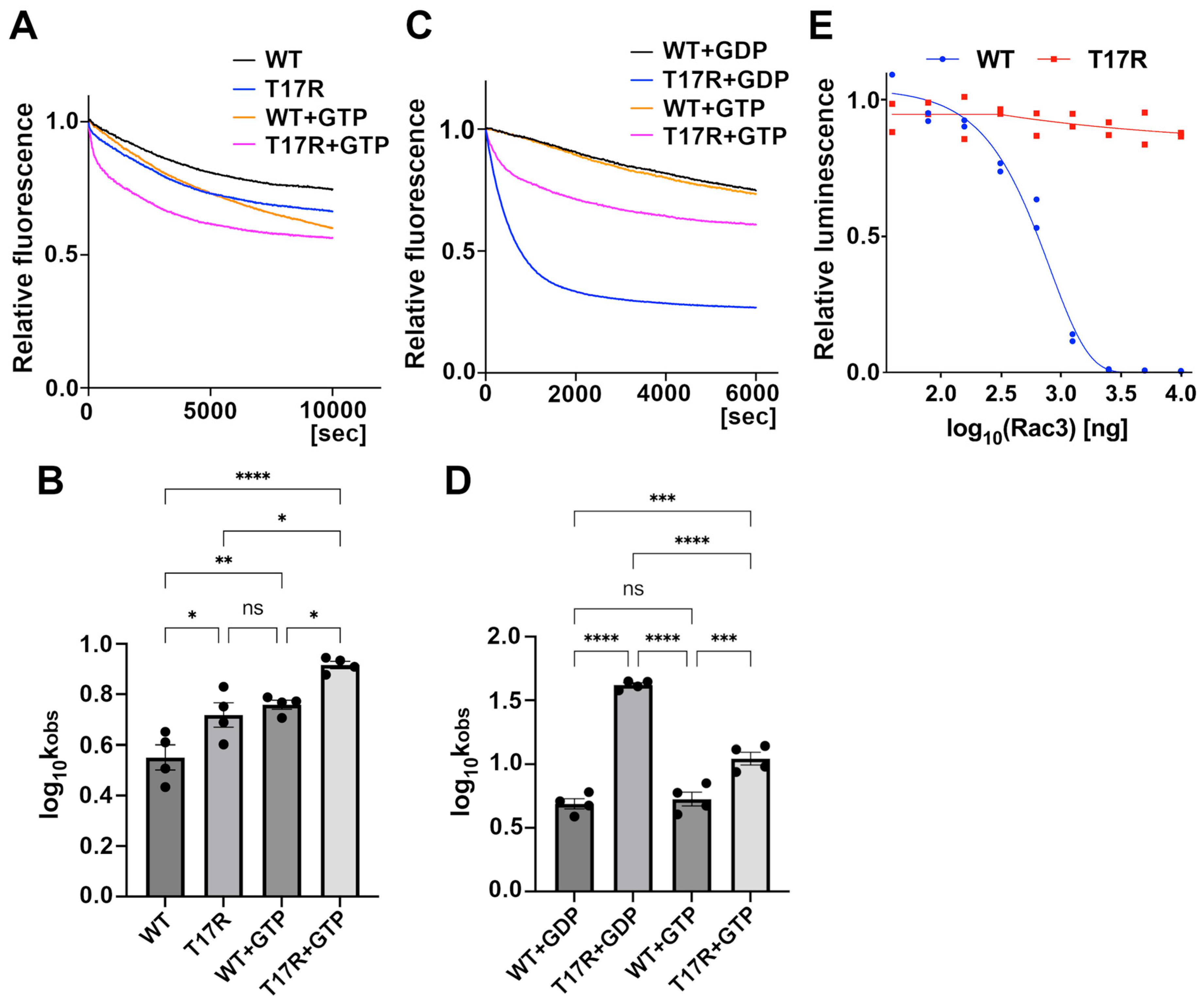
Impact of the p.T17R variant on the affinity of RAC3 proteins for GDP and GTP, and on GTP hydrolysis activity. (A–D) Mant-GDP-dissociation assay. (A) Fluorescently labeled mant-GDP-loaded His-tagged RAC3 (WT) and RAC3-T17R (T17R) were incubated with or without a non-hydrolysable GTP analog, GppNHp, and fluorescence changes were monitored over time to assess exchange activity. (B) The dissociation kinetics of mant-GDP from WT and T17R were evaluated, and the rate constants (kobs [×10−5/s]) were calculated based on the data from panel A. The data were analyzed using one-way ANOVA with Tukey’s post hoc test and shown as interleaved scatter with bars (p < 0.05). Sample sizes: WT, n = 4; T17R, n = 4; WT + GTP, n = 4; T17R + GTP; n = 4. WT vs. T17R, p = 0.031; WT vs. WT + GTP, p = 0.0078; WT vs. T17R + GTP, p < 0.0001; T17R vs. WT + GTP, p = 0.86, T17R vs. T17R + GTP, p = 0.011; WT + GTP vs. T17R + GTP, p = 0.043. ns: not significant, * p < 0.05, ** p < 0.01, **** p < 0.0001. (C) Mant-GDP-loaded WT and T17R were incubated with GDP or GppNHp, and fluorescence changes were then monitored over time. (D) The dissociation kinetics of mant-GDP from WT and T17R were evaluated, and the rate constants (kobs [×10−5/s]) were calculated based on the data from panel C. The data were analyzed using one-way ANOVA with Tukey’s post hoc test and shown with interleaved scatter with bars (p < 0.05). Samples sizes: WT, n = 4; T17R, n = 4; WT + GTP, n = 4; T17R + GTP; n = 4. WT + GDP vs. T17R + GDP, p < 0.0001; WT + GDP vs. WT + GTP, 0.92; WT + GDP vs. T17R + GTP, p = 0.0004; T17R + GDP vs. WT + GTP, p < 0.0001, T17R + GDP vs. T17R + GTP, p < 0.0001; WT + GTP vs. T17R + GTP, p = 0.001. ns: not significant, *** p < 0.001, **** p < 0.0001. (E) GTP hydrolysis assay. GTPase activity of WT and T17R was measured using the GTPase-Glo assay kit, which quantifies changes in GTP concentration. Sample sizes: WT, n = 3; T17R, n = 3. The sigmoidal fitting curve could not be generated for the T17R variant, preventing the determination of the half-maximal effective concentration (EC50). Statistical comparisons were not performed.
Figure 4.
Impact of the p.T17R variant on the affinity of RAC3 proteins for GDP and GTP, and on GTP hydrolysis activity. (A–D) Mant-GDP-dissociation assay. (A) Fluorescently labeled mant-GDP-loaded His-tagged RAC3 (WT) and RAC3-T17R (T17R) were incubated with or without a non-hydrolysable GTP analog, GppNHp, and fluorescence changes were monitored over time to assess exchange activity. (B) The dissociation kinetics of mant-GDP from WT and T17R were evaluated, and the rate constants (kobs [×10−5/s]) were calculated based on the data from panel A. The data were analyzed using one-way ANOVA with Tukey’s post hoc test and shown as interleaved scatter with bars (p < 0.05). Sample sizes: WT, n = 4; T17R, n = 4; WT + GTP, n = 4; T17R + GTP; n = 4. WT vs. T17R, p = 0.031; WT vs. WT + GTP, p = 0.0078; WT vs. T17R + GTP, p < 0.0001; T17R vs. WT + GTP, p = 0.86, T17R vs. T17R + GTP, p = 0.011; WT + GTP vs. T17R + GTP, p = 0.043. ns: not significant, * p < 0.05, ** p < 0.01, **** p < 0.0001. (C) Mant-GDP-loaded WT and T17R were incubated with GDP or GppNHp, and fluorescence changes were then monitored over time. (D) The dissociation kinetics of mant-GDP from WT and T17R were evaluated, and the rate constants (kobs [×10−5/s]) were calculated based on the data from panel C. The data were analyzed using one-way ANOVA with Tukey’s post hoc test and shown with interleaved scatter with bars (p < 0.05). Samples sizes: WT, n = 4; T17R, n = 4; WT + GTP, n = 4; T17R + GTP; n = 4. WT + GDP vs. T17R + GDP, p < 0.0001; WT + GDP vs. WT + GTP, 0.92; WT + GDP vs. T17R + GTP, p = 0.0004; T17R + GDP vs. WT + GTP, p < 0.0001, T17R + GDP vs. T17R + GTP, p < 0.0001; WT + GTP vs. T17R + GTP, p = 0.001. ns: not significant, *** p < 0.001, **** p < 0.0001. (E) GTP hydrolysis assay. GTPase activity of WT and T17R was measured using the GTPase-Glo assay kit, which quantifies changes in GTP concentration. Sample sizes: WT, n = 3; T17R, n = 3. The sigmoidal fitting curve could not be generated for the T17R variant, preventing the determination of the half-maximal effective concentration (EC50). Statistical comparisons were not performed.
![Cells 14 01499 g004 Cells 14 01499 g004]()
Figure 5.
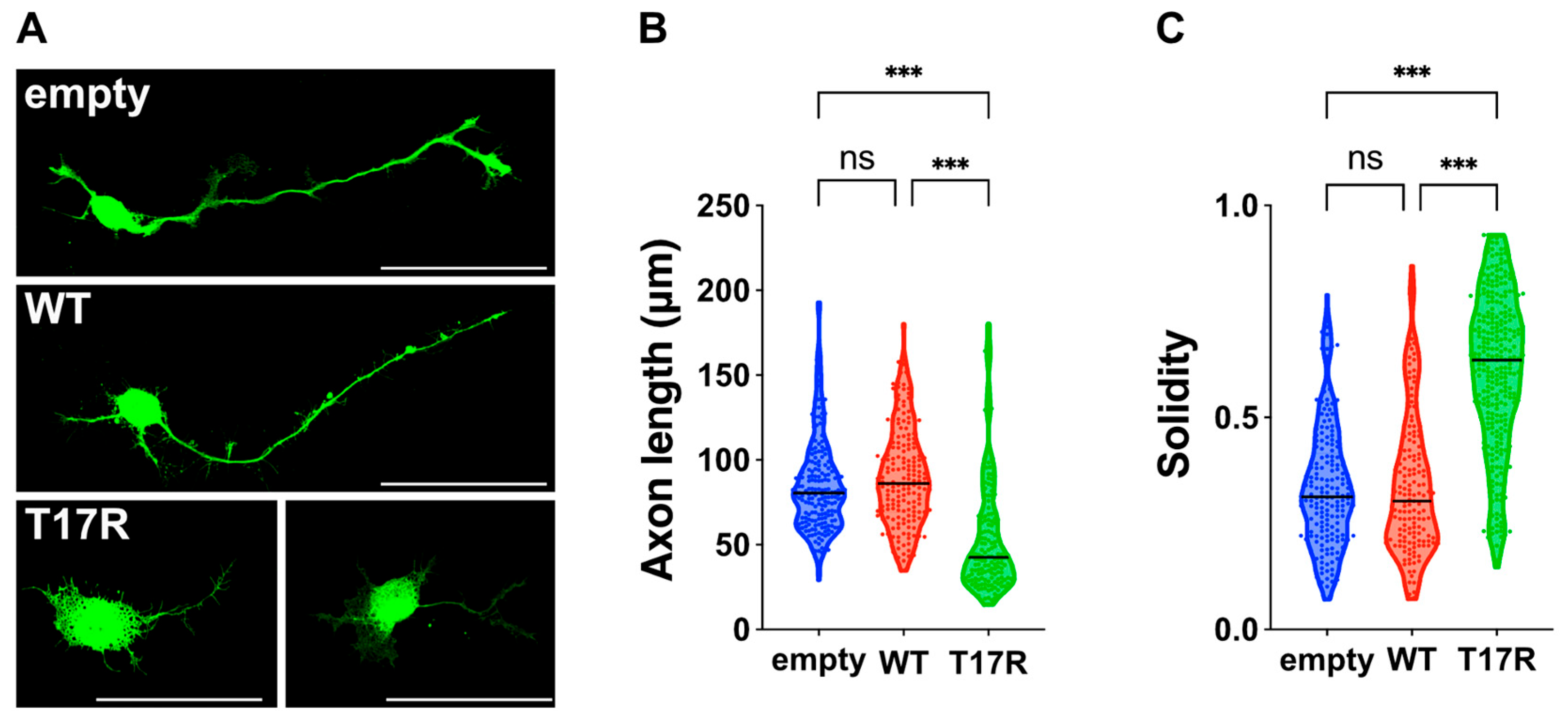
Effects of the p.(T17R) variant on neuron morphology in vitro. (
A) Primary hippocampal neurons were isolated from E16 embryos and electroporated with pCAG-EGFP (0.1 μg) along with either pCAG-Myc (empty), pCAG-Myc-RAC3 (WT), or pCAG-Myc-RAC3-T17R (0.3 μg each). After 3 days in culture, neurons were fixed and immunostained using anti-GFP (green). Scale bars, 50 μm. (
B,
C) Morphometric analysis of neurons shown in (
A). (
B) Axon length, defined as the longest neurite in each GFP-labeled neuron, was quantified and presented as violin plots with dots. Neuron counts: empty,
n = 196; WT,
n = 197; T17R,
n = 160. Statistical significance between vector (empty), WT, and T17R was determined using one-way ANOVA with Tukey’s post hoc test (
p < 0.05). empty vs. WT,
p = 0.3; empty vs. T17R,
p < 0.001; WT vs. T17R,
p < 0.001. (
C) Cell solidity was measured in GFP-positive neurons and shown as violin plots with boxplots. “Solidity” is the ratio of the area of a cell to the area of a convex hull of the cell [
16]. Neuron counts: empty,
n = 181; WT,
n = 157; T17R,
n = 290. Statistical significance between vector (empty), WT, and T17R was determined using one-way ANOVA with Tukey’s post hoc test (
p < 0.05). empty vs. WT,
p = 0.97; empty vs. T17R,
p < 0.001; WT vs. T17R,
p < 0.001. ns: not significant, ***
p < 0.001.
Figure 5.
Effects of the p.(T17R) variant on neuron morphology in vitro. (
A) Primary hippocampal neurons were isolated from E16 embryos and electroporated with pCAG-EGFP (0.1 μg) along with either pCAG-Myc (empty), pCAG-Myc-RAC3 (WT), or pCAG-Myc-RAC3-T17R (0.3 μg each). After 3 days in culture, neurons were fixed and immunostained using anti-GFP (green). Scale bars, 50 μm. (
B,
C) Morphometric analysis of neurons shown in (
A). (
B) Axon length, defined as the longest neurite in each GFP-labeled neuron, was quantified and presented as violin plots with dots. Neuron counts: empty,
n = 196; WT,
n = 197; T17R,
n = 160. Statistical significance between vector (empty), WT, and T17R was determined using one-way ANOVA with Tukey’s post hoc test (
p < 0.05). empty vs. WT,
p = 0.3; empty vs. T17R,
p < 0.001; WT vs. T17R,
p < 0.001. (
C) Cell solidity was measured in GFP-positive neurons and shown as violin plots with boxplots. “Solidity” is the ratio of the area of a cell to the area of a convex hull of the cell [
16]. Neuron counts: empty,
n = 181; WT,
n = 157; T17R,
n = 290. Statistical significance between vector (empty), WT, and T17R was determined using one-way ANOVA with Tukey’s post hoc test (
p < 0.05). empty vs. WT,
p = 0.97; empty vs. T17R,
p < 0.001; WT vs. T17R,
p < 0.001. ns: not significant, ***
p < 0.001.
![Cells 14 01499 g005 Cells 14 01499 g005]()
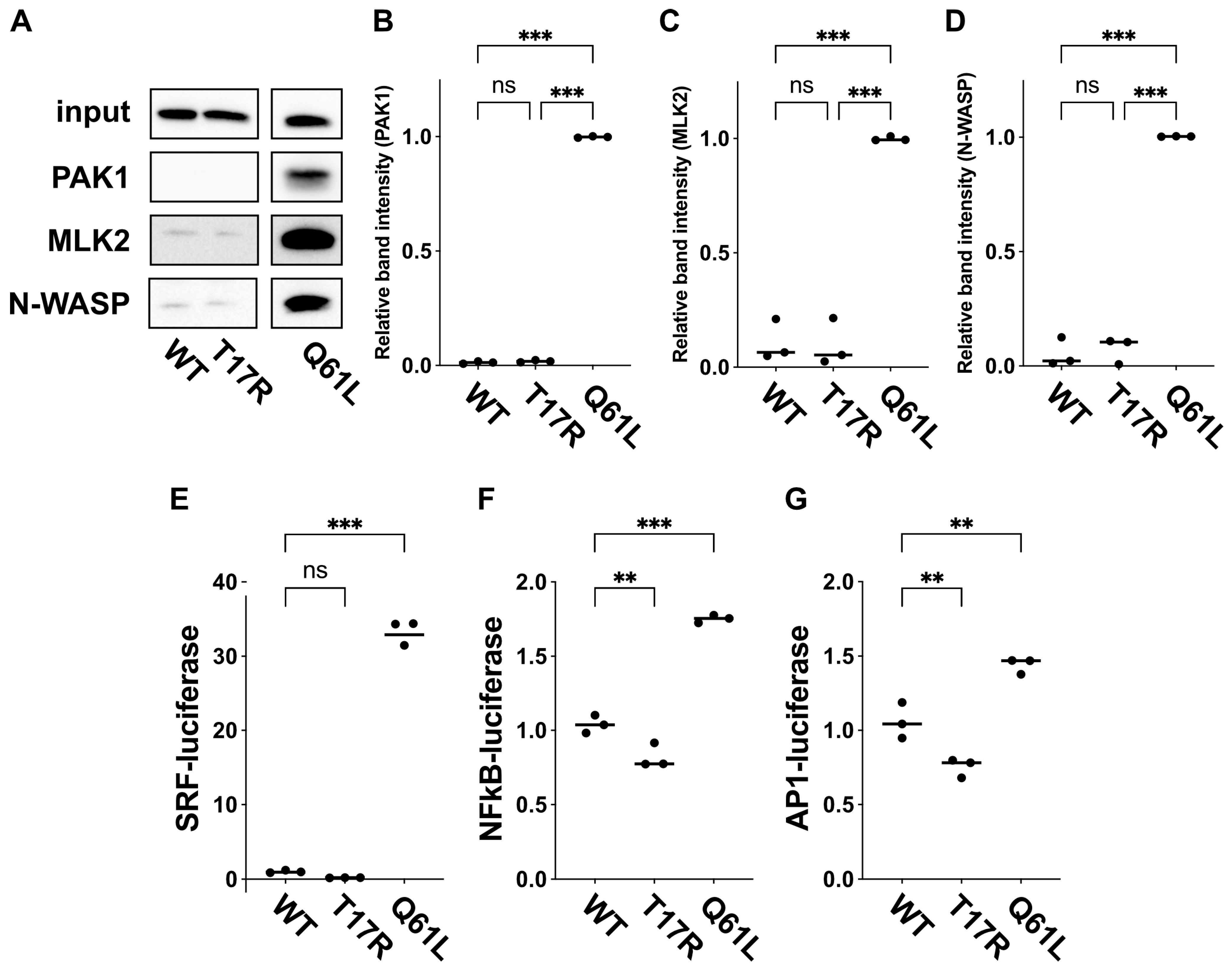
Figure 6.
Effects of the p.(T17R) variant on interaction with downstream effectors. (
A) Evaluation of binding to the RBRs of PAK1, MLK2, and N-WASP. COS7 cells were transfected with pCAG-Myc-RAC3 (WT), -RAC3-T17R, or -RAC3-Q61L (0.3 μg each). GST pull-down assays were conducted using GST-tagged RBRs from PAK1, MLK2, and N-WASP (5 μg each) as in “Materials and methods”. Bound RAC3 proteins were detected via Western blot using anti-Myc. Input samples were also analyzed to verify equivalent expression levels. Full Western blot data of this figure and blot data with molecular marker bands are provided in
Supplementary Figures S2 and S3, respectively. (
B–
D) The intensity of RAC3 band bound to the GST-fused RBR of PAK1 (
B), MLK2 (
C), or N-WASP (
D) was quantified. The relative band intensity is displayed, with RAC3-Q61L set at 1.0. Samples sizes:
n = 3 replicates. Statistical significance between WT and each variant was assessed using one-way ANOVA with Tukey’s post hoc test (
p < 0.05). (B) WT vs. T17R,
p = 0.32; WT vs. Q61L,
p < 0.001; T17R vs. Q61L,
p < 0.001. (C) WT vs. T17R,
p = 0.99; WT vs. Q61L,
p < 0.001; T17R vs. Q61L,
p < 0.001. (D) WT vs. T17R,
p = 0.86; WT vs. Q61L,
p < 0.001; T17R vs. Q61L,
p < 0.001. ns: not significant, ***
p < 0.001. (
E–
G) Effects of p.(T17R) on SRF-, NFkB- and AP1-dependent gene transcription. COS7 cells were transfected with pCAG-Myc-RAC3 (WT) or -RAC3-T17R (0.1 μg each) along with luciferase reporter constructs for SRF (
E), NFkB (
F), or AP1 (
G) (0.05 μg each). Reporter activity was measured, normalized to WT (set at 1.0), and presented as scatter plots with bars. Samples sizes:
n = 3 replicates. Statistical significance was determined using one-way ANOVA with Dunnett’s post hoc test (
p < 0.05). (E) WT vs. T17R,
p = 0.52; WT vs. Q61L,
p < 0.001. (F) WT vs. T17R,
p = 0.008; WT vs. Q61L,
p < 0.001. (G) WT vs. T17R,
p = 0.008; WT vs. Q61L,
p = 0.003. ns: not significant, **
p < 0.01, ***
p < 0.001.
Figure 6.
Effects of the p.(T17R) variant on interaction with downstream effectors. (
A) Evaluation of binding to the RBRs of PAK1, MLK2, and N-WASP. COS7 cells were transfected with pCAG-Myc-RAC3 (WT), -RAC3-T17R, or -RAC3-Q61L (0.3 μg each). GST pull-down assays were conducted using GST-tagged RBRs from PAK1, MLK2, and N-WASP (5 μg each) as in “Materials and methods”. Bound RAC3 proteins were detected via Western blot using anti-Myc. Input samples were also analyzed to verify equivalent expression levels. Full Western blot data of this figure and blot data with molecular marker bands are provided in
Supplementary Figures S2 and S3, respectively. (
B–
D) The intensity of RAC3 band bound to the GST-fused RBR of PAK1 (
B), MLK2 (
C), or N-WASP (
D) was quantified. The relative band intensity is displayed, with RAC3-Q61L set at 1.0. Samples sizes:
n = 3 replicates. Statistical significance between WT and each variant was assessed using one-way ANOVA with Tukey’s post hoc test (
p < 0.05). (B) WT vs. T17R,
p = 0.32; WT vs. Q61L,
p < 0.001; T17R vs. Q61L,
p < 0.001. (C) WT vs. T17R,
p = 0.99; WT vs. Q61L,
p < 0.001; T17R vs. Q61L,
p < 0.001. (D) WT vs. T17R,
p = 0.86; WT vs. Q61L,
p < 0.001; T17R vs. Q61L,
p < 0.001. ns: not significant, ***
p < 0.001. (
E–
G) Effects of p.(T17R) on SRF-, NFkB- and AP1-dependent gene transcription. COS7 cells were transfected with pCAG-Myc-RAC3 (WT) or -RAC3-T17R (0.1 μg each) along with luciferase reporter constructs for SRF (
E), NFkB (
F), or AP1 (
G) (0.05 μg each). Reporter activity was measured, normalized to WT (set at 1.0), and presented as scatter plots with bars. Samples sizes:
n = 3 replicates. Statistical significance was determined using one-way ANOVA with Dunnett’s post hoc test (
p < 0.05). (E) WT vs. T17R,
p = 0.52; WT vs. Q61L,
p < 0.001. (F) WT vs. T17R,
p = 0.008; WT vs. Q61L,
p < 0.001. (G) WT vs. T17R,
p = 0.008; WT vs. Q61L,
p = 0.003. ns: not significant, **
p < 0.01, ***
p < 0.001.
![Cells 14 01499 g006 Cells 14 01499 g006]()
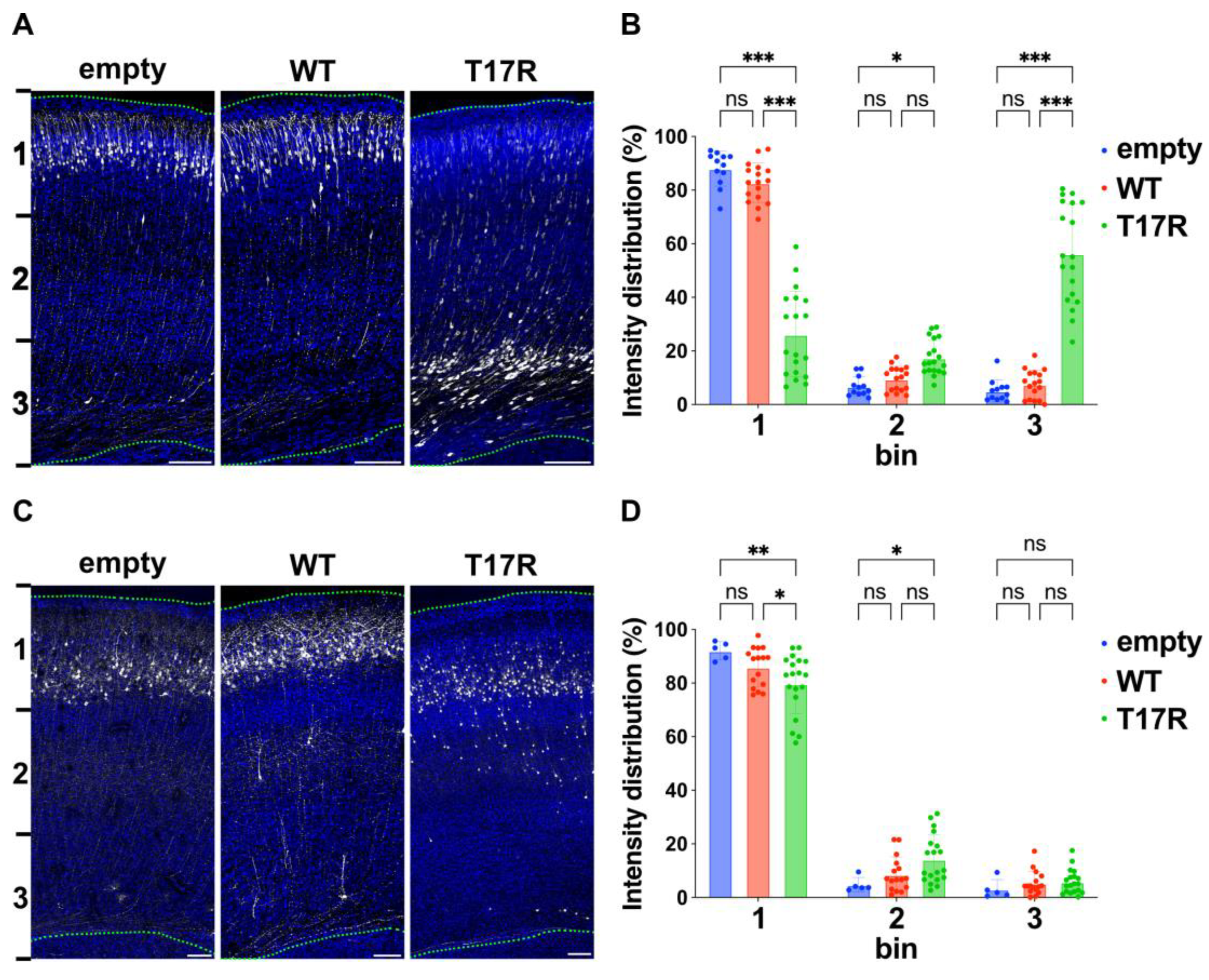
Figure 7.
Effects of the p.(T17R) variant on excitatory neuron migration during corticogenesis. (A,C) Representative images of migration defects of neurons expressing the p.(T17R) variant. E14.5 VZ progenitor cells were electroporated in utero with pCAG-EGFP (0.5 μg) along with either pCAG-Myc (empty), pCAG-Myc-RAC3 (WT), or pCAG-RAC3-T17R (0.1 μg each). Coronal sections were collected at P0 (A) and P7 (C), and stained with anti-GFP (white) and DAPI (blue). Scale bars, 100 μm. (B,D) Quantification of (A,C). Number of replicates, n ≥ 5. Statistical significance between WT and the variant was determined using two-way ANOVA with Tukey’s post hoc test and shown with interleaved scatter with bars (p < 0.05). (B) bin 1: empty vs. WT, p = 0.36; empty vs. T17R, p < 0.001; WT vs. T17R, p < 0.001. bin 2: empty vs. WT, p = 0.76; empty vs. T17R, p = 0.01; WT vs. T17R, p = 0.06. bin 3: empty vs. WT, p = 0.80; empty vs. T17R, p < 0.001; WT vs. T17R, p < 0.001. (D) bin 1: empty vs. WT, p = 0.23; empty vs. T17R, p = 0.003; WT vs. T17R, p = 0.04. bin 2: empty vs. WT, p = 0.55; empty vs. T17R, p = 0.03; WT vs. T17R, p = 0.06. bin 3: empty vs. WT, p = 0.82; empty vs. T17R, p = 0.73; WT vs. T17R, p = 0.98. ns: not significant, * p < 0.05, ** p < 0.01, *** p < 0.001.
Figure 7.
Effects of the p.(T17R) variant on excitatory neuron migration during corticogenesis. (A,C) Representative images of migration defects of neurons expressing the p.(T17R) variant. E14.5 VZ progenitor cells were electroporated in utero with pCAG-EGFP (0.5 μg) along with either pCAG-Myc (empty), pCAG-Myc-RAC3 (WT), or pCAG-RAC3-T17R (0.1 μg each). Coronal sections were collected at P0 (A) and P7 (C), and stained with anti-GFP (white) and DAPI (blue). Scale bars, 100 μm. (B,D) Quantification of (A,C). Number of replicates, n ≥ 5. Statistical significance between WT and the variant was determined using two-way ANOVA with Tukey’s post hoc test and shown with interleaved scatter with bars (p < 0.05). (B) bin 1: empty vs. WT, p = 0.36; empty vs. T17R, p < 0.001; WT vs. T17R, p < 0.001. bin 2: empty vs. WT, p = 0.76; empty vs. T17R, p = 0.01; WT vs. T17R, p = 0.06. bin 3: empty vs. WT, p = 0.80; empty vs. T17R, p < 0.001; WT vs. T17R, p < 0.001. (D) bin 1: empty vs. WT, p = 0.23; empty vs. T17R, p = 0.003; WT vs. T17R, p = 0.04. bin 2: empty vs. WT, p = 0.55; empty vs. T17R, p = 0.03; WT vs. T17R, p = 0.06. bin 3: empty vs. WT, p = 0.82; empty vs. T17R, p = 0.73; WT vs. T17R, p = 0.98. ns: not significant, * p < 0.05, ** p < 0.01, *** p < 0.001.
![Cells 14 01499 g007 Cells 14 01499 g007]()
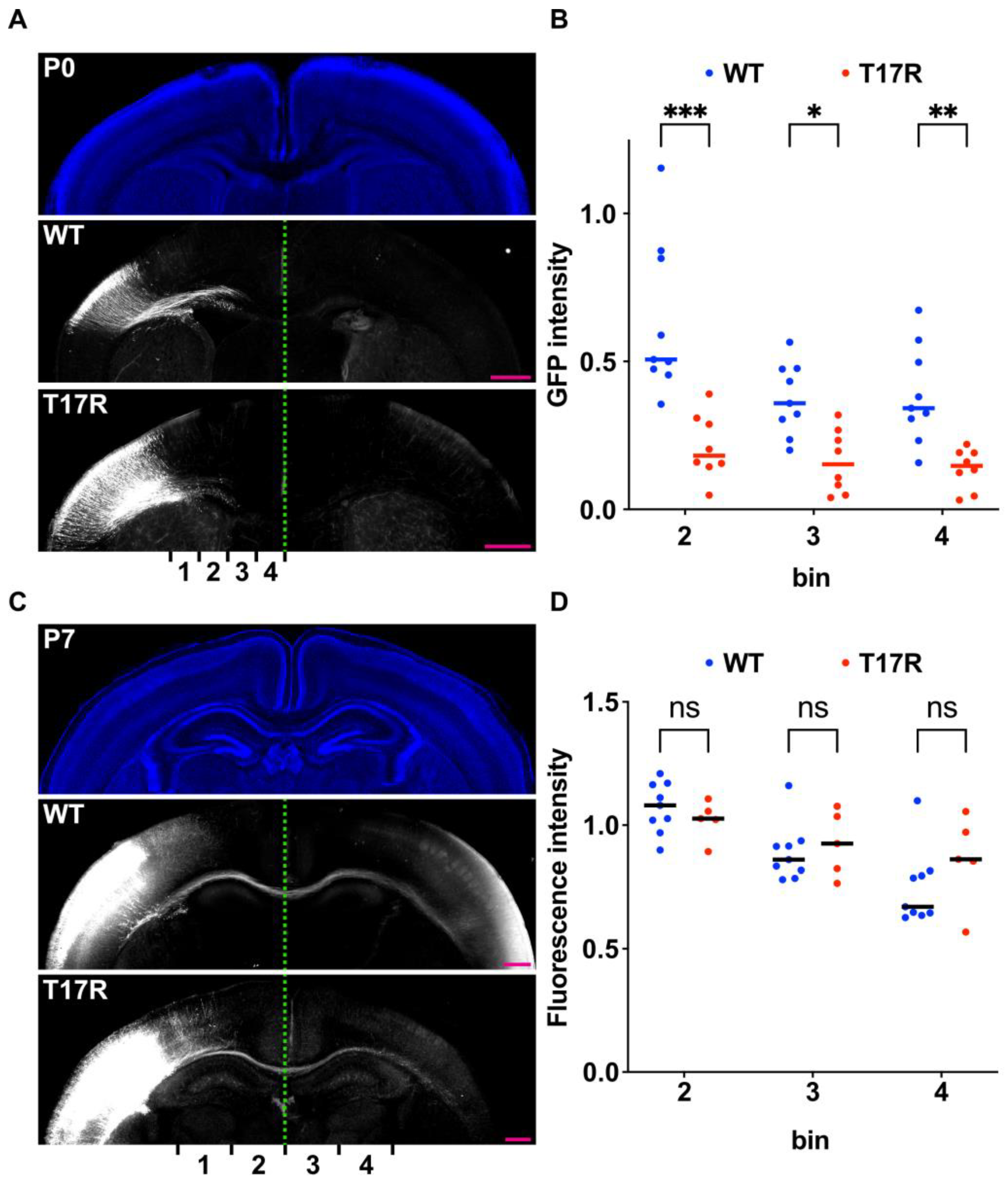
Figure 8.
Effects of the p.(T17R) variant on axonal extension during corticogenesis in vivo. (A,C) pCAG-GFP was co-electroporated at E14 with either pCAG-Myc-RAC3 (WT) or -RAC3-T17R (0.1 μg each). Coronal sections were obtained at P0 (A) or P7 (C), and GFP fluorescence was used to visualize callosal axons (white). Nuclei were stained with DAPI (blue). Scale bars, 500 μm. (B,D) Intensity of GFP-labeled callosal axons was quantified at P0 (B) and P7 (D) across different cortical regions (bins 1–4). Intensities of bins were normalized with bin 1 as 1.0. Statistical significance was assessed using two-way ANOVA with Šídák’s post hoc test and shown with interleaved scatter plots (p < 0.05). (B) The number of brains analyzed was as follows: WT, n = 9; T17R, n = 8. bin 2, p < 0.001; bin 3, p = 0.02; bin 4, p = 0.005. (D) The number of brains was as follows: WT, n = 9; T17R, n = 5. bin 2, p = 0.86; bin 3, p = 0.95; bin 4, p = 0.32. ns: not significant, * p < 0.05, ** p < 0.01, *** p < 0.001.
Figure 8.
Effects of the p.(T17R) variant on axonal extension during corticogenesis in vivo. (A,C) pCAG-GFP was co-electroporated at E14 with either pCAG-Myc-RAC3 (WT) or -RAC3-T17R (0.1 μg each). Coronal sections were obtained at P0 (A) or P7 (C), and GFP fluorescence was used to visualize callosal axons (white). Nuclei were stained with DAPI (blue). Scale bars, 500 μm. (B,D) Intensity of GFP-labeled callosal axons was quantified at P0 (B) and P7 (D) across different cortical regions (bins 1–4). Intensities of bins were normalized with bin 1 as 1.0. Statistical significance was assessed using two-way ANOVA with Šídák’s post hoc test and shown with interleaved scatter plots (p < 0.05). (B) The number of brains analyzed was as follows: WT, n = 9; T17R, n = 8. bin 2, p < 0.001; bin 3, p = 0.02; bin 4, p = 0.005. (D) The number of brains was as follows: WT, n = 9; T17R, n = 5. bin 2, p = 0.86; bin 3, p = 0.95; bin 4, p = 0.32. ns: not significant, * p < 0.05, ** p < 0.01, *** p < 0.001.
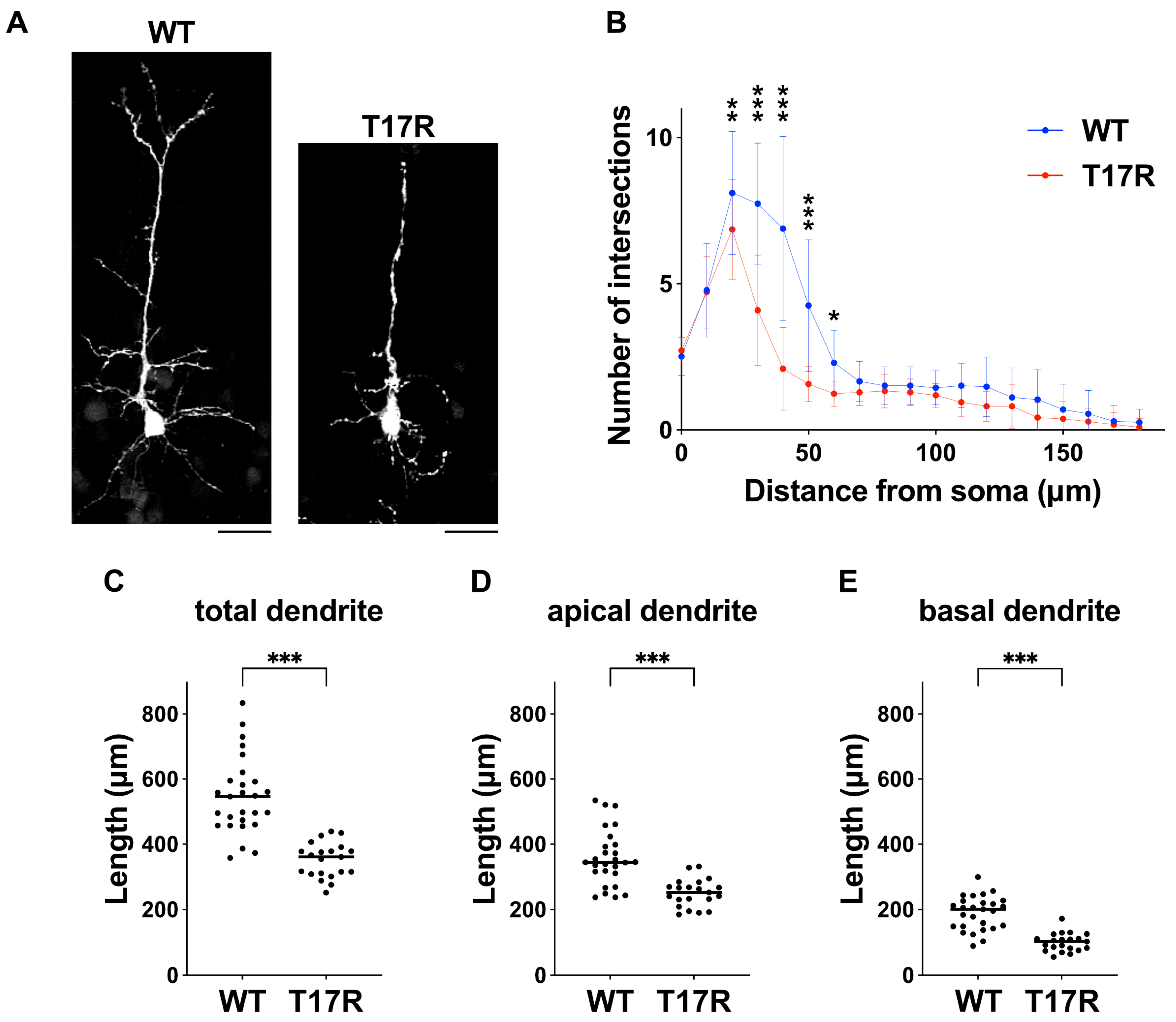
Figure 9.
Effects of the p.(T17R) variant on dendritic growth in vivo. (A) pCAG-GFP was co-electroporated at E14 with either pCAG-Myc-RAC3 (WT) or pCAG-Myc-RAC3-T17R (0.1 μg each). Coronal sections were obtained at P7, and GFP fluorescence was used to visualize callosal axons (white). Representative average Z-stack projection images of GFP-labeled cortical neurons in the upper cortical plate are shown. Scale bars, 30 μm. (B) Dendritic branching in (A) was quantified using Sholl analysis. Six brains were analyzed per condition. Number of replicates: n ≥ 21. Error bars represent SD. Statistical significance was assessed using two-way ANOVA followed by Šídák’s post hoc test. * p < 0.05, ** p < 0.01, *** p < 0.001. (C–E) Dendritic length measurements of neurons shown in (A) are presented as scatter plots for (C) total, (D) apical, and (E) basal dendrites. Statistical significance was calculated using unpaired t test (p < 0.05). (C) *** p < 0.001. (D) *** p < 0.001. (E) *** p < 0.001.
Figure 9.
Effects of the p.(T17R) variant on dendritic growth in vivo. (A) pCAG-GFP was co-electroporated at E14 with either pCAG-Myc-RAC3 (WT) or pCAG-Myc-RAC3-T17R (0.1 μg each). Coronal sections were obtained at P7, and GFP fluorescence was used to visualize callosal axons (white). Representative average Z-stack projection images of GFP-labeled cortical neurons in the upper cortical plate are shown. Scale bars, 30 μm. (B) Dendritic branching in (A) was quantified using Sholl analysis. Six brains were analyzed per condition. Number of replicates: n ≥ 21. Error bars represent SD. Statistical significance was assessed using two-way ANOVA followed by Šídák’s post hoc test. * p < 0.05, ** p < 0.01, *** p < 0.001. (C–E) Dendritic length measurements of neurons shown in (A) are presented as scatter plots for (C) total, (D) apical, and (E) basal dendrites. Statistical significance was calculated using unpaired t test (p < 0.05). (C) *** p < 0.001. (D) *** p < 0.001. (E) *** p < 0.001.