mRNA Multipeptide-HLA Class II Immunotherapy for Melanoma
Abstract
1. Introduction
1.1. Melanoma
1.2. Melanoma Cancer Antigens
1.3. Melanoma Treatment
Melanoma Vaccines
1.4. Human Leukocyte Antigen (HLA)
1.4.1. Peptide-HLA Binding
1.4.2. Binding of pHLA to T Cell Receptor (TCR)
1.5. HLA Expression in Melanoma Cells
1.5.1. General
1.5.2. Influence of HLA Melanoma Expression on Outcomes of Immunotherapy
1.5.3. HLA and Melanoma Vaccines: The Present Study
2. Materials and Methods
2.1. Melanoma/Cancer Antigens
2.2. In Silico Determination of Predicted Best Binding Affinities (PBBA) to Melanoma Antigens
2.3. Statistical Analysis
3. Results
3.1. Predicted Antigenicity of Melanoma-Associated Epitopes
3.1.1. Peptide Epitopes
3.1.2. HLA-II Alleles
4. Discussion
4.1. Methodological Considerations
4.2. Antigenicity and Immunogenicity
4.3. Antigenic pHLA-II Complexes for Melanoma
4.4. Implications for Multipeptide HLA-II Restricted Melanoma Vaccines
4.5. Melanoma Immunotherapy Based on the Synthesis of New HLA Molecules
4.5.1. HLA-II Molecules
4.5.2. HLA-I Molecules
4.5.3. Tumor Microenvironment
4.5.4. Challenges
5. Limitations
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Centeno, P.P.; Pavet, V.; Marais, R. The journey from melanocytes to melanoma. Nat. Rev. Cancer 2023, 23, 372–390. [Google Scholar] [CrossRef]
- American Cancer Society. Cancer Facts and Figures 2025; American Cancer Society: Atlanta, GA, USA, 2025. [Google Scholar]
- Elder, D.E.; Bastian, B.C.; Cree, I.A.; Massi, D.; Scolyer, R.A. The 2018 World Health Organization classification of cutaneous, mucosal, and uveal melanoma: Detailed analysis of 9 distinct subtypes defined by their evolutionary pathway. Arch. Pathol. Lab. Med. 2020, 144, 500–522. [Google Scholar] [CrossRef]
- Patel, V.R.; Roberson, M.L.; Pignone, M.P.; Adamson, A.S. Risk of mortality after a diagnosis of melanoma in situ. JAMA Dermatol. 2023, 159, 703–710. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Shen, Y.; Liu, Q.; Zhang, H.; Jia, L.; Chai, Y.; Jiang, H.; Wu, M.; Li, Y. Global trends in melanoma burden: A comprehensive analysis from the Global Burden of Disease Study, 1990–2021. J. Am. Acad. Dermatol. 2025, 92, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Waseh, S.; Lee, J.B. Advances in melanoma: Epidemiology, diagnosis, and prognosis. Front. Med. 2023, 10, 1268479. [Google Scholar] [CrossRef] [PubMed]
- Pitcovski, J.; Shahar, E.; Aizenshtein, E.; Gorodetsky, R. Melanoma antigens and related immunological markers. Crit. Rev. Oncol. Hematol. 2017, 115, 36–49. [Google Scholar] [CrossRef]
- Lawrence, M.S.; Stojanov, P.; Polak, P.; Kryukov, G.V.; Cibulskis, K.; Sivachenko, A.; Carter, S.L.; Stewart, C.; Mermel, C.H.; Roberts, S.A.; et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013, 499, 214–218. [Google Scholar] [CrossRef]
- Jiang, T.; Shi, T.; Zhang, H.; Song, Y.; Wei, J.; Ren, S.; Zhou, C. Tumor neoantigens: From basic research to clinical applications. J. Hematol. Oncol. 2019, 12, 93. [Google Scholar] [CrossRef]
- Xie, N.; Shen, G.; Gao, W.; Huang, Z.; Huang, C.; Fu, L. Neoantigens: Promising targets for cancer therapy. Sig. Transduct. Target Ther. 2023, 8, 9. [Google Scholar] [CrossRef]
- Kalaora, S.; Nagler, A.; Wargo, J.A.; Samuels, Y. Mechanisms of immune activation and regulation: Lessons from melanoma. Nat. Rev. Cancer 2022, 22, 195–207. [Google Scholar] [CrossRef]
- Cui, C.; Ott, P.A.; Wu, C.J. Advances in vaccines for melanoma. Hematol. Oncol. Clin. North Am. 2024, 38, 1045–1060. [Google Scholar] [CrossRef]
- Svedman, F.C.; Pillas, D.; Taylor, A.; Kaur, M.; Linder, R.; Hansson, J. Stage-specific survival and recurrence in patients with cutaneous malignant melanoma in Europe—A systematic review of the literature. Clin. Epidemiol. 2016, 8, 109–122. [Google Scholar] [CrossRef]
- Huang, A.C.; Zappasodi, R. A decade of checkpoint blockade immunotherapy in melanoma: Understanding the molecular basis for immune sensitivity and resistance. Nat. Immunol. 2022, 23, 660–670. [Google Scholar] [CrossRef]
- Knight, A.; Karapetyan, L.; Kirkwood, J.M. Immunotherapy in melanoma: Recent advances and future directions. Cancers 2023, 15, 1106. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ma, S.; Zhu, S.; Zhu, L.; Guo, W. Advances in immunotherapy and targeted therapy of malignant melanoma. Biomedicines 2025, 13, 225. [Google Scholar] [CrossRef] [PubMed]
- Larkin, J.; Chiarion-Sileni, V.; Gonzalez, R.; Grob, J.-J.; Rutkowski, P.; Lao, C.D.; Cowey, L.; Schadendorf, D.; Wagstaff, J.; Drummer, R.; et al. Five-year survival with combined nivolumab and ipilimumab in advanced melanoma. N. Engl. J. Med. 2019, 381, 1535–1546. [Google Scholar] [CrossRef] [PubMed]
- Seta, T.; Nakamura, S.; Oura, M.; Yokoyoma, K.; Nishikawa, Y.; Hoshino, N.; Ninomiya, K.; Shimoi, T.; Hotta, K.; Nakayama, T. Efficacy and safety of cancer vaccine therapy in malignant melanoma: A systematic review. Int. J. Clin. Oncol. 2025, 30, 1080–1097. [Google Scholar] [CrossRef]
- Khaddour, K.; Buchbinder, E.I. Individualized neoantigen-directed melanoma therapy. Am. J. Clin. Dermatol. 2025, 26, 225–235. [Google Scholar] [CrossRef]
- Liao, H.C.; Liu, S.J. Advances in nucleic acid-based cancer vaccines. J. Biomed. Sci. 2025, 32, 10. [Google Scholar] [CrossRef]
- Yaremenko, A.V.; Khan, M.M.; Zhen, X.; Tang, Y.; Tao, W. Clinical advances of mRNA vaccines for cancer immunotherapy. Med 2025, 6, 100562. [Google Scholar] [CrossRef]
- Chekaoui, A.; Garofalo, M.; Gad, B.; Staniszewska, M.; Chiaro, J.; Pancer, K.; Cryciuk, A.; Cerullo, V.; Salmaso, S.; Caliceti, P.; et al. Cancer vaccines: An update on recent achievements and prospects for cancer therapy. Clin. Exp. Med. 2024, 25, 24. [Google Scholar] [CrossRef]
- Medhasi, S.; Chantratita, N. Human leukocyte antigen (HLA) system: Genetics and association with bacterial and viral infections. J. Immunol. Res. 2022, 2022, 9710376. [Google Scholar] [CrossRef]
- Neefjes, J.; Jongsma, M.; Paul, P.; Bakke, O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol. 2011, 11, 823–836. [Google Scholar] [CrossRef] [PubMed]
- Cerundolo, V.; Elliott, T.; Elvin, J.; Bastin, J.; Rammensee, H.G.; Townsend, A. The binding affinity and dissociation rates of peptides for class I major histocompatibility complex molecules. Eur. J. Immunol. 1991, 21, 2069–2075. [Google Scholar] [CrossRef] [PubMed]
- Chicz, R.M.; Urban, R.G.; Lane, W.S.; Gorga, J.C.; Stern, L.J.; Vignali, D.A.; Strominger, J.L. Predominant naturally processed peptides bound to HLA-DR1 are derived from MHC-related molecules and are heterogeneous in size. Nature 1992, 358, 764–768. [Google Scholar] [CrossRef] [PubMed]
- Trowsdale, J.; Knight, J.C. Major histocompatibility complex genomics and human disease. Annu. Rev. Genomics Hum. Genet. 2013, 14, 301–323. [Google Scholar] [CrossRef]
- Dendrou, C.A.; Petersen, J.; Rossjohn, J.; Fugger, L. HLA variation and disease. Nat. Rev. Immunol. 2018, 18, 325–339. [Google Scholar] [CrossRef]
- Sette, A.; Vitiello, A.; Reherman, B.; Fowler, P.; Nayersina, R.; Kast, W.M.; Melief, C.J.; Oseroff, C.; Yuan, L.; Ruppert, J.; et al. The relationship between class I binding affinity and immunogenicity of potential cytotoxic T cell epitopes. J. Immunol. 1994, 153, 5586–5592. [Google Scholar] [CrossRef]
- Hov, J.R.; Kosmoliaptsis, V.; Traherne, J.A.; Olsson, M.; Bobreg, K.M.; Bergquist, A.; Schrumpf, E.; Bradley, J.A.; Taylor, C.J.; Lie, B.A.; et al. Electrostatic modifications of the human leukocyte antigen-DR P9 peptide-binding pocket and susceptibility to primary sclerosing cholangitis. Hepatology 2011, 53, 1967–1976. [Google Scholar] [CrossRef]
- Davenport, M.P.; Quinn, C.L.; Chicz, R.M.; Green, B.N.; Willis, A.C.; Lane, W.S.; Bell, J.I.; Hill, A.V. Naturally processed peptides from two disease-resistance-associated HLA-DR13 alleles show related sequence motifs and the effects of the dimorphism at position 86 of the HLA-DR beta chain. Proc. Natl. Acad. Sci. USA 1995, 92, 6567–6571. [Google Scholar] [CrossRef]
- van Deutekom, H.W.; Keşmir, C. Zooming into the binding groove of HLA molecules: Which positions and which substitutions change peptide binding most? Immunogenetics 2015, 67, 425–436. [Google Scholar] [CrossRef]
- Paul, S.; Weiskopf, D.; Angelo, M.A.; Sidney, J.; Peters, B.; Sette, A. HLA class I alleles are associated with peptide-binding repertoires of different size, affinity, and immunogenicity. J. Immunol. 2013, 191, 5831–5839. [Google Scholar] [CrossRef]
- Margulies, D.H.; Corr, M.; Boyd, L.F.; Khilko, S.N. MHC class I/peptide interactions: Binding specificity and kinetics. J. Mol. Recognit. 1993, 6, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Sykulev, Y.; Joo, M.; Vturina, I.; Tsomides, T.J.; Eisen, H.N. Evidence that a single peptide-MHC complex on a target cell can elicit a cytolytic T cell response. Immunity 1996, 4, 565–571. [Google Scholar] [CrossRef] [PubMed]
- Irvine, D.J.; Purbhoo, M.A.; Krogsgaard, M.; Davis, M.M. Direct observation of ligand recognition by T cells. Nature 2002, 419, 845–849. [Google Scholar] [CrossRef] [PubMed]
- Taramelli, D.; Fossati, G.; Mazzocchi, A.; Delia, D.; Ferrone, S.; Parmiani, G. Classes I and II HLA and melanoma-associated antigen expression and modulation on melanoma cells isolated from primary and metastatic lesions. Cancer Res. 1986, 46, 433–939. [Google Scholar]
- Johnson, D.B.; Estrada, M.V.; Salgado, R.; Sanchez, V.; Doxie, D.B.; Opalenik, S.R.; Vilgelm, A.E.; Feld, E.; Johnson, A.S.; Greenplate, A.R.; et al. Melanoma-specific MHC-II expression represents a tumour-autonomous phenotype and predicts response to anti-PD-1/PD-L1 therapy. Nat. Commun. 2016, 7, 10582. [Google Scholar] [CrossRef]
- Chen, Y.-Y.; Chang, W.-A.; Lin, E.-S.; Chen, Y.-J.; Kuo, P.-L. Expressions of HLA Class II genes in cutaneous melanoma were associated with clinical outcome: Bioinformatics approaches and systematic analysis of public microarray and RNA-Seq datasets. Diagnostics 2019, 9, 59. [Google Scholar] [CrossRef]
- Costantini, F.; Barbieri, G. The HLA-DR mediated signalling increases the migration and invasion of melanoma cells, the expression and lipid raft recruitment of adhesion receptors, PD-L1 and signal transduction proteins. Cell. Signal. 2017, 36, 189–203. [Google Scholar] [CrossRef]
- Amrane, K.; Le Meur, C.; Besse, B.; Hemon, P.; Le Noac’h, P.; Pradier, O.; Berthou, C.; Abgral, R.; Uguen, A. HLA-DR expression in melanoma: From misleading therapeutic target to potential immunotherapy biomarker. Front. Immunol. 2024, 14, 1285895. [Google Scholar] [CrossRef]
- Mendez, R.; Aptsiauri, N.; Del Campo, A.; Maleno, I.; Cabrera, T.; Ruiz-Cabello, F.; Garrido, F.; Garcia-Lora, A. HLA and melanoma: Multiple alterations in HLA class I and II expression in human melanoma cell lines from ESTDAB cell bank. Cancer Immunol. Immunother. 2009, 58, 1507–1515. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Yu, Y.; Wu, D.; Wang, S.; Fang, Y.; Miao, H.; Ma, P.; Huang, H.; Zhang, M.; Zhang, Y.; et al. HLA and tumour immunology: Immune escape, immunotherapy and immune-related adverse events. J. Cancer Res. Clin. Oncol. 2023, 149, 737–747. [Google Scholar] [CrossRef] [PubMed]
- Chowell, D.; Morris, L.G.T.; Grigg, C.M.; Weber, J.K.; Samstein, R.M.; Makarov, V.; Kuo, F.; Kendall, S.M.; Requena, D.; Riaz, N.; et al. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science 2018, 359, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Georgopoulos, A.P.; James, L.M.; Charonis, S.A.; Sanders, M. Melanoma and Human Leukocyte Antigen (HLA): Immunogenicity of 69 HLA Class I Alleles With 11 Antigens Expressed in Melanoma Tumors. Cancer Inform. 2023, 22, 11769351231172604. [Google Scholar] [CrossRef]
- Ott, P.A.; Hu, Z.; Keskin, D.B.; Shukla, S.A.; Sun, J.; Bozym, D.J.; Zhang, W.; Luoma, A.; Giobbie-Hurder, A.; Peter, L.; et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 2017, 547, 217–221. [Google Scholar] [CrossRef]
- Sahin, U.; Derhovanessian, E.; Miller, M.; Kloke, B.P.; Simon, P.; Löwer, M.; Bukur, V.; Tadmor, A.D.; Luxemburger, U.; Schrörs, B.; et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 2017, 547, 222–226. [Google Scholar] [CrossRef]
- James, L.M. HLA-Based Immunotherapy for Cancer. In Cancer Immunology—Cellular Mechanisms, Therapeutic Advances and Emerging Frontiers; Scheffel, T.B., Ed.; IntechOpen: London, UK, 2025. [Google Scholar] [CrossRef]
- Georgopoulos, A.P. Human leukocyte antigen (HLA) and cancer immunotherapy. Explor. Immunol. 2025, submitted.
- Fan, T.; Zhang, M.; Yang, J.; Zhu, Z.; Cao, W.; Dong, C. Therapeutic cancer vaccines: Advancements, challenges and prospects. Signal Transduct. Target. Ther. 2023, 8, 450. [Google Scholar] [CrossRef]
- Sahin, U.; Türeci, Ö. Personalized vaccines for cancer immunotherapy. Science 2018, 359, 1355–1360. [Google Scholar] [CrossRef]
- Reynisson, B.; Alvarez, B.; Paul, S.; Peters, B.; Nielsen, M. NetMHCpan-4.1 and NetMHCIIpan-4.0: Improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020, 48, W449–W454. [Google Scholar] [CrossRef]
- Hurley, C.K.; Kempenich, J.; Wadsworth, K.; Sauter, J.; Hofmann, J.A.; Schefzyk, D.; Schmidt, A.H.; Galarza, P.; Cardozo, M.B.R.; Dudkiewicz, M.; et al. Common, intermediate and well-documented HLA alleles in world populations: CIWD version 3.0.0. HLA 2020, 95, 516–531. [Google Scholar] [CrossRef]
- Charonis, S.; Tsilibary, E.P.; Georgopoulos, A. SARS-CoV-2 virus and Human Leukocyte Antigen (HLA) Class II: Investigation in silico of binding affinities for COVID-19 protection and vaccine development. J. Immunol. Sci. 2020, 4, 12–23. [Google Scholar] [CrossRef]
- Charonis, S.A.; Tsilibary, E.P.; Georgopoulos, A.P. In silico investigation of binding affinities between human leukocyte antigen class I molecules and SARS-CoV-2 virus spike and ORF1ab proteins. Explor. Immunol. 2021, 1, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Engelhard, V.H. Structure of peptides associated with class I and class II MHC molecules. Annu. Rev. Immunol. 1994, 12, 181–207. [Google Scholar] [CrossRef] [PubMed]
- Omran, A.; Amberg, A.; Ecker, G.F. Exploring diverse approaches for predicting interferon-gamma release: Utilizing MHC class II and peptide sequences. Brief Bioinform. 2025, 26, bbaf101. [Google Scholar] [CrossRef] [PubMed]
- Istrail, S.; Florea, L.; Halldórsson, B.V.; Kohlbacher, O.; Schwartz, R.S.; Yap, V.B.; Yewdell, J.W.; Hoffman, S.L. Comparative immunopeptidomics of humans and their pathogens. Proc. Natl. Acad. Sci. USA 2004, 101, 13268–13272. [Google Scholar] [CrossRef]
- Zeng, X.; Bai, G.; Sun, C.; Ma, B. Recent progress in antibody epitope prediction. Antibodies 2023, 12, 52. [Google Scholar] [CrossRef]
- Wang, Y.; Kim, M.; Su, S.; Halwatura, L.; You, S.; Kim, H.L. Using major histocompatibility complex (MHC) II expression to predict antitumor response to CD4 + lymphocyte depletion. Sci. Rep. 2025, 15, 5469. [Google Scholar] [CrossRef]
- Alvarez-Dominguez, C.; Calderón-Gonzalez, R.; Terán-Navarro, H.; Salcines-Cuevas, D.; Garcia-Castaño, A.; Freire, J.; Gomez-Roman, J.; Rivera, F. Dendritic cell therapy in melanoma. Ann. Transl. Med. 2017, 5, 386. [Google Scholar] [CrossRef]
- Castellino, F.; Germain, R.N. Cooperation between CD4+ and CD8+ T cells: When, where, and how. Annu. Rev. Immunol. 2006, 24, 519–540. [Google Scholar] [CrossRef]
- Forthal, D.N. Functions of antibodies. Microbiol. Spectr. 2014, 2, AID-0019-2014. [Google Scholar] [CrossRef]
- Borrow, P. Mechanisms of viral clearance and persistence. J. Viral Hepat. 1997, 4 (Suppl. S2), 16–24. [Google Scholar] [CrossRef]
- Burton, D.R. Antiviral neutralizing antibodies: From in vitro to in vivo activity. Nat. Rev. Immunol. 2023, 23, 720–734. [Google Scholar] [CrossRef] [PubMed]
- Reed, C.M.; Cresce, N.D.; Mauldin, I.S.; Slingluff, C.L., Jr.; Olson, W.C. Vaccination with melanoma helper peptides induces antibody responses associated with improved overall survival. Clin. Cancer Res. 2015, 21, 3879–3887. [Google Scholar] [CrossRef] [PubMed]
- Ostrand-Rosenberg, S.; Thakur, A.; Clements, V. Rejection of mouse sarcoma cells after transfection of MHC class II genes. J. Immunol. 1990, 144, 4068–4071. [Google Scholar] [CrossRef] [PubMed]
- Georgopoulos, A.P.; James, L.M. Direct administration of human leucocyte antigen (dHLA) molecules into tumour sites: Proposal for a new immunotherapy for cancer. BJC Rep. 2023, 1, 17. [Google Scholar] [CrossRef]
- Wennhold, K.; Shimabukuro-Vornhagen, A.; von Bergwelt-Baildon, M. B cell-based cancer immunotherapy. Transfus. Med. Hemother. 2019, 46, 36–46. [Google Scholar] [CrossRef]
- Georgopoulos, A.P.; James, L.M.; Sanders, M. Nine human leukocyte antigen (HLA) class I alleles are omnipotent against 11 antigens expressed in melanoma tumors. Cancer Inform. 2024, 23, 11769351241274160. [Google Scholar] [CrossRef]
- Anderson, N.M.; Simon, M.C. The tumor microenvironment. Curr. Biol. 2020, 30, R921–R925. [Google Scholar] [CrossRef]
- Georgopoulos, A.P.; James, L.M. Solid tumor rejection by personalized incompatible Human Leukocyte Antigen (HLA) and ABH antigens. Explor. Immunol. 2025. submitted. [Google Scholar]
- Cozzi, E.; Colpo, A.; De Silvestro, G. The mechanisms of rejection in solid organ transplantation. Transfus. Apher. Sci. 2017, 56, 498–505. [Google Scholar] [CrossRef]
- Gfeller, D.; Bassani-Sternberg, M. Predicting Antigen Presentation-What Could We Learn from a Million Peptides? Front. Immunol. 2018, 9, 1716. [Google Scholar] [CrossRef] [PubMed]
- Andersen, R.S.; Thrue, C.A.; Junker, N.; Lyngaa, R.; Donia, M.; Ellebæk, E.; Svane, I.M.; Schumacher, T.N.; Thor Straten, P.; Hadrup, S.R. Dissection of T-cell antigen specificity in human melanoma. Cancer Res. 2012, 72, 1642–1650. [Google Scholar] [CrossRef]
- Linnemann, C.; van Buuren, M.M.; Bies, L.; Verdegaal, E.M.; Schotte, R.; Calis, J.J.; Behjati, S.; Velds, A.; Hilkmann, H.; Atmioui, D.E.; et al. High-throughput epitope discovery reveals frequent recognition of neo-antigens by CD4+ T cells in human melanoma. Nat. Med. 2015, 21, 81–85. [Google Scholar] [CrossRef]
- Paolino, G.; Huber, V.; Camerini, S.; Casella, M.; Macone, A.; Bertuccini, L.; Iosi, F.; Moliterni, E.; Cecchetti, S.; Ruspantini, I.; et al. The fatty acid and protein profiles of circulating CD81-positive small extracellular vesicles are associated with disease stage in melanoma patients. Cancers 2021, 13, 4157. [Google Scholar] [CrossRef]
- Gomase, V.S.; Sharma, R.; Dhamane, S.P. Immunoinformatics approach for optimization of targeted vaccine design: New paradigm in clinical trials and healthcare management. Rev. Recent Clin. Trials 2025. [Google Scholar] [CrossRef]
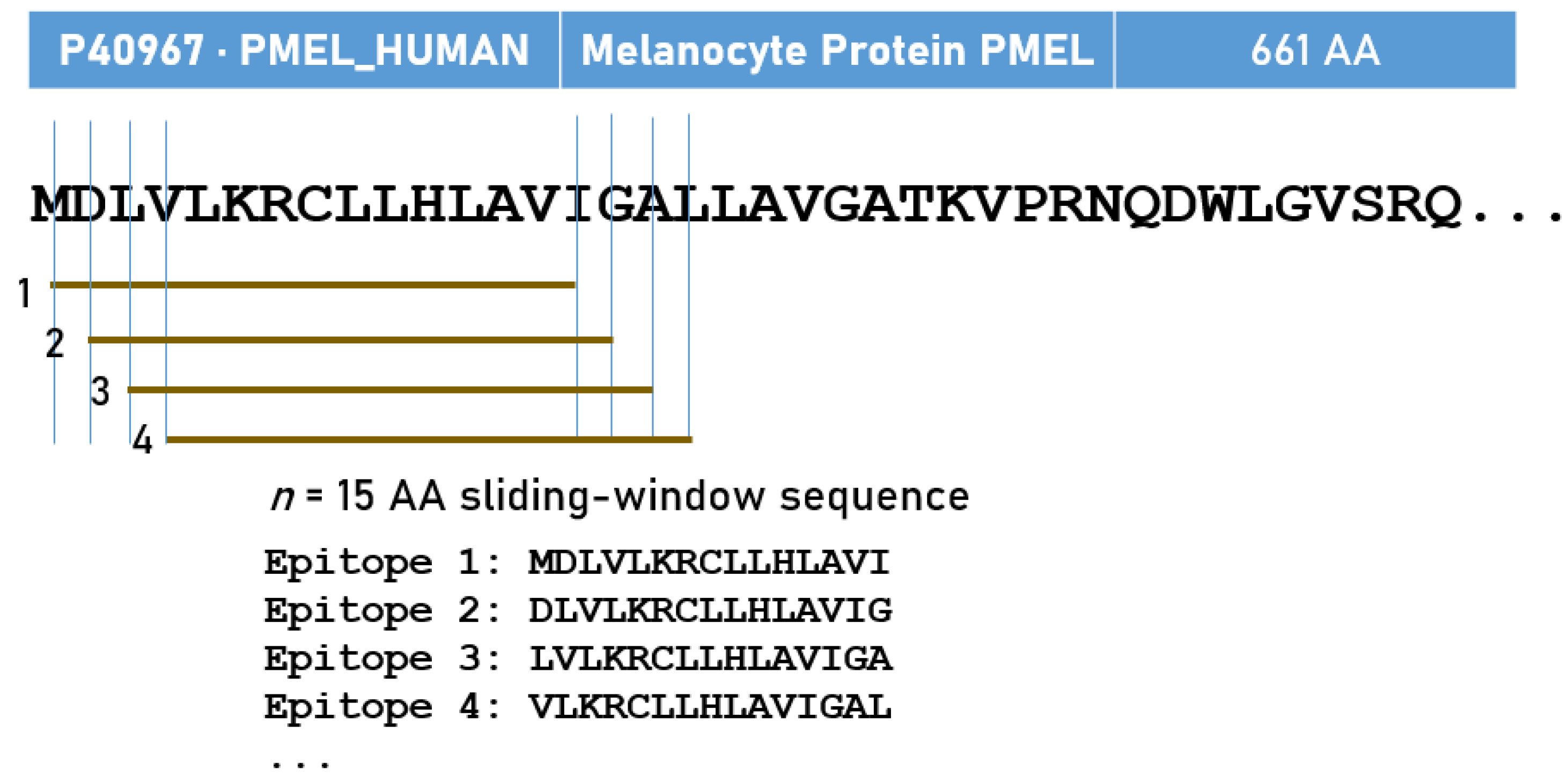
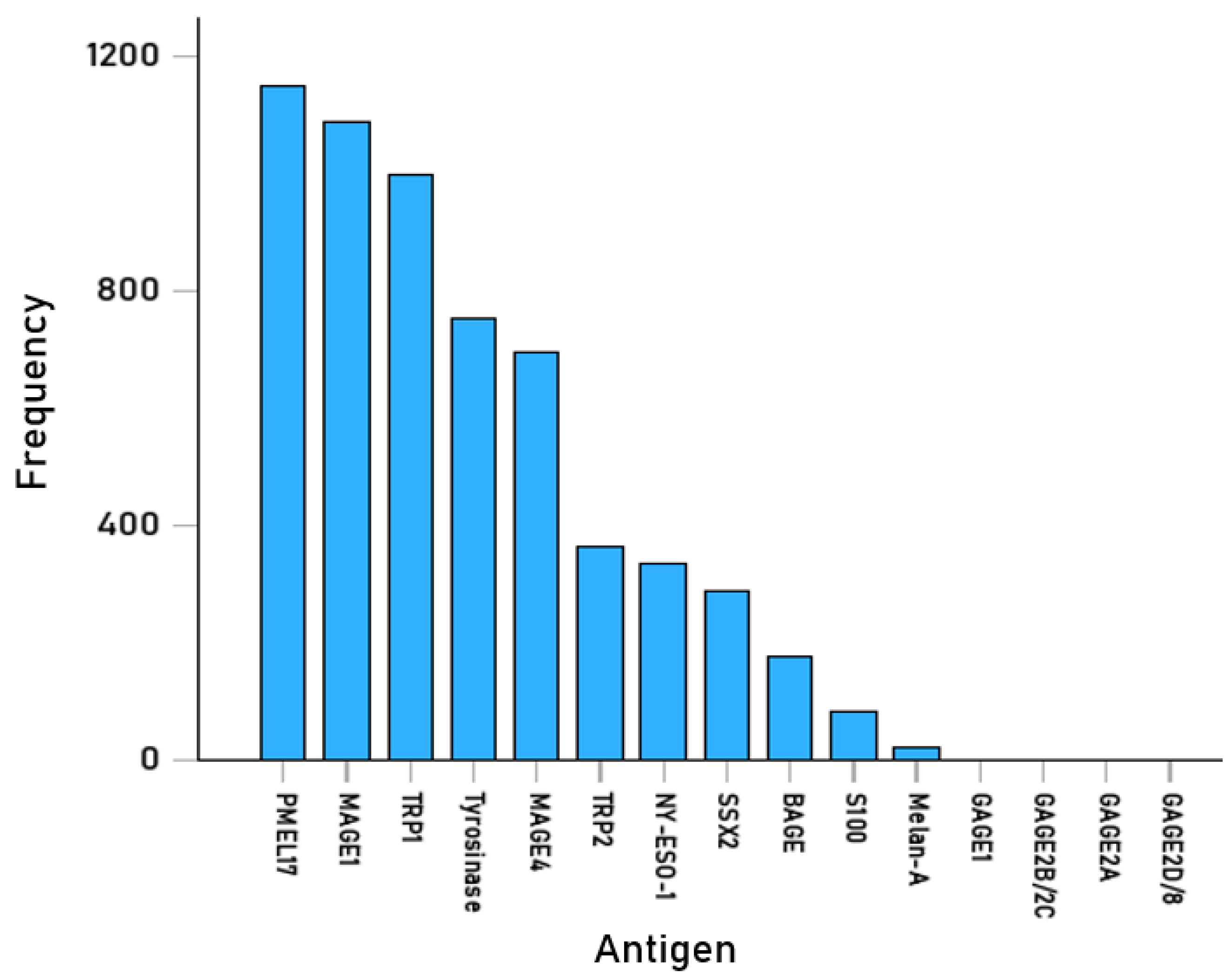

| Uniprot | Cancer Antigen | N (AA) | N (15-AA) | N Tested | Hits | % Hits | |
|---|---|---|---|---|---|---|---|
| 1 | O75767 | TRP2 | 237 | 223 | 42,816 | 364 | 0.85 |
| 2 | P04271 | S100 | 92 | 78 | 14,976 | 83 | 0.55 |
| 3 | P0DTW1 | GAGE1 [CTA] | 117 | 103 | 19,776 | 0 | 0.00 |
| 4 | P14679 | Tyrosinase | 529 | 515 | 98,880 | 751 | 0.76 |
| 5 | P17643 | TRP1 | 537 | 523 | 100,416 | 998 | 0.99 |
| 6 | P40967 | PMEL17/gp100 | 661 | 647 | 124,224 | 1151 | 0.93 |
| 7 | P43355 | MAGE1 [CTA] | 309 | 295 | 56,640 | 1087 | 1.92 |
| 8 | P43358 | MAGE4 [CTA] | 317 | 303 | 58,176 | 695 | 1.19 |
| 9 | P78358 | NY-ESO-1 [CTA] | 180 | 166 | 31,872 | 333 | 1.04 |
| 10 | Q13066 | GAGE2B/2C | 116 | 102 | 19,584 | 0 | 0.00 |
| 11 | Q13072 | BAGE [CTA] | 43 | 29 | 5568 | 174 | 3.13 |
| 12 | Q16385 | SSX2 [CTA] | 188 | 174 | 33,408 | 286 | 0.86 |
| 13 | Q16655 | Melan-A/MART-1 | 118 | 104 | 19,968 | 19 | 0.10 |
| 14 | Q6NT46 | GAGE2A [CTA] | 116 | 102 | 19,584 | 0 | 0.00 |
| 15 | Q9UEU5 | GAGE2D/GAGE8 [CTA] | 116 | 102 | 19,584 | 0 | 0.00 |
| Total | 3466 | 665,472 | 5941 | ||||
| Allele | N | Allele | N | Allele | N | Allele | N |
|---|---|---|---|---|---|---|---|
| DRB1*01:01 | 11 | DRB1*11:62 | 8 | DPB1*46:01 | 5 | DRB1*04:10 | 2 |
| DRB1*01:18 | 11 | DRB1*11:65 | 8 | DPB1*47:01 | 5 | DRB1*08:02 | 2 |
| DRB1*01:24 | 11 | DRB1*11:74 | 8 | DPB1*72:01 | 5 | DRB1*08:30 | 2 |
| DRB1*01:29 | 11 | DRB1*13:01 | 8 | DPB1*81:01 | 5 | DRB1*11:27 | 2 |
| DRB1*10:01 | 11 | DRB1*13:05 | 8 | DRB1*04:01 | 5 | DRB1*11:54 | 2 |
| DRB1*01:11 | 10 | DRB1*13:11 | 8 | DRB1*11:37 | 5 | DRB1*12:03 | 2 |
| DRB1*01:20 | 10 | DRB1*13:14 | 8 | DRB1*13:07 | 5 | DRB1*14:04 | 2 |
| DPB1*33:01 | 9 | DRB1*13:50 | 8 | DRB1*15:03 | 5 | DRB1*14:38 | 2 |
| DPB1*71:01 | 9 | DRB1*14:32 | 8 | DRB1*15:07 | 5 | DPB1*04:02 | 1 |
| DRB1*07:01 | 9 | DRB1*15:01 | 8 | DRB1*16:05 | 5 | DPB1*105:0 | 1 |
| DRB1*11:13 | 9 | DRB1*15:06 | 8 | DRB1*14:12 | 4 | DPB1*16:01 | 1 |
| DRB1*11:14 | 9 | DRB1*16:09 | 8 | DRB1*15:37 | 4 | DPB1*19:01 | 1 |
| DRB1*11:42 | 9 | DRB1*01:02 | 7 | DPB1*01:01 | 3 | DPB1*34:01 | 1 |
| DRB1*13:02 | 9 | DRB1*08:04 | 7 | DRB1*04:04 | 3 | DPB1*41:01 | 1 |
| DRB1*13:23 | 9 | DRB1*11:03 | 7 | DRB1*04:05 | 3 | DPB1*49:01 | 1 |
| DRB1*13:97 | 9 | DRB1*11:84 | 7 | DRB1*04:72 | 3 | DPB1*55:01 | 1 |
| DRB1*16:02 | 9 | DRB1*13:96 | 7 | DRB1*08:01 | 3 | DRB1*01:03 | 1 |
| DRB1*03:11 | 8 | DRB1*15:02 | 7 | DRB1*08:24 | 3 | DRB1*03:15 | 1 |
| DRB1*09:01 | 8 | DRB1*15:15 | 7 | DRB1*11:11 | 3 | DRB1*04:44 | 1 |
| DRB1*11:01 | 8 | DRB1*16:01 | 7 | DRB1*11:19 | 3 | DRB1*11:06 | 1 |
| DRB1*11:02 | 8 | DRB1*04:08 | 6 | DRB1*12:16 | 3 | DRB1*11:07 | 1 |
| DRB1*11:04 | 8 | DRB1*13:21 | 6 | DRB1*13:61 | 3 | DRB1*12:02 | 1 |
| DRB1*11:08 | 8 | DRB1*14:06 | 6 | DRB1*13:66 | 3 | DRB1*13:33 | 1 |
| DRB1*11:10 | 8 | DPB1*02:01 | 5 | DRB1*14:01 | 3 | DRB1*14:02 | 1 |
| DRB1*11:12 | 8 | DPB1*02:02 | 5 | DRB1*14:54 | 3 | DRB1*14:05 | 1 |
| DRB1*11:28 | 8 | DPB1*04:01 | 5 | DRB1*16:04 | 3 | DRB1*14:07 | 1 |
| DRB1*11:29 | 8 | DPB1*126:0 | 5 | DPB1*40:01 | 2 | DRB1*14:23 | 1 |
| DRB1*11:46 | 8 | DPB1*15:01 | 5 | DRB1*03:01 | 2 | ||
| DRB1*11:49 | 8 | DPB1*23:01 | 5 | DRB1*03:04 | 2 | ||
| DRB1*11:58 | 8 | DPB1*39:01 | 5 | DRB1*03:13 | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Georgopoulos, A.P.; James, L.M.; Sanders, M. mRNA Multipeptide-HLA Class II Immunotherapy for Melanoma. Cells 2025, 14, 1430. https://doi.org/10.3390/cells14181430
Georgopoulos AP, James LM, Sanders M. mRNA Multipeptide-HLA Class II Immunotherapy for Melanoma. Cells. 2025; 14(18):1430. https://doi.org/10.3390/cells14181430
Chicago/Turabian StyleGeorgopoulos, Apostolos P., Lisa M. James, and Matthew Sanders. 2025. "mRNA Multipeptide-HLA Class II Immunotherapy for Melanoma" Cells 14, no. 18: 1430. https://doi.org/10.3390/cells14181430
APA StyleGeorgopoulos, A. P., James, L. M., & Sanders, M. (2025). mRNA Multipeptide-HLA Class II Immunotherapy for Melanoma. Cells, 14(18), 1430. https://doi.org/10.3390/cells14181430





