Ago2-Mediated Recruitment of HP1a on Transposable Elements in Drosophila Brain
Abstract
1. Introduction
2. Materials and Methods
2.1. Fly Crossing and Transgenic Lines
2.2. DamID with HP1a in Brain
2.3. Bioinformatics Analysis of DamID Data
2.4. Immunostaining of Drosophila Brain
2.5. RT-qPCR Analysis
2.6. Statistical Analysis
3. Results
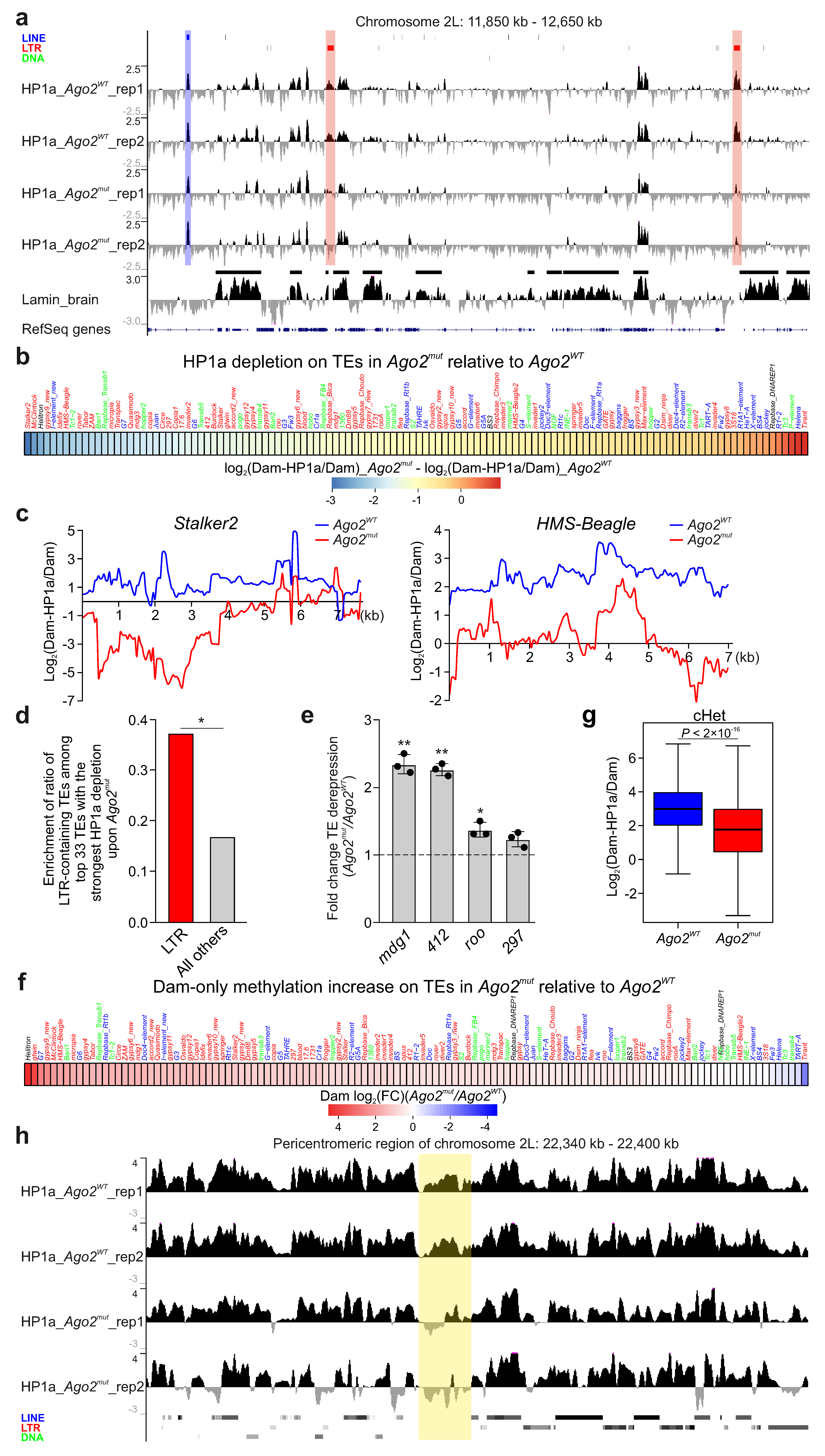
3.1. HP1a Is Drastically Decreased on TEs and in cHet Regions in Ago2-Mutant Drosophila Brain
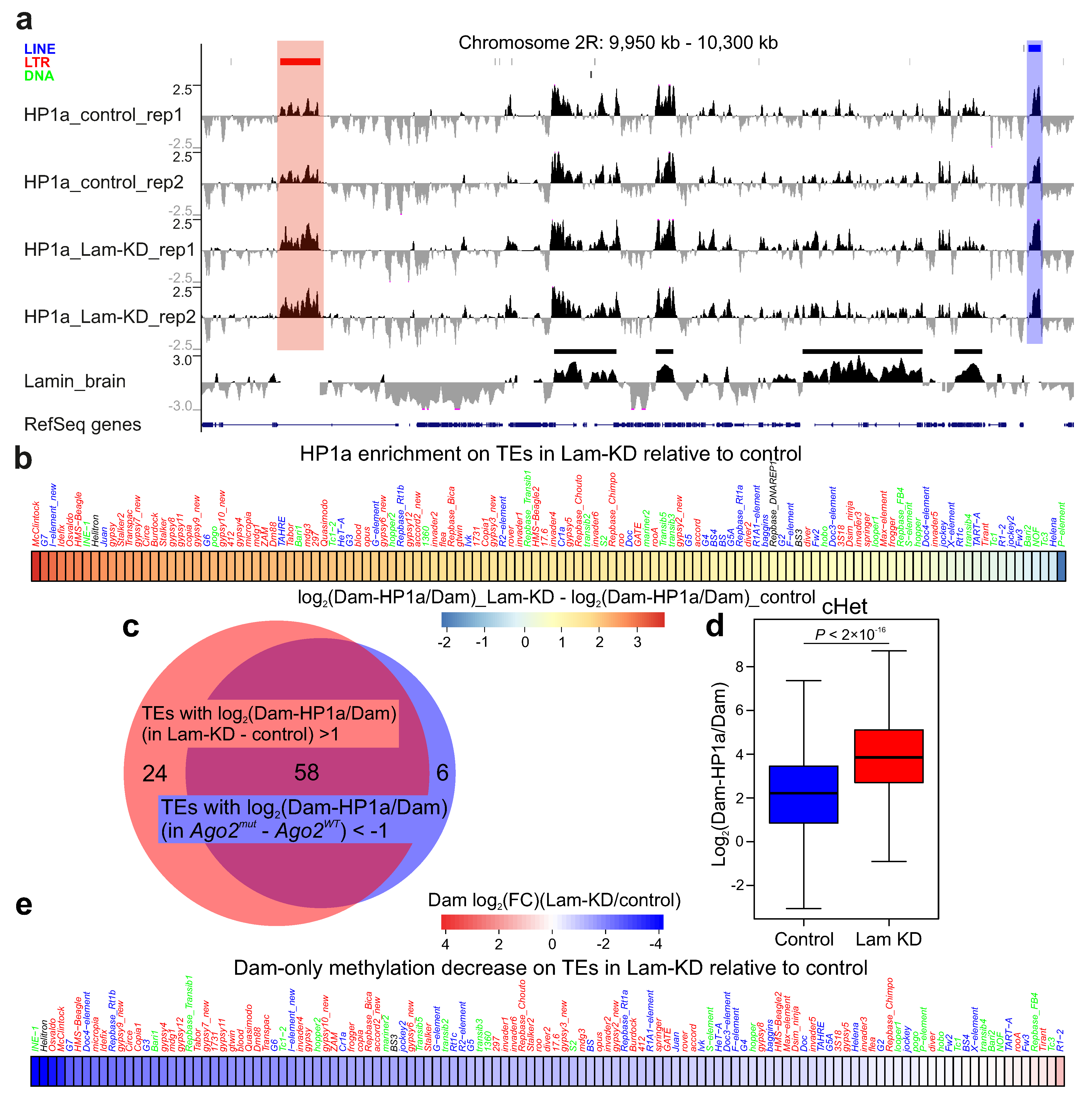
3.2. HP1a Is Mainly Increased on the LTR-Containing Retrotransposons and in cHet Regions upon Lam-KD in Neurons
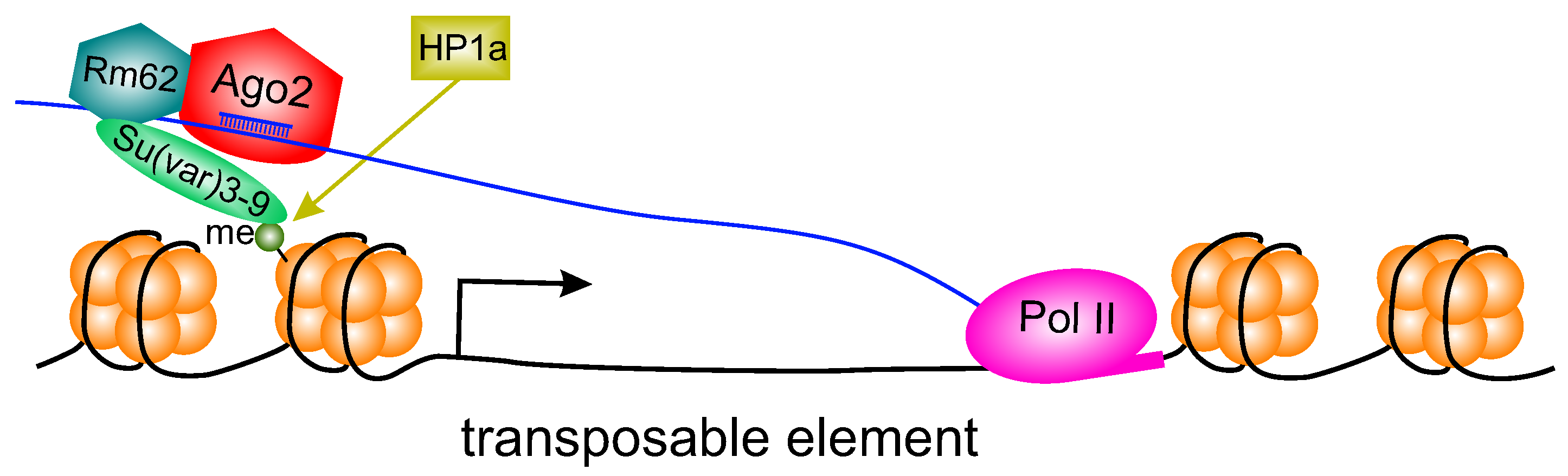
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Ago2 | Argonaute 2 |
| ChIP | Chromatin immunoprecipitation |
| cHet | Constitutive heterochromatin |
| CPMs | Counts per million |
| DamID | Dam identification |
| fHet | Facultative heterochromatin |
| HP1a | Heterochromatin protein 1a |
| Lam-KD | Knock-down of lamin Dm0 |
| LAD | Lamina-associated domain |
| LTR | Long terminal repeat |
| NL | Nuclear lamina |
| piRNA | Piwi-interacting RNA |
| RT-qPCR | Quantitative polymerase chain reaction after reverse transcription |
| siRNA | Small interfering RNA |
| TE | Transposable element |
| WT | Wild-type |
References
- Grewal, S.I.S.; Jia, S. Heterochromatin revisited. Nat. Rev. Genet. 2007, 8, 35–46. [Google Scholar] [CrossRef]
- Solovei, I.; Wang, A.S.; Thanisch, K.; Schmidt, C.S.; Krebs, S.; Zwerger, M.; Cohen, T.V.; Devys, D.; Foisner, R.; Peichl, L.; et al. LBR and Lamin A/C Sequentially Tether Peripheral Heterochromatin and Inversely Regulate Differentiation. Cell 2013, 152, 584–598. [Google Scholar] [CrossRef]
- Guelen, L.; Pagie, L.; Brasset, E.; Meuleman, W.; Faza, M.B.; Talhout, W.; Eussen, B.H.; de Klein, A.; Wessels, L.; de Laat, W.; et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 2008, 453, 948–951. [Google Scholar] [CrossRef]
- Filion, G.J.; van Bemmel, J.G.; Braunschweig, U.; Talhout, W.; Kind, J.; Ward, L.D.; Brugman, W.; de Castro, I.J.; Kerkhoven, R.M.; Bussemaker, H.J.; et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 2010, 143, 212–224. [Google Scholar] [CrossRef]
- Peric-Hupkes, D.; Meuleman, W.; Pagie, L.; Bruggeman, S.W.; Solovei, I.; Brugman, W.; Gräf, S.; Flicek, P.; Kerkhoven, R.M.; van Lohuizen, M.; et al. Molecular Maps of the Reorganization of Genome-Nuclear Lamina Interactions during Differentiation. Mol. Cell 2010, 38, 603–613. [Google Scholar] [CrossRef]
- van Bemmel, J.G.; Pagie, L.; Braunschweig, U.; Brugman, W.; Meuleman, W.; Kerkhoven, R.M.; van Steensel, B.; Veenstra, G.J.C. The Insulator Protein SU(HW) Fine-Tunes Nuclear Lamina Interactions of the Drosophila Genome. PLoS ONE 2010, 5, e15013. [Google Scholar] [CrossRef]
- Ikegami, K.; A Egelhofer, T.; Strome, S.; Lieb, J.D. Caenorhabditis elegans chromosome arms are anchored to the nuclear membrane via discontinuous association with LEM-2. Genome Biol. 2010, 11, R120. [Google Scholar] [CrossRef] [PubMed]
- Gruenbaum, Y.; Foisner, R. Lamins: Nuclear Intermediate Filament Proteins with Fundamental Functions in Nuclear Mechanics and Genome Regulation. Annu. Rev. Biochem. 2015, 84, 131–164. [Google Scholar] [CrossRef]
- Romano, N.C.; Fanti, L. Transposable Elements: Major Players in Shaping Genomic and Evolutionary Patterns. Cells 2022, 11, 1048. [Google Scholar] [CrossRef] [PubMed]
- Sienski, G.; Batki, J.; Senti, K.-A.; Dönertas, D.; Tirian, L.; Meixner, K.; Brennecke, J. Silencio/CG9754 connects the Piwi–piRNA complex to the cellular heterochromatin machinery. Genes Dev. 2015, 29, 2258–2271. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Gu, J.; Jin, Y.; Luo, Y.; Preall, J.B.; Ma, J.; Czech, B.; Hannon, G.J. Panoramix enforces piRNA-dependent cotranscrip-tional silencing. Science 2015, 350, 339–342. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, Y.W.; Murano, K.; Ishizu, H.; Shibuya, A.; Iyoda, Y.; Siomi, M.C.; Siomi, H.; Saito, K. Piwi Modulates Chromatin Accessibility by Regulating Multiple Factors Including Histone H1 to Repress Transposons. Mol. Cell 2016, 63, 408–419. [Google Scholar] [CrossRef]
- Sato, K.; Siomi, M.C. The piRNA pathway in Drosophila ovarian germ and somatic cells. Proc. Jpn. Acad. Ser. B 2020, 96, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.C.; Wood, J.G.; Chang, C.; Tam, A.D.; Franklin, M.J.; Siegel, E.R.; Helfand, S.L. A somatic piRNA pathway in the Drosophila fat body ensures metabolic homeostasis and normal lifespan. Nat. Commun. 2016, 7, 13856. [Google Scholar] [CrossRef]
- Czech, B.; Malone, C.D.; Zhou, R.; Stark, A.; Schlingeheyde, C.; Dus, M.; Perrimon, N.; Kellis, M.; Wohlschlegel, J.A.; Sachidanandam, R.; et al. An endogenous small interfering RNA pathway in Drosophila. Nature 2008, 453, 798–802. [Google Scholar] [CrossRef]
- Ghildiyal, M.; Seitz, H.; Horwich, M.D.; Li, C.; Du, T.; Lee, S.; Xu, J.; Kittler, E.L.; Zapp, M.L.; Weng, Z.; et al. Endogenous siRNAs Derived from Transposons and mRNAs in Drosophila Somatic Cells. Science 2008, 320, 1077–1081. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, Y.; Saito, K.; Kin, T.; Ono, Y.; Asai, K.; Sunohara, T.; Okada, T.N.; Siomi, M.C.; Siomi, H. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature 2008, 453, 793–797. [Google Scholar] [CrossRef]
- Czech, B.; Hannon, G.J. Small RNA sorting: Matchmaking for Argonautes. Nat. Rev. Genet. 2011, 12, 19–31. [Google Scholar] [CrossRef]
- Moshkovich, N.; Lei, E.P.; Lee, J.T. HP1 Recruitment in the Absence of Argonaute Proteins in Drosophila. PLoS Genet. 2010, 6, e1000880. [Google Scholar] [CrossRef]
- Moshkovich, N.; Nisha, P.; Boyle, P.J.; Thompson, B.A.; Dale, R.K.; Lei, E.P. RNAi-independent role for Argonaute2 in CTCF/CP190 chromatin insulator function. Genes Dev. 2011, 25, 1686–1701. [Google Scholar] [CrossRef]
- Deshpande, G.; Calhoun, G.; Schedl, P. Drosophila argonaute-2 is required early in embryogenesis for the assembly of centric/centromeric heterochromatin, nuclear division, nuclear migration, and germ-cell formation. Genes Dev. 2005, 19, 1680–1685. [Google Scholar] [CrossRef]
- Fagegaltier, D.; Bougé, A.-L.; Berry, B.; Poisot, É.; Sismeiro, O.; Coppée, J.-Y.; Théodore, L.; Voinnet, O.; Antoniewski, C. The endogenous siRNA pathway is involved in heterochromatin formation in Drosophila. Proc. Natl. Acad. Sci. USA 2009, 106, 21258–21263. [Google Scholar] [CrossRef]
- Cernilogar, F.M.; Onorati, M.C.; Kothe, G.O.; Burroughs, A.M.; Parsi, K.M.; Breiling, A.; Sardo, F.L.; Saxena, A.; Miyoshi, K.; Siomi, H.; et al. Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila. Nature 2011, 480, 391–395. [Google Scholar] [CrossRef]
- Taliaferro, J.M.; Aspden, J.L.; Bradley, T.; Marwha, D.; Blanchette, M.; Rio, D.C. Two new and distinct roles for Drosophila Argonaute-2 in the nucleus: Alternative pre-mRNA splicing and transcriptional repression. Genes Dev. 2013, 27, 378–389. [Google Scholar] [CrossRef]
- Lee, S.K.; Xue, Y.; Shen, W.; Zhang, Y.; Joo, Y.; Ahmad, M.; Chinen, M.; Ding, Y.; Ku, W.L.; De, S.; et al. Topoisomerase 3β interacts with RNAi machinery to promote heterochromatin formation and transcriptional silencing in Drosophila. Nat. Commun. 2018, 9, 4946. [Google Scholar] [CrossRef]
- Pindyurin, A.V.; Pagie, L.; Kozhevnikova, E.N.; van Arensbergen, J.; van Steensel, B. Inducible DamID systems for genomic mapping of chromatin proteins in Drosophila. Nucleic Acids Res. 2016, 44, 5646–5657. [Google Scholar] [CrossRef]
- Pindyurin, A.V.; Ilyin, A.A.; Ivankin, A.V.; Tselebrovsky, M.V.; Nenasheva, V.V.; Mikhaleva, E.A.; Pagie, L.; van Steensel, B.; Shevelyov, Y.Y. The large fraction of heterochromatin in Drosophila neurons is bound by both B-type lamin and HP1a. Epigenet. Chromatin 2018, 11, 65. [Google Scholar] [CrossRef] [PubMed]
- Hain, D.; Bettencourt, B.R.; Okamura, K.; Csorba, T.; Meyer, W.; Jin, Z.; Biggerstaff, J.; Siomi, H.; Hutvagner, G.; Lai, E.C.; et al. Natural Variation of the Amino-Terminal Glutamine-Rich Domain in Drosophila Argonaute2 Is Not Associated with Developmental Defects. PLoS ONE 2010, 5, e15264. [Google Scholar] [CrossRef] [PubMed]
- Okamura, K.; Ishizuka, A.; Siomi, H.; Siomi, M.C. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004, 18, 1655–1666. [Google Scholar] [CrossRef]
- Vogel, M.J.; Peric-Hupkes, D.; van Steensel, B. Detection of in vivo protein–DNA interactions using DamID in mammalian cells. Nat. Protoc. 2007, 2, 1467–1478. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, F.; Ryan, D.P.; Grüning, B.; Bhardwaj, V.; Kilpert, F.; Richter, A.S.; Heyne, S.; Dündar, F.; Manke, T. deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016, 44, W160–W165. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Ilyin, A.A.; Ryazansky, S.S.; Doronin, S.A.; Olenkina, O.M.; Mikhaleva, E.A.; Yakushev, E.Y.; Abramov, Y.A.; Belyakin, S.N.; Ivankin, A.V.; Pindyurin, A.V.; et al. Piwi interacts with chromatin at nuclear pores and promiscuously binds nuclear transcripts in Drosophila ovarian somatic cells. Nucleic Acids Res. 2017, 45, 7666–7680. [Google Scholar] [CrossRef]
- Stuurman, N.; Maus, N.; Fisher, P.A. Interphase phosphorylation of the Drosophila nuclear lamin: Site-mapping using a monoclonal antibody. J. Cell Sci. 1995, 108, 3137–3144. [Google Scholar] [CrossRef]
- Ayyanathan, K.; Lechner, M.S.; Bell, P.; Maul, G.G.; Schultz, D.C.; Yamada, Y.; Tanaka, K.; Torigoe, K.; Rauscher, F.J. Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: A mammalian cell culture model of gene variegation. Genes Dev. 2003, 17, 1855–1869. [Google Scholar] [CrossRef]
- Li, Y.; Danzer, J.R.; Alvarez, P.; Belmont, A.S.; Wallrath, L.L. Effects of tethering HP1 to euchromatic regions of the Drosophila genome. Development 2003, 130, 1817–1824. [Google Scholar] [CrossRef]
- Brueckner, L.; van Arensbergen, J.; Akhtar, W.; Pagie, L.; van Steensel, B. High-throughput assessment of context-dependent effects of chromatin proteins. Epigenet. Chromatin 2016, 9, 38. [Google Scholar] [CrossRef] [PubMed]
- Canzio, D.; Liao, M.; Naber, N.; Pate, E.; Larson, A.; Wu, S.; Marina, D.B.; Garcia, J.F.; Madhani, H.D.; Cooke, R.; et al. A conformational switch in HP1 releases auto-inhibition to drive heterochromatin assembly. Nature 2013, 496, 377–381. [Google Scholar] [CrossRef]
- Azzaz, A.M.; Vitalini, M.W.; Thomas, A.S.; Price, J.P.; Blacketer, M.J.; Cryderman, D.E.; Zirbel, L.N.; Woodcock, C.L.; Elcock, A.H.; Wallrath, L.L.; et al. Human heterochromatin protein 1α promotes nucleosome associations that drive chromatin con-densation. J. Biol. Chem. 2014, 289, 6850–6861. [Google Scholar] [CrossRef]
- Larson, A.G.; Elnatan, D.; Keenen, M.M.; Trnka, M.J.; Johnston, J.B.; Burlingame, A.L.; Agard, D.A.; Redding, S.; Narlikar, G.J. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 2017, 547, 236–240. [Google Scholar] [CrossRef]
- Wines, D.R.; Talbert, P.B.; Clark, D.V.; Henikoff, S. Introduction of a DNA methyltransferase into Drosophila to probe chro-matin structure in vivo. Chromosoma 1996, 104, 332–340. [Google Scholar] [CrossRef]
- Aughey, G.N.; Gomez, A.E.; Thomson, J.; Yin, H.; Southall, T.D. CATaDa reveals global remodelling of chromatin accessibility during stem cell differentiation in vivo. eLife 2018, 7, e32341. [Google Scholar] [CrossRef] [PubMed]
- Hoskins, R.A.; Carlson, J.W.; Kennedy, C.; Acevedo, D.; Evans-Holm, M.; Frise, E.; Wan, K.H.; Park, S.; Mendez-Lago, M.; Rossi, F.; et al. Sequence Finishing and Mapping of Drosophila melanogaster Heterochromatin. Science 2007, 316, 1625–1628. [Google Scholar] [CrossRef]
- Nazer, E.; Dale, R.K.; Chinen, M.; Radmanesh, B.; Lei, E.P.; Miska, E.A. Argonaute2 and LaminB modulate gene expression by controlling chromatin topology. PLoS Genet. 2018, 14, e1007276. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zheng, X.; Xiao, D.; Zheng, Y. Age-associated de-repression of retrotransposons in the Drosophila fat body, its potential cause and consequence. Aging Cell 2016, 15, 542–552. [Google Scholar] [CrossRef]
- Hall, I.M.; Shankaranarayana, G.D.; Noma, K.; Ayoub, N.; Cohen, A.; Grewal, S.I. Establishment and maintenance of a het-erochromatin domain. Science 2002, 297, 2232–2237. [Google Scholar] [CrossRef]
- Volpe, T.A.; Kidner, C.; Hall, I.M.; Teng, G.; Grewal, S.I.S.; Martienssen, R.A. Regulation of Heterochromatic Silencing and Histone H3 Lysine-9 Methylation by RNAi. Science 2002, 297, 1833–1837. [Google Scholar] [CrossRef]
- Bühler, M.; Verdel, A.; Moazed, D. Tethering RITS to a Nascent Transcript Initiates RNAi- and Heterochromatin-Dependent Gene Silencing. Cell 2006, 125, 873–886. [Google Scholar] [CrossRef]
- Grishok, A.; Sinskey, J.L.; Sharp, P.A. Transcriptional silencing of a transgene by RNAi in the soma of C. elegans. Genes Dev. 2005, 19, 683–696. [Google Scholar] [CrossRef] [PubMed]
- Guang, S.; Bochner, A.F.; Burkhart, K.B.; Burton, N.; Pavelec, D.M.; Kennedy, S. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature 2010, 465, 1097–1101. [Google Scholar] [CrossRef]
- Zilberman, D.; Cao, X.; Jacobsen, S.E. ARGONAUTE4 Control of Locus-Specific siRNA Accumulation and DNA and Histone Methylation. Science 2003, 299, 716–719. [Google Scholar] [CrossRef] [PubMed]
- Herr, A.J.; Jensen, M.B.; Dalmay, T.; Baulcombe, D.C. RNA Polymerase IV Directs Silencing of Endogenous DNA. Science 2005, 308, 118–120. [Google Scholar] [CrossRef]
- Morris, K.V.; Chan, S.W.-L.; Jacobsen, S.E.; Looney, D.J. Small Interfering RNA-Induced Transcriptional Gene Silencing in Human Cells. Science 2004, 305, 1289–1292. [Google Scholar] [CrossRef] [PubMed]
- Ting, A.H.; E Schuebel, K.; Herman, J.G.; Baylin, S.B. Short double-stranded RNA induces transcriptional gene silencing in human cancer cells in the absence of DNA methylation. Nat. Genet. 2005, 37, 906–910. [Google Scholar] [CrossRef]
- Kim, D.H.; Villeneuve, L.M.; Morris, K.V.; Rossi, J.J. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat. Struct. Mol. Biol. 2006, 13, 793–797. [Google Scholar] [CrossRef]
- Sala, L.; Kumar, M.; Prajapat, M.; Chandrasekhar, S.; Cosby, R.L.; La Rocca, G.; Macfarlan, T.S.; Awasthi, P.; Chari, R.; Kruhlak, M.; et al. AGO2 silences mobile transposons in the nucleus of quiescent cells. Nat. Struct. Mol. Biol. 2023, 30, 1985–1995. [Google Scholar] [CrossRef]
- Deshpande, N.; Meller, V.H. Chromatin That Guides Dosage Compensation Is Modulated by the siRNA Pathway in Drosophila melanogaster. Genetics 2018, 209, 1085–1097. [Google Scholar] [CrossRef]
- Nazer, E.; Dale, R.K.; Palmer, C.; Lei, E.P. Argonaute2 attenuates active transcription by limiting RNA Polymerase II elongation in Drosophila melanogaster. Sci. Rep. 2018, 8, 15685. [Google Scholar] [CrossRef]
- Ilyin, Y.; Chrneliauskaite, V.; Georgiev, G. Double-stranded sequences in RNA of Drosophila metanogaster: Relation to mobile dispersed genes. Nucleic Acids Res. 1980, 8, 3439–3457. [Google Scholar] [CrossRef] [PubMed]
- Russo, J.; Harrington, A.W.; Steiniger, M. Antisense Transcription of Retrotransposons in Drosophila: An Origin of Endogenous Small Interfering RNA Precursors. Genetics 2015, 202, 107–121. [Google Scholar] [CrossRef]
- Peng, J.C.; Valouev, A.; Liu, N.; Lin, H. Piwi maintains germline stem cells and oogenesis in Drosophila through negative regulation of Polycomb group proteins. Nat. Genet. 2016, 48, 283–291. [Google Scholar] [CrossRef]
- Ishizuka, A.; Siomi, M.C.; Siomi, H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 2002, 16, 2497–2508. [Google Scholar] [CrossRef] [PubMed]
- Boeke, J.; Bag, I.; Ramaiah, M.J.; Vetter, I.; Kremmer, E.; Pal-Bhadra, M.; Bhadra, U.; Imhof, A.; Tora, L. The RNA Helicase Rm62 Cooperates with SU(VAR)3-9 to Re-Silence Active Transcription in Drosophila melanogaster. PLoS ONE 2011, 6, e20761. [Google Scholar] [CrossRef] [PubMed]
- Rea, S.; Eisenhaber, F.; O’Carroll, D.; Strahl, B.D.; Sun, Z.-W.; Schmid, M.; Opravil, S.; Mechtler, K.; Ponting, C.P.; Allis, C.D.; et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 2000, 406, 593–599. [Google Scholar] [CrossRef]
- Bannister, A.J.; Zegerman, P.; Partridge, J.F.; Miska, E.A.; Thomas, J.O.; Allshire, R.C.; Kouzarides, T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 2001, 410, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Lachner, M.; O'CArroll, D.; Rea, S.; Mechtler, K.; Jenuwein, T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 2001, 410, 116–120. [Google Scholar] [CrossRef]
- Gu, T.; Elgin, S.C.R.; Brennecke, J. Maternal Depletion of Piwi, a Component of the RNAi System, Impacts Heterochromatin Formation in Drosophila. PLoS Genet. 2013, 9, e1003780. [Google Scholar] [CrossRef]
- Klenov, M.S.; Lavrov, S.A.; Stolyarenko, A.D.; Ryazansky, S.S.; Aravin, A.A.; Tuschl, T.; Gvozdev, V.A. Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res. 2007, 35, 5430–5438. [Google Scholar] [CrossRef]
- Beek, M.v.D.; da Silva, B.; Pouch, J.; Chaouche, M.e.A.A.; Carré, C.; Antoniewski, C. Dual-layer transposon repression in heads of Drosophila melanogaster. RNA 2018, 24, 1749–1760. [Google Scholar] [CrossRef]
- Li, W.; Prazak, L.; Chatterjee, N.; Grüninger, S.; Krug, L.; Theodorou, D.; Dubnau, J. Activation of transposable elements during aging and neuronal decline in Drosophila. Nat. Neurosci. 2013, 16, 529–531. [Google Scholar] [CrossRef] [PubMed]
- Erwin, J.A.; Marchetto, M.C.; Gage, F.H. Mobile DNA elements in the generation of diversity and complexity in the brain. Nat. Rev. Neurosci. 2014, 15, 497–506. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olenkina, O.M.; Simonov, R.A.; Ivannikova, A.Y.; Abramov, Y.A.; Sivkina, A.L.; Ulianov, S.V.; Shevelyov, Y.Y. Ago2-Mediated Recruitment of HP1a on Transposable Elements in Drosophila Brain. Cells 2025, 14, 1361. https://doi.org/10.3390/cells14171361
Olenkina OM, Simonov RA, Ivannikova AY, Abramov YA, Sivkina AL, Ulianov SV, Shevelyov YY. Ago2-Mediated Recruitment of HP1a on Transposable Elements in Drosophila Brain. Cells. 2025; 14(17):1361. https://doi.org/10.3390/cells14171361
Chicago/Turabian StyleOlenkina, Oxana M., Ruslan A. Simonov, Anna Y. Ivannikova, Yuri A. Abramov, Anastasiia L. Sivkina, Sergey V. Ulianov, and Yuri Y. Shevelyov. 2025. "Ago2-Mediated Recruitment of HP1a on Transposable Elements in Drosophila Brain" Cells 14, no. 17: 1361. https://doi.org/10.3390/cells14171361
APA StyleOlenkina, O. M., Simonov, R. A., Ivannikova, A. Y., Abramov, Y. A., Sivkina, A. L., Ulianov, S. V., & Shevelyov, Y. Y. (2025). Ago2-Mediated Recruitment of HP1a on Transposable Elements in Drosophila Brain. Cells, 14(17), 1361. https://doi.org/10.3390/cells14171361




