BQ323636.1 Employs the AR-CCRK Axis to Modulate the Expression of KU70 to Interfere with Non-Homologous End Joining Mediated DNA Repair Mechanism †
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture, Transfection, siRNA, and Stable Cell Line Establishment
2.2. Molecular Cloning
2.3. Chemicals
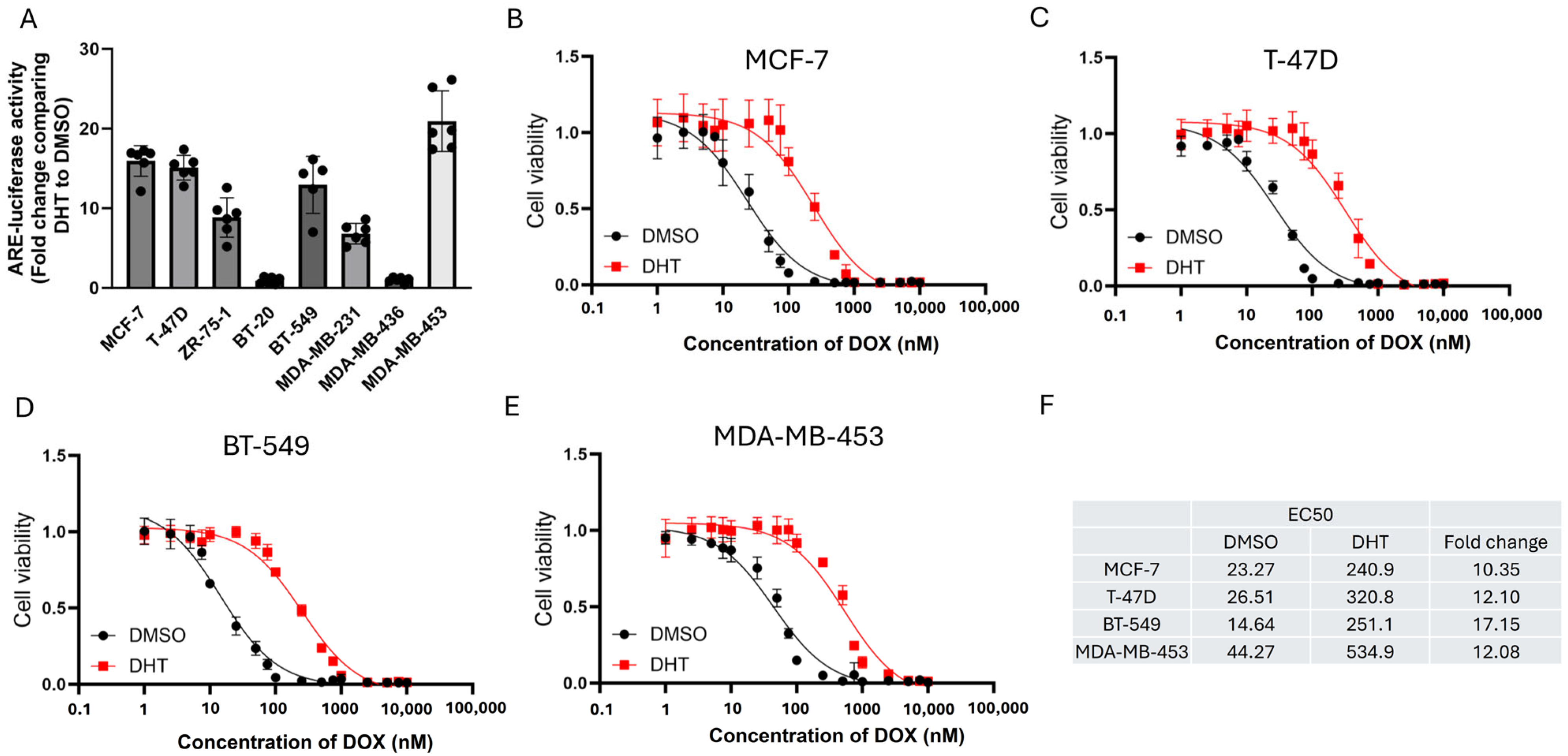
2.4. ARE-Luciferase Reporter Assay
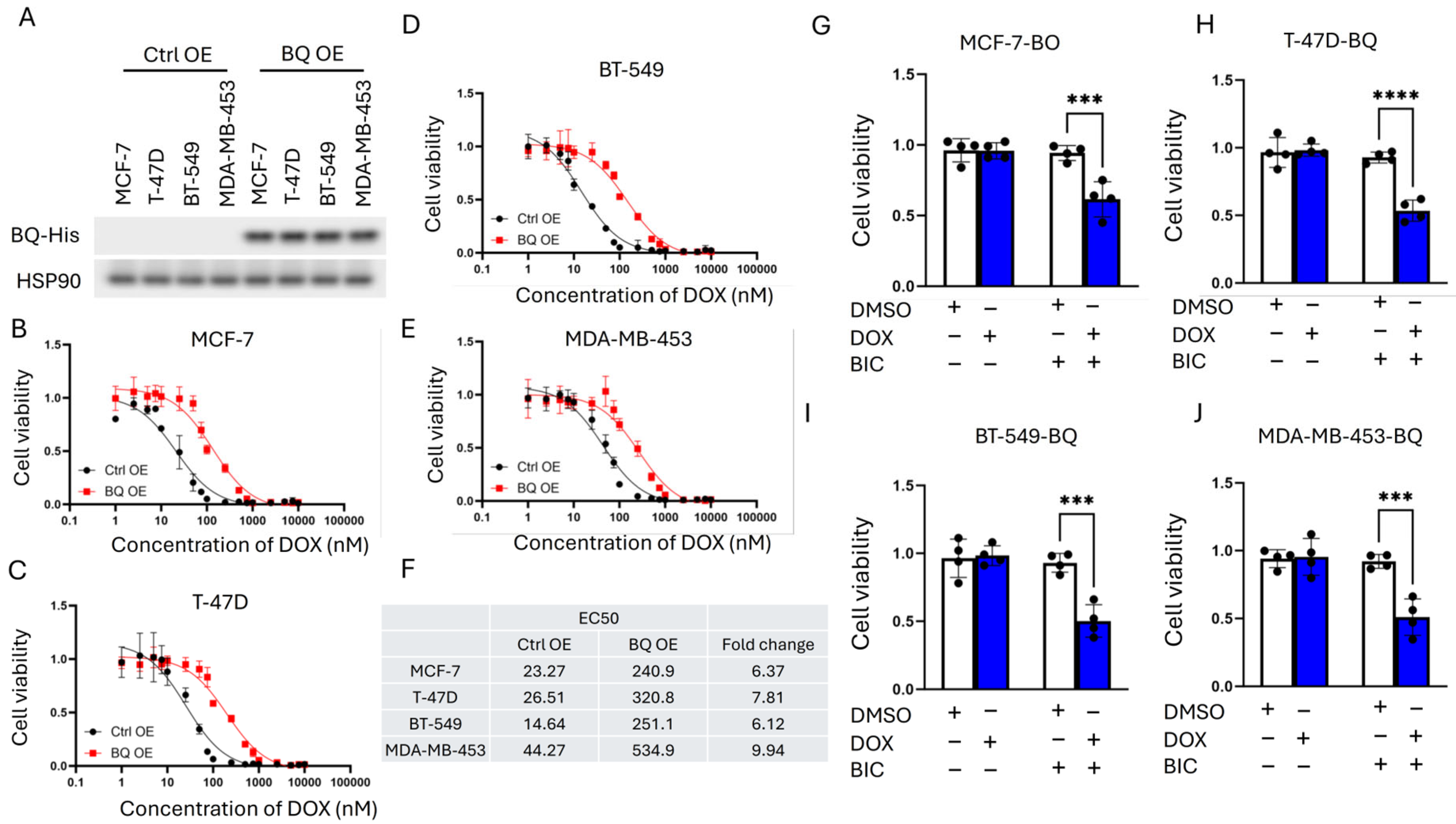
2.5. Cell Viability Assay
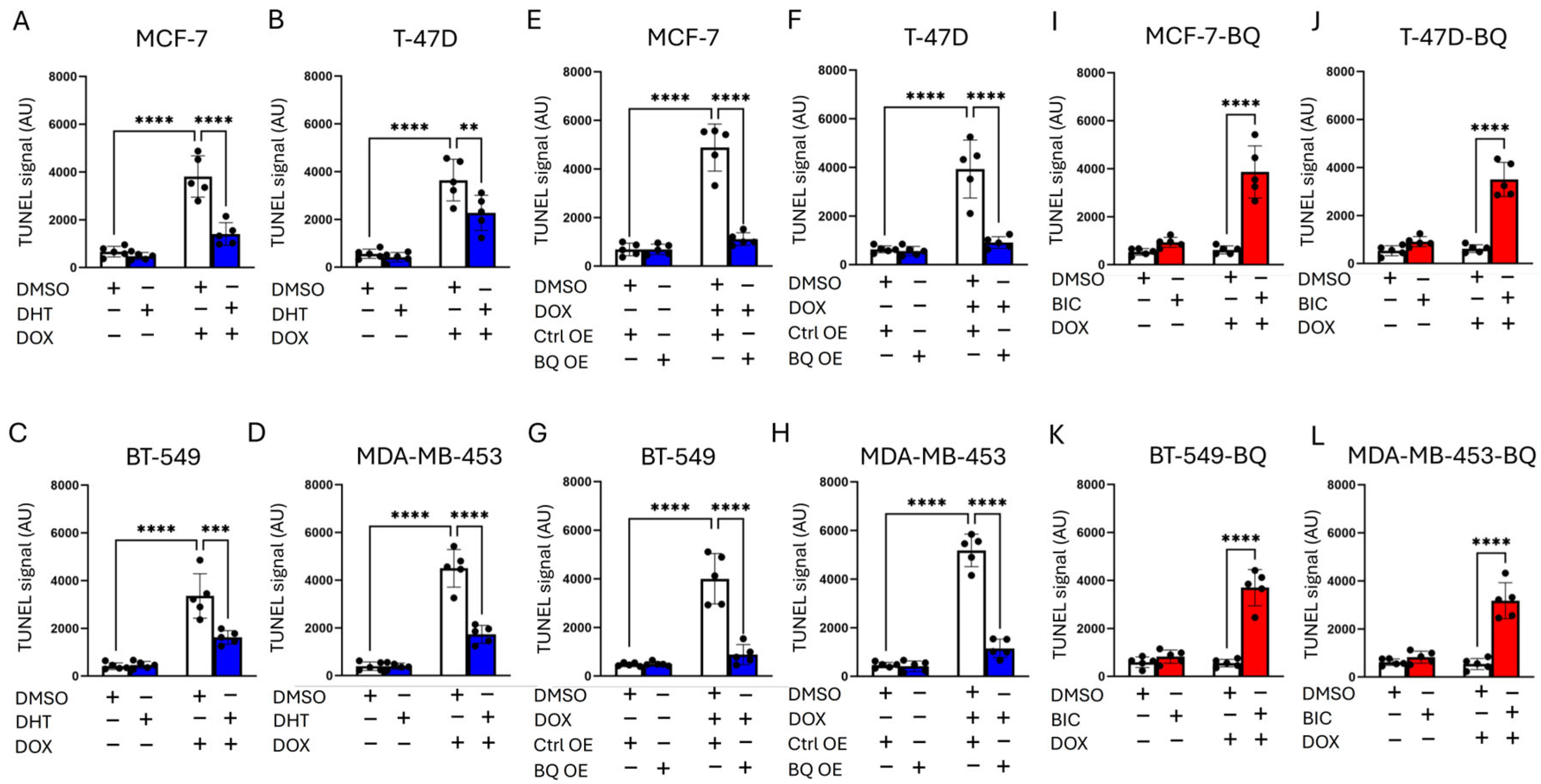
2.6. TUNEL Assay and Comet Assay
2.7. RT-qPCR
2.8. Western Blot
2.9. Co-Immunoprecipitation
2.10. HR and NHEJ Assays
2.11. Gene Expression and Pathway Enrichment Analyses
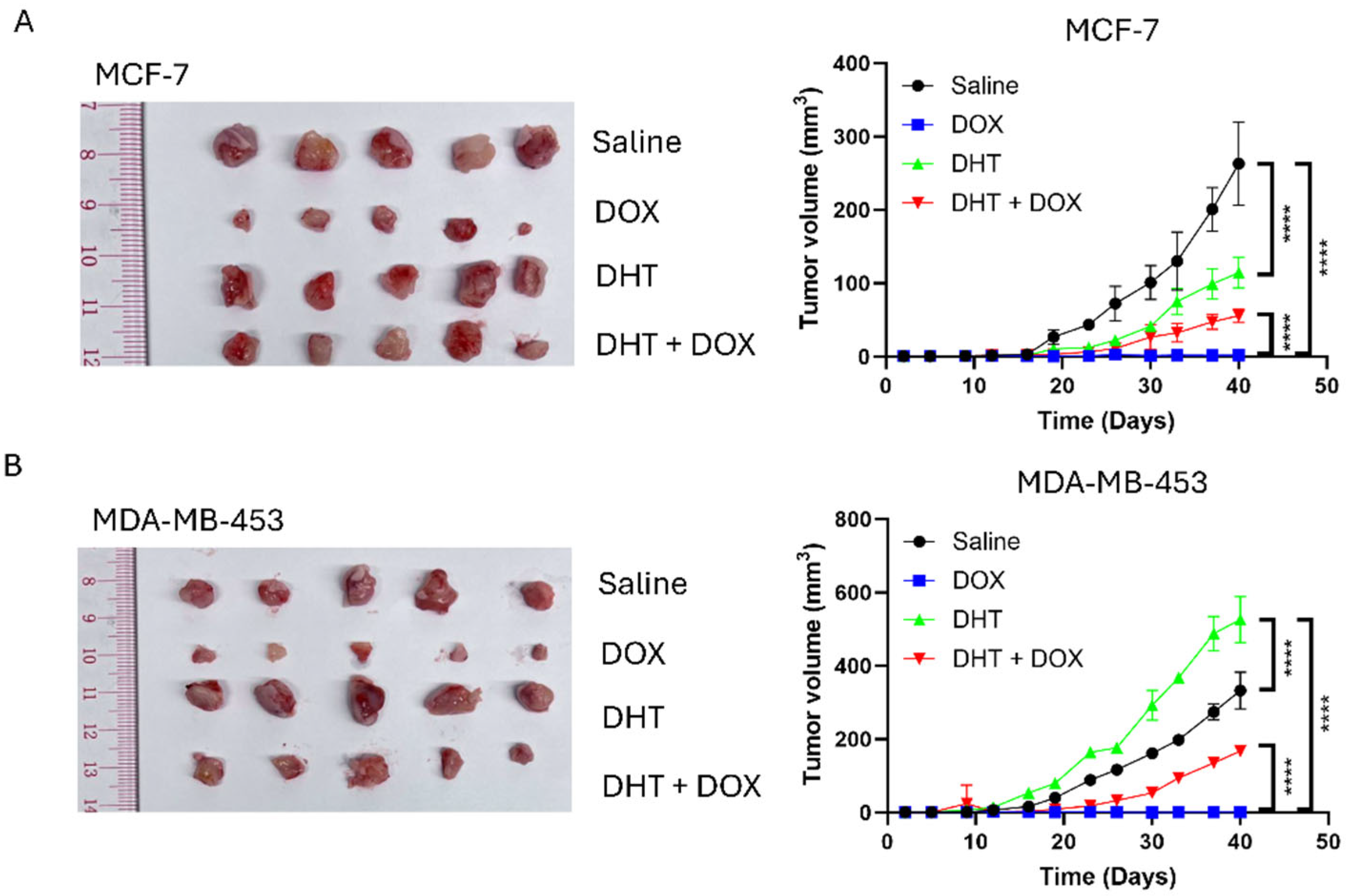
2.12. Xenograft
2.13. Immunohistochemistry and TMA
2.14. Statistical Analysis and Graphic Illustration
3. Results
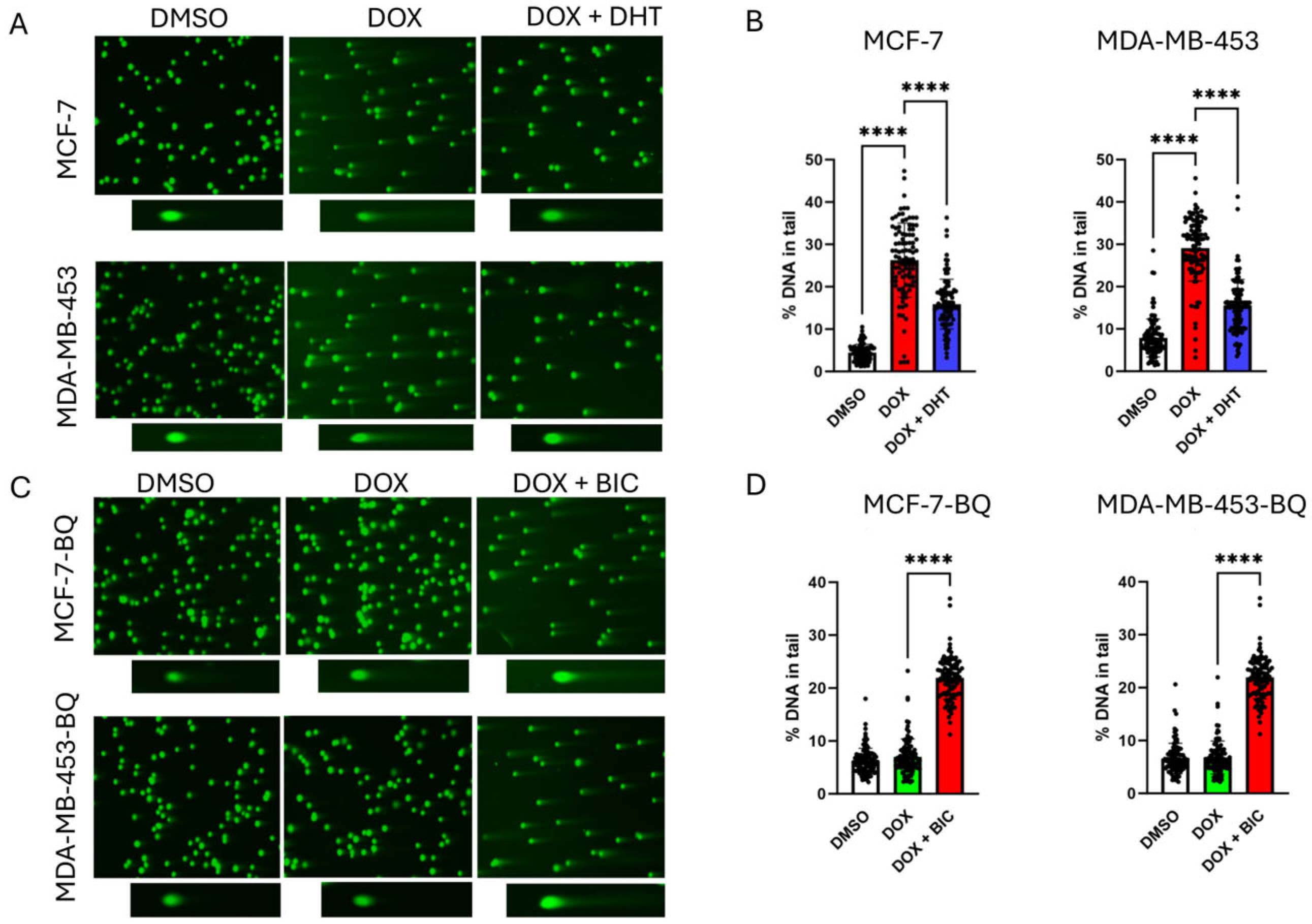
3.1. Doxorubicin (DOX) Resistance Mediated by BQ Overexpression Was AR-Dependent
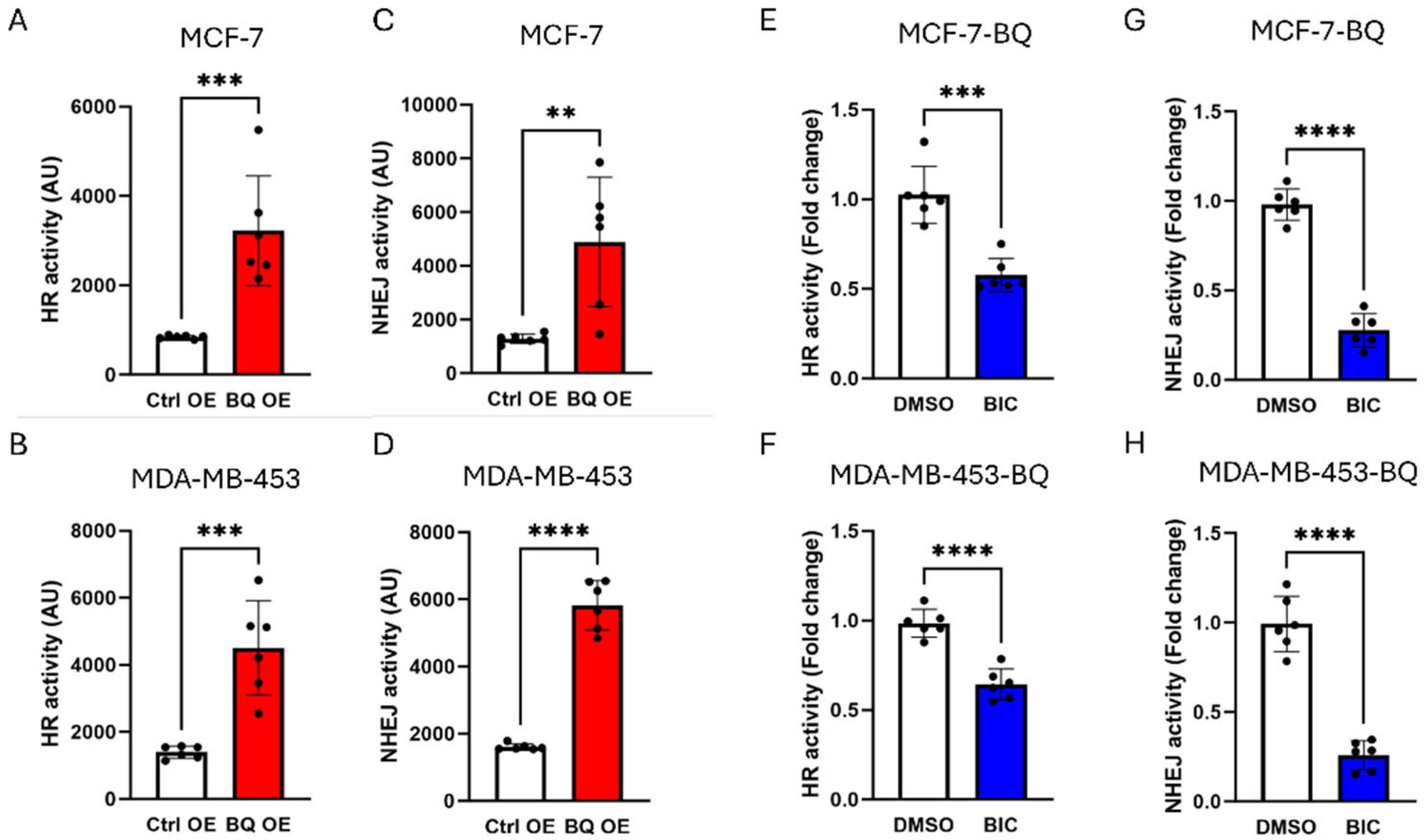
3.2. BQ Overexpression Enhanced DNA Repair Capacity
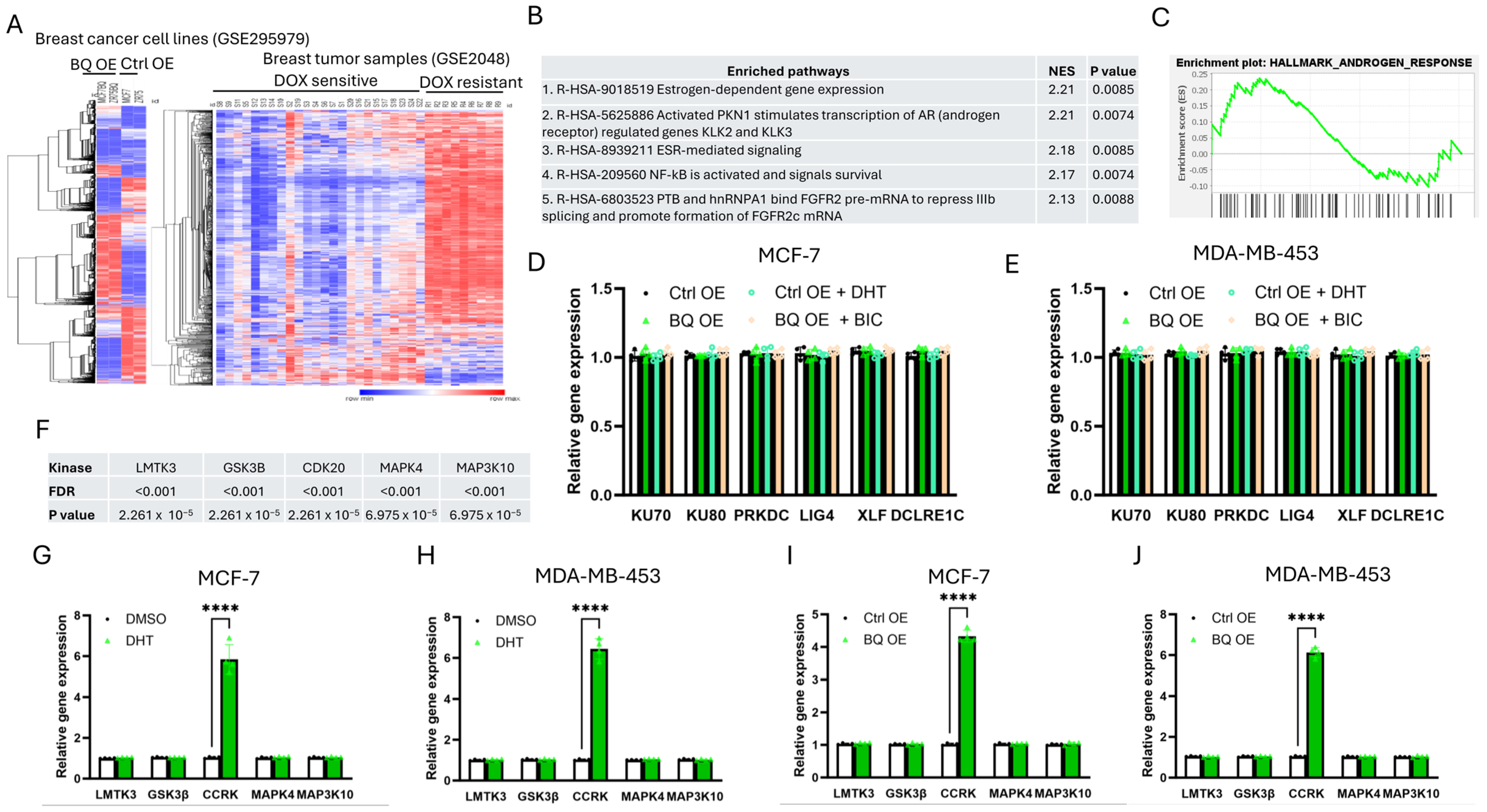
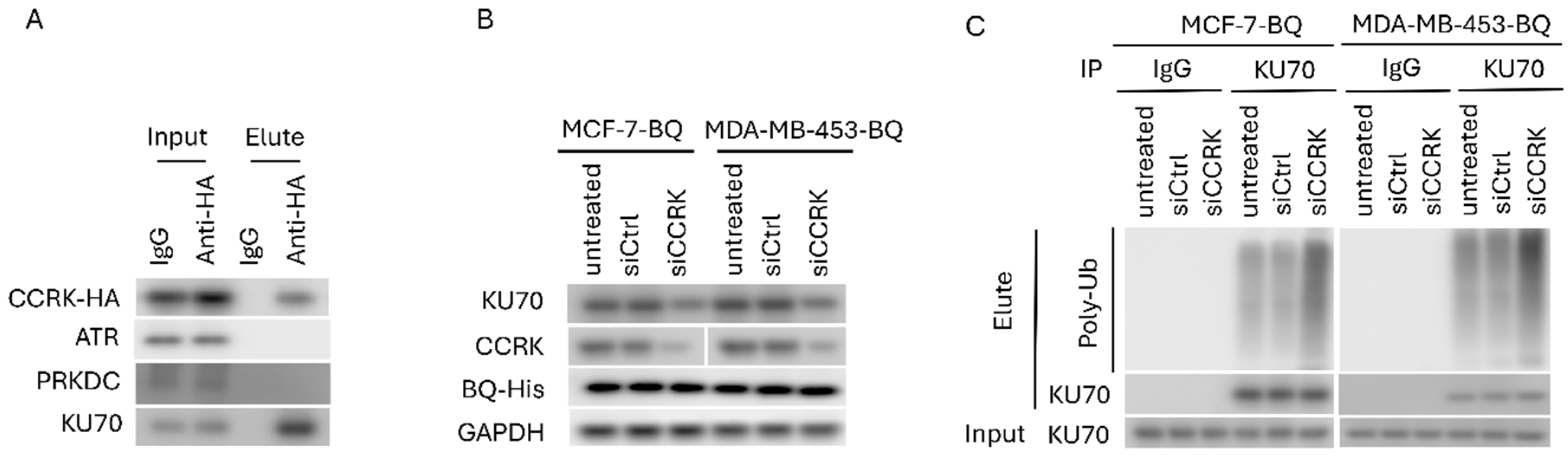
3.3. Identification of CCRK as a Downstream Modulator of AR
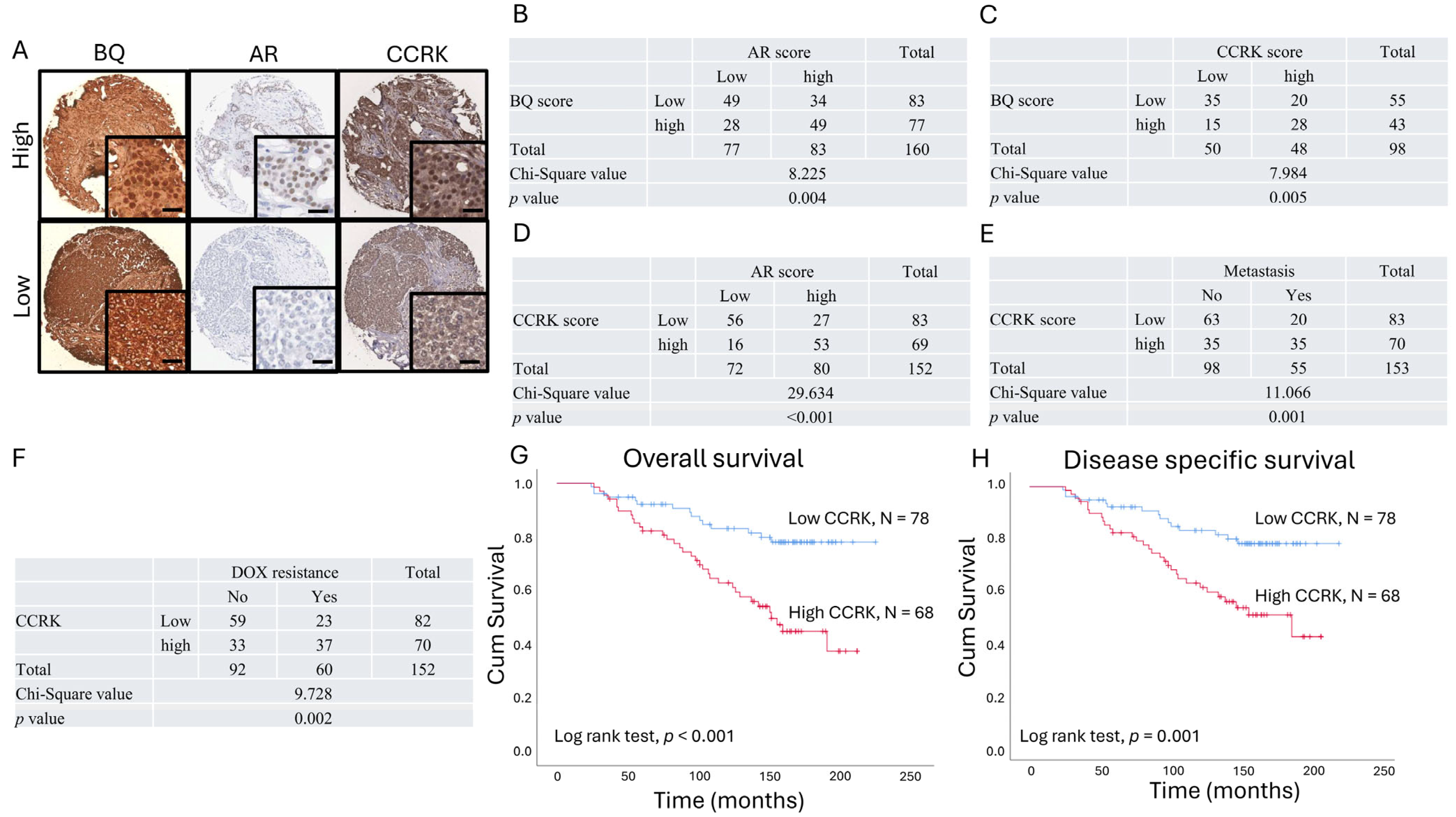
3.4. Clinical Significance of AR, BQ, and CCRK in ER-Positive Breast Cancer
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Szymiczek, A.; Lone, A.; Akbari, M.R. Molecular intrinsic versus clinical subtyping in breast cancer: A comprehensive review. Clin. Genet. 2021, 99, 613–637. [Google Scholar] [CrossRef]
- Thennavan, A.; Beca, F.; Xia, Y.; Recio, S.G.; Allison, K.; Collins, L.C.; Tse, G.M.; Chen, Y.Y.; Schnitt, S.J.; Hoadley, K.A.; et al. Molecular analysis of TCGA breast cancer histologic types. Cell Genom. 2021, 1, 100067. [Google Scholar] [CrossRef] [PubMed]
- Buus, R.; Sestak, I.; Kronenwett, R.; Ferree, S.; Schnabel, C.A.; Baehner, F.L.; Mallon, E.A.; Cuzick, J.; Dowsett, M. Molecular Drivers of Oncotype DX, Prosigna, EndoPredict, and the Breast Cancer Index: A TransATAC Study. J. Clin. Oncol. 2021, 39, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Waks, A.G.; Winer, E.P. Breast Cancer Treatment: A Review. JAMA 2019, 321, 288–300. [Google Scholar] [CrossRef]
- Park, Y.H.; Senkus-Konefka, E.; Im, S.A.; Pentheroudakis, G.; Saji, S.; Gupta, S.; Iwata, H.; Mastura, M.Y.; Dent, R.; Lu, Y.S.; et al. Pan-Asian adapted ESMO Clinical Practice Guidelines for the management of patients with early breast cancer: A KSMO-ESMO initiative endorsed by CSCO, ISMPO, JSMO, MOS, SSO and TOS. Ann. Oncol. 2020, 31, 451–469. [Google Scholar] [CrossRef]
- Vasan, N.; Baselga, J.; Hyman, D.M. A view on drug resistance in cancer. Nature 2019, 575, 299–309. [Google Scholar] [CrossRef]
- Smid, M.; Rodriguez-Gonzalez, F.G.; Sieuwerts, A.M.; Salgado, R.; Prager-Van der Smissen, W.J.C.; van der Vlugt-Daane, M.; van Galen, A.; Nik-Zainal, S.; Staaf, J.; Brinkman, A.B.; et al. Breast cancer genome and transcriptome integration implicates specific mutational signatures with immune cell infiltration. Nat. Commun. 2016, 7, 12910. [Google Scholar] [CrossRef]
- Brueffer, C.; Gladchuk, S.; Winter, C.; Vallon-Christersson, J.; Hegardt, C.; Hakkinen, J.; George, A.M.; Chen, Y.; Ehinger, A.; Larsson, C.; et al. The mutational landscape of the SCAN-B real-world primary breast cancer transcriptome. Embo Mol. Med. 2020, 12, e12118. [Google Scholar] [CrossRef]
- Duffy, M.J.; Synnott, N.C.; Crown, J. Mutant p53 in breast cancer: Potential as a therapeutic target and biomarker. Breast Cancer Res. Treat. 2018, 170, 213–219. [Google Scholar] [CrossRef]
- Mukohara, T. PI3K mutations in breast cancer: Prognostic and therapeutic implications. Breast Cancer-Target 2015, 7, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Martorana, F.; Motta, G.; Pavone, G.; Motta, L.; Stella, S.; Vitale, S.R.; Manzella, L.; Vigneri, P. AKT Inhibitors: New Weapons in the Fight Against Breast Cancer? Front. Pharmacol. 2021, 12, 662232. [Google Scholar] [CrossRef]
- Liu, R.; Chen, Y.W.; Liu, G.Z.; Li, C.X.; Song, Y.R.; Cao, Z.W.; Li, W.; Hu, J.H.; Lu, C.; Liu, Y.Y. PI3K/AKT pathway as a key link modulates the multidrug resistance of cancers. Cell Death Dis. 2020, 11, 797. [Google Scholar] [CrossRef]
- Dong, C.; Wu, J.; Chen, Y.; Nie, J.N.; Chen, C.S. Activation of PI3K/AKT/mTOR Pathway Causes Drug Resistance in Breast Cancer. Front. Pharmacol. 2021, 12, 628690. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Z.; Horn, J.L.; Banda, K.; Goodman, A.Z.; Lim, Y.; Jana, S.; Arora, S.; Germanos, A.A.; Wen, L.X.C.; Hardin, W.R.; et al. The androgen receptor regulates a druggable translational regulon in advanced prostate cancer. Sci. Transl. Med. 2019, 11, eaaw4993. [Google Scholar] [CrossRef] [PubMed]
- Obinata, D.; Lawrence, M.G.; Takayama, K.; Choo, N.; Risbridger, G.P.; Takahashi, S.; Inoue, S. Recent Discoveries in the Androgen Receptor Pathway in Castration-Resistant Prostate Cancer. Front. Oncol. 2020, 10, 581515. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.H.; van Gent, D.C.; Incrocci, L.; van Weerden, W.M.; Nonnekens, J. Role of the DNA damage response in prostate cancer formation, progression and treatment. Prostate Cancer Prostatic Dis. 2020, 23, 24–37, Erratum in Prostate Cancer Prostatic Dis. 2020, 23, 194. [Google Scholar] [CrossRef]
- Polkinghorn, W.R.; Parker, J.S.; Lee, M.X.; Kass, E.M.; Spratt, D.E.; Iaquinta, P.J.; Arora, V.K.; Yen, W.F.; Cai, L.; Zheng, D.Y.; et al. Androgen Receptor Signaling Regulates DNA Repair in Prostate Cancers. Cancer Discov. 2013, 3, 1245–1253. [Google Scholar] [CrossRef]
- Cannan, W.J.; Pederson, D.S. Mechanisms and Consequences of Double-Strand DNA Break Formation in Chromatin. J. Cell Physiol. 2016, 231, 3–14. [Google Scholar] [CrossRef]
- Li, L.Y.; Guan, Y.D.; Chen, X.S.; Yang, J.M.; Cheng, Y. DNA Repair Pathways in Cancer Therapy and Resistance. Front. Pharmacol. 2021, 11, 629266. [Google Scholar] [CrossRef]
- Jette, N.; Lees-Miller, S.P. The DNA-dependent protein kinase: A multifunctional protein kinase with roles in DNA double strand break repair and mitosis. Prog. Biophys. Mol. Bio. 2015, 117, 194–205. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, H.; Liu, T.; Huang, M.; Butter, P.P.; Li, C.; Zhang, L.; Kao, G.D.; Gong, Y.; Maity, A.; et al. Temporal DNA-PK activation drives genomic instability and therapy resistance in glioma stem cells. JCI Insight 2018, 3, e98096. [Google Scholar] [CrossRef]
- Li, L.K.; Karanika, S.; Yang, G.; Wang, J.X.; Park, S.; Broom, B.M.; Manyam, G.C.; Wu, W.H.; Luo, Y.; Basourakos, S.; et al. Androgen receptor inhibitor-induced “BRCAness” and PARP inhibition are synthetically lethal for castration-resistant prostate cancer. Sci. Signal 2017, 10, eaam7479. [Google Scholar] [CrossRef] [PubMed]
- Asim, M.; Tarish, F.; Zecchini, H.I.; Sanjiv, K.; Gelali, E.; Massie, C.E.; Baridi, A.; Warren, A.Y.; Zhao, W.F.; Ogris, C.; et al. Synthetic lethality between androgen receptor signalling and the PARP pathway in prostate cancer. Nat. Commun. 2017, 8, 374. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, P.; Schweizer, M.T.; Lucas, J.M.; Coleman, I.; Nyquist, M.D.; Frank, S.B.; Tharakan, R.; Mostaghel, E.; Luo, J.; Pritchard, C.C.; et al. Supraphysiological androgens suppress prostate cancer growth through androgen receptor-mediated DNA damage. J. Clin. Investig. 2019, 129, 4245–4260. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.Y.; Yang, Y.B.; Xu, K.; Li, L.L.; Huang, J.; Qiu, F.M. Androgen Receptor in Breast Cancer: From Bench to Bedside. Front. Endocrinol. 2020, 11, 573. [Google Scholar] [CrossRef]
- Hu, R.; Dawood, S.; Holmes, M.D.; Collins, L.C.; Schnitt, S.J.; Cole, K.; Marotti, J.D.; Hankinson, S.E.; Colditz, G.A.; Tamimi, R.M. Androgen Receptor Expression and Breast Cancer Survival in Postmenopausal Women. Clin. Cancer Res. 2011, 17, 1867–1874. [Google Scholar] [CrossRef]
- Kensler, K.H.; Regan, M.M.; Heng, Y.J.J.; Baker, G.M.; Pyle, M.E.; Schnitt, S.J.; Hazra, A.; Kammler, R.; Thurlimann, B.; Colleoni, M.; et al. Prognostic and predictive value of androgen receptor expression in postmenopausal women with estrogen receptor-positive breast cancer: Results from the Breast International Group Trial 1-98. Breast Cancer Res. 2019, 21, 30. [Google Scholar] [CrossRef]
- Hickey, T.E.; Selth, L.A.; Chia, K.M.; Laven-Law, G.; Milioli, H.H.; Roden, D.; Jindal, S.; Hui, M.; Finlay-Schultz, J.; Ebrahimie, E.; et al. The androgen receptor is a tumor suppressor in estrogen receptor-positive breast cancer. Nat. Med. 2021, 27, 310–320. [Google Scholar] [CrossRef]
- Ni, M.; Chen, Y.W.; Lim, E.; Wimberly, H.; Bailey, S.T.; Imai, Y.; Rimm, D.L.; Liu, X.S.; Brown, M. Targeting Androgen Receptor in Estrogen Receptor-Negative Breast Cancer. Cancer Cell 2011, 20, 119–131. [Google Scholar] [CrossRef]
- Zhang, L.; Gong, C.; Lau, S.L.; Yang, N.; Wong, O.G.; Cheung, A.N.; Tsang, J.W.; Chan, K.Y.; Khoo, U.S. SpliceArray profiling of breast cancer reveals a novel variant of NCOR2/SMRT that is associated with tamoxifen resistance and control of ERalpha transcriptional activity. Cancer Res. 2013, 73, 246–255. [Google Scholar] [CrossRef]
- Gong, C.; Man, E.P.S.; Tsoi, H.; Lee, T.K.W.; Lee, P.; Ma, S.T.; Wong, L.S.; Luk, M.Y.; Rakha, E.A.; Green, A.R.; et al. BQ323636.1, a Novel Splice Variant to NCOR2, as a Predictor for Tamoxifen-Resistant Breast Cancer. Clin. Cancer Res. 2018, 24, 3681–3691. [Google Scholar] [CrossRef]
- Tsoi, H.; Lok, J.; Man, E.P.; Cheng, C.N.; Leung, M.H.; You, C.P.; Chan, S.Y.; Chan, W.L.; Khoo, U.S. Overexpression of BQ323636.1 contributes to anastrozole resistance in AR+ve/ER+ve breast cancer. J. Pathol. 2023, 261, 156–168. [Google Scholar] [CrossRef] [PubMed]
- Leung, M.H.; Tsoi, H.; Gong, C.; Man, E.P.; Zona, S.; Yao, S.; Lam, E.W.; Khoo, U.S. A Splice Variant of NCOR2, BQ323636.1, Confers Chemoresistance in Breast Cancer by Altering the Activity of NRF2. Cancers 2020, 12, 533. [Google Scholar] [CrossRef] [PubMed]
- Gyori, B.M.; Venkatachalam, G.; Thiagarajan, P.S.; Hsu, D.; Clement, M.V. Corrigendum to OpenComet: An automated tool for comet assay image analysis [Redox Biol. Volume 2, 2014, Pages 457–465]. Redox Biol. 2021, 40, 101876. [Google Scholar] [CrossRef]
- Pierce, A.J.; Johnson, R.D.; Thompson, L.H.; Jasin, M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes. Dev. 1999, 13, 2633–2638. [Google Scholar] [CrossRef] [PubMed]
- Bennardo, N.; Gunn, A.; Cheng, A.; Hasty, P.; Stark, J.M. Limiting the persistence of a chromosome break diminishes its mutagenic potential. PLoS Genet. 2009, 5, e1000683. [Google Scholar] [CrossRef]
- Richardson, C.; Moynahan, M.E.; Jasin, M. Double-strand break repair by interchromosomal recombination: Suppression of chromosomal translocations. Genes. Dev. 1998, 12, 3831–3842. [Google Scholar] [CrossRef]
- Folgueira, M.A.; Brentani, H.; Katayama, M.L.; Patrao, D.F.; Carraro, D.M.; Mourao Netto, M.; Barbosa, E.M.; Caldeira, J.R.; Abreu, A.P.; Lyra, E.C.; et al. Gene expression profiling of clinical stages II and III breast cancer. Braz. J. Med. Biol. Res. 2006, 39, 1101–1113. [Google Scholar] [CrossRef][Green Version]
- Tsoi, H.; You, C.-P.; Cheung, K.H.-L.; Tse, Y.-S.; Khoo, U.-S. The Splice Variant of the NCOR2 Gene BQ323636.1 Modulates ACSL4 Expression to Enhance Fatty Acid Metabolism and Support of Tumor Growth in Breast Cancer. Int. J. Mol. Sci. 2025, 26, 4989. [Google Scholar] [CrossRef]
- Ge, S.X.; Son, E.W.; Yao, R. iDEP: An integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinform. 2018, 19, 534. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Matsuura, Y.; Ishiguro-Watanabe, M. KEGG: Biological systems database as a model of the real world. Nucleic Acids Res. 2025, 53, D672–D677. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Yu, B.; Liu, Y.; Luo, H.; Fu, J.; Li, Y.; Shao, C. Androgen receptor splicing variant 7 (ARV7) inhibits docetaxel sensitivity by inactivating the spindle assembly checkpoint. J. Biol. Chem. 2021, 296, 100276. [Google Scholar] [CrossRef]
- Luo, H.; Liu, Y.; Li, Y.; Zhang, C.; Yu, B.; Shao, C. Androgen receptor splicing variant 7 (ARv7) promotes DNA damage response in prostate cancer cells. FASEB J. 2022, 36, e22495. [Google Scholar] [CrossRef]
- Nesic, K.; Parker, P.; Swisher, E.M.; Krais, J.J. DNA repair and the contribution to chemotherapy resistance. Genome Med. 2025, 17, 62. [Google Scholar] [CrossRef]
- Davis, C.; Spaller, B.L.; Matouschek, A. Mechanisms of substrate recognition by the 26S proteasome. Curr. Opin. Struct. Biol. 2021, 67, 161–169. [Google Scholar] [CrossRef]
- Linders, A.N.; Dias, I.B.; Lopez Fernandez, T.; Tocchetti, C.G.; Bomer, N.; Van der Meer, P. A review of the pathophysiological mechanisms of doxorubicin-induced cardiotoxicity and aging. npj Aging 2024, 10, 9. [Google Scholar] [CrossRef]
- Katsumata, N.; Watanabe, T.; Minami, H.; Aogi, K.; Tabei, T.; Sano, M.; Masuda, N.; Andoh, J.; Ikeda, T.; Shibata, T.; et al. Phase III trial of doxorubicin plus cyclophosphamide (AC), docetaxel, and alternating AC and docetaxel as front-line chemotherapy for metastatic breast cancer: Japan Clinical Oncology Group trial (JCOG9802). Ann. Oncol. 2009, 20, 1210–1215. [Google Scholar] [CrossRef] [PubMed]
- Baldassarre, G.; L de la Serna, I.; Vallette, F.M. Death-ision: The link between cellular resilience and cancer resistance to treatments. Mol. Cancer 2025, 24, 144. [Google Scholar] [CrossRef] [PubMed]
- Khoo, U.S.; Man, E.P.S.; So, Z.Q.; Tsoi, H. BQ323636.1 modulates the AR-CDK20 axis to confer doxorubicin resistance in breast cancer (ENA24-0047). In Proceedings of the 36th EORTC-NCI-AACR Symposium, Barcelona, Spain, 23–25 October 2024. [Google Scholar]
- de Klein, B.; Eickhoff, N.; Zwart, W. The emerging regulatory interface between DNA repair and steroid hormone receptors in cancer. Trends Mol. Med. 2025, 31, 718–730. [Google Scholar] [CrossRef]
- Hussain, M.; Daignault-Newton, S.; Twardowski, P.W.; Albany, C.; Stein, M.N.; Kunju, L.P.; Siddiqui, J.; Wu, Y.M.; Robinson, D.; Lonigro, R.J.; et al. Targeting Androgen Receptor and DNA Repair in Metastatic Castration-Resistant Prostate Cancer: Results from NCI 9012. J. Clin. Oncol. 2018, 36, 991–999. [Google Scholar] [CrossRef]
- Sun, H.; Yang, W.; Tian, Y.; Zeng, X.; Zhou, J.; Mok, M.T.S.; Tang, W.; Feng, Y.; Xu, L.; Chan, A.W.H.; et al. An inflammatory-CCRK circuitry drives mTORC1-dependent metabolic and immunosuppressive reprogramming in obesity-associated hepatocellular carcinoma. Nat. Commun. 2018, 9, 5214. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Zhou, J.; Xiong, Z.; Sun, H.; Yang, W.; Mok, M.T.S.; Wang, J.; Li, J.; Liu, M.; Tang, W.; et al. Cell cycle-related kinase reprograms the liver immune microenvironment to promote cancer metastasis. Cell Mol. Immunol. 2021, 18, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Cheng, A.S.; Tsang, D.P.; Li, M.S.; Go, M.Y.; Cheung, Y.S.; Zhao, G.J.; Ng, S.S.; Lin, M.C.; Yu, J.; et al. Cell cycle-related kinase is a direct androgen receptor-regulated gene that drives beta-catenin/T cell factor-dependent hepatocarcinogenesis. J. Clin. Investig. 2011, 121, 3159–3175. [Google Scholar] [CrossRef] [PubMed]
| Clinical Characters | Number of Cases | Percentage (%) | |
|---|---|---|---|
| ER-positive breast cancer patients | 170 | 100 | |
| Age | <50 | 86 | 50.6 |
| ≥50 | 77 | 45.3 | |
| T stage | I, II | 146 | 85.9 |
| III, IV | 17 | 10.0 | |
| Lymph Node status | Positive | 133 | 78.2 |
| Negative | 35 | 20.6 | |
| Tumor Grade | 1, 2 | 119 | 70.0 |
| 3 | 40 | 23.5 | |
| Tumor Size | <2 cm | 13 | 7.6 |
| ≥2 cm | 117 | 68.8 | |
| Progesterone receptor status | Positive | 125 | 73.5 |
| Negative | 44 | 25.9 | |
| HER2 receptor status | Positive | 26 | 15.3 |
| Negative | 106 | 62.4 | |
| CCRK nucleus score | Positive | 153 | 90.0 |
| Negative | 0 | 0.0 | |
| AR nucleus score | Positive | 152 | 89.4 |
| Positive | 163 | 95.9 | |
| BQ nucleus score | Positive | 163 | 95.9 |
| Negative | 0 | 0.0 |
| Clinical-Pathological Parameters | Univariate Analysis | |
|---|---|---|
| RR (95% CI) | p value | |
| Age (N = 163) | 1.335 (0.781, 2.283) | 0.290 |
| T-stage (N = 158) | 2.393 (1.161, 4.931) | 0.018 |
| Lymph-node involvement (N = 162) | 2.931 (1.165, 7.376) | 0.022 |
| Tumor-Grade (N = 154) | 1.768 (0.994, 3.142) | 0.052 |
| Histological type (N = 157) | 1.003 (0.427, 2.354) | 0.995 |
| PR status (N = 162) | 0.545 (0.314, 0.948) | 0.031 |
| HER2 status (N = 127) | 1.975 (1.007, 3.871) | 0.048 |
| Tumor size (N = 126) | 1.507 (0.463, 4.912) | 0.496 |
| Cases with high CCRK nucleus score (N = 146) | 3.056 (1.661, 5.621) | <0.001 |
| Cases with high AR nucleus score (N = 155) | 3.420 (1.783, 6.562) | <0.001 |
| Cases with high BQ nucleus score (N = 156) | 2.175 (1.210, 3.911) | 0.009 |
| Clinical-Pathological Parameters | Univariate Analysis | |
|---|---|---|
| RR (95% CI) | p value | |
| Age (N = 163) | 1.212 (0.694, 2.117) | 0.499 |
| T-stage (N = 158) | 2.604 (1.255, 5.403) | 0.010 |
| Lymph-node involvement (N =1 62) | 2.633 (1.042, 6.651) | 0.041 |
| Tumor-Grade (N = 154) | 1.839 (1.013, 3.339) | 0.045 |
| Histological type (N = 157) | 0.916 (0.388, 2.161) | 0.841 |
| PR status (N = 162) | 0.524 (0.296, 0.928) | 0.027 |
| HER2 status (N = 127) | 1.812 (0.905, 3.629) | 0.093 |
| Tumor size (N = 126) | 1.303 (0.397, 4.274) | 0.662 |
| Cases with high CCRK nucleus score (N = 146) | 2.731 (1.472, 5.067) | <0.001 |
| Cases with high AR nucleus score (N = 155) | 3.092 (1.601, 5.970) | <0.001 |
| Cases with high BQ nucleus score (N = 156) | 2.145 (1.172, 3.928) | 0.013 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsoi, H.; So, Z.-Q.; Man, E.P.S.; You, C.-P.; Cheung, K.H.-L.; Tse, Y.-S.; Chan, W.-L.; Khoo, U.-S. BQ323636.1 Employs the AR-CCRK Axis to Modulate the Expression of KU70 to Interfere with Non-Homologous End Joining Mediated DNA Repair Mechanism. Cells 2025, 14, 1341. https://doi.org/10.3390/cells14171341
Tsoi H, So Z-Q, Man EPS, You C-P, Cheung KH-L, Tse Y-S, Chan W-L, Khoo U-S. BQ323636.1 Employs the AR-CCRK Axis to Modulate the Expression of KU70 to Interfere with Non-Homologous End Joining Mediated DNA Repair Mechanism. Cells. 2025; 14(17):1341. https://doi.org/10.3390/cells14171341
Chicago/Turabian StyleTsoi, Ho, Zi-Qing So, Ellen P. S. Man, Chan-Ping You, Koei Ho-Lam Cheung, Yin-Suen Tse, Wing-Lok Chan, and Ui-Soon Khoo. 2025. "BQ323636.1 Employs the AR-CCRK Axis to Modulate the Expression of KU70 to Interfere with Non-Homologous End Joining Mediated DNA Repair Mechanism" Cells 14, no. 17: 1341. https://doi.org/10.3390/cells14171341
APA StyleTsoi, H., So, Z.-Q., Man, E. P. S., You, C.-P., Cheung, K. H.-L., Tse, Y.-S., Chan, W.-L., & Khoo, U.-S. (2025). BQ323636.1 Employs the AR-CCRK Axis to Modulate the Expression of KU70 to Interfere with Non-Homologous End Joining Mediated DNA Repair Mechanism. Cells, 14(17), 1341. https://doi.org/10.3390/cells14171341








