Highly Oligomeric DRP1 Strategic Positioning at Mitochondria–Sarcoplasmic Reticulum Contacts in Adult Murine Heart Through ACTIN Anchoring
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Isolation of Mouse Cardiomyocytes
2.3. Electrical Field Stimulation on Isolated Adult Mouse Cardiomyocytes
2.4. Isolation of Subcellular Fractions from Murine Heart
2.5. Protein Analysis and Western Blot
2.6. First Dimension Light Blue Native Polyacrylamide Gel Electrophoresis (LBN-PAGE)
2.7. Second-Dimension SDS-PAGE
2.8. Immunofluorescence (IF) and Imaging Analysis
2.9. Pre-Embedding Immunogold (IG) and Transmission Electron Microscopy (TEM)
2.10. Langendorff Perfusion of Rat Hearts
2.11. Citrate Cynthase Activity Measurements
2.12. Statistical Analysis
3. Results
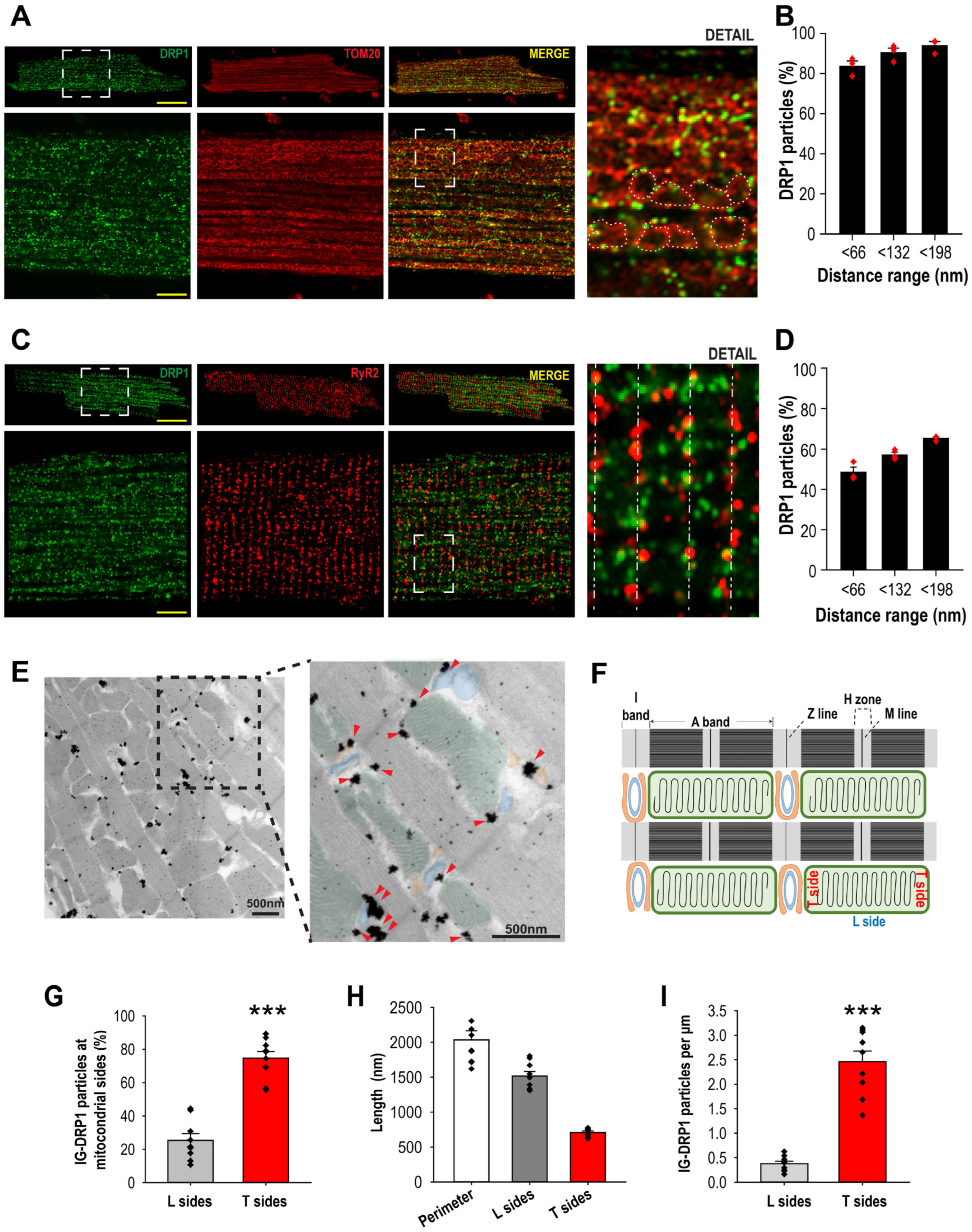
3.1. DRP1 Is Preferentially Located at the Mitochondria–jSR Contacts in Primary Adult Cardiomyocytes
3.2. Mitochondria–SR Contacts Harbor the Membrane-Bound DRP1
3.3. DRP1 Localized at the SR and MAM Fractions at High Oligomeric State
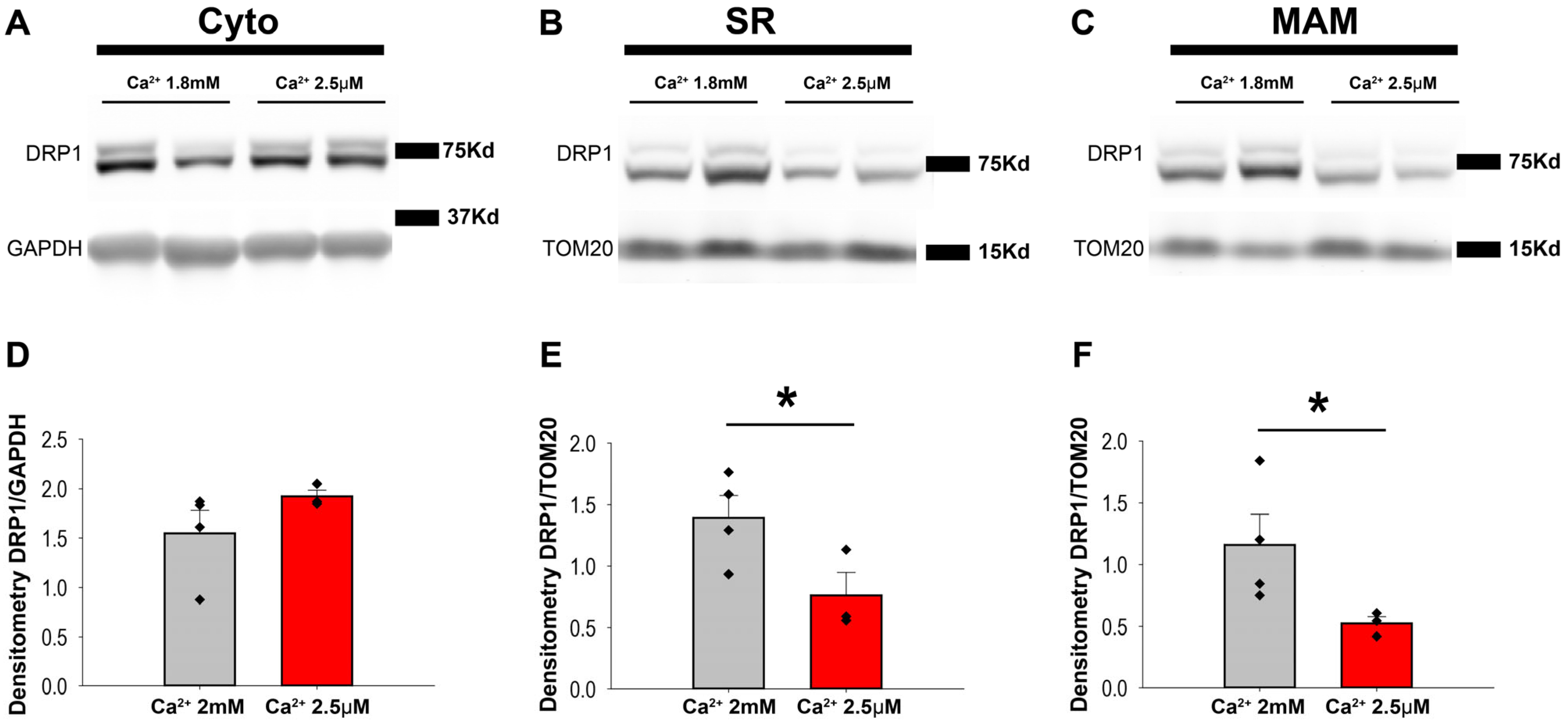
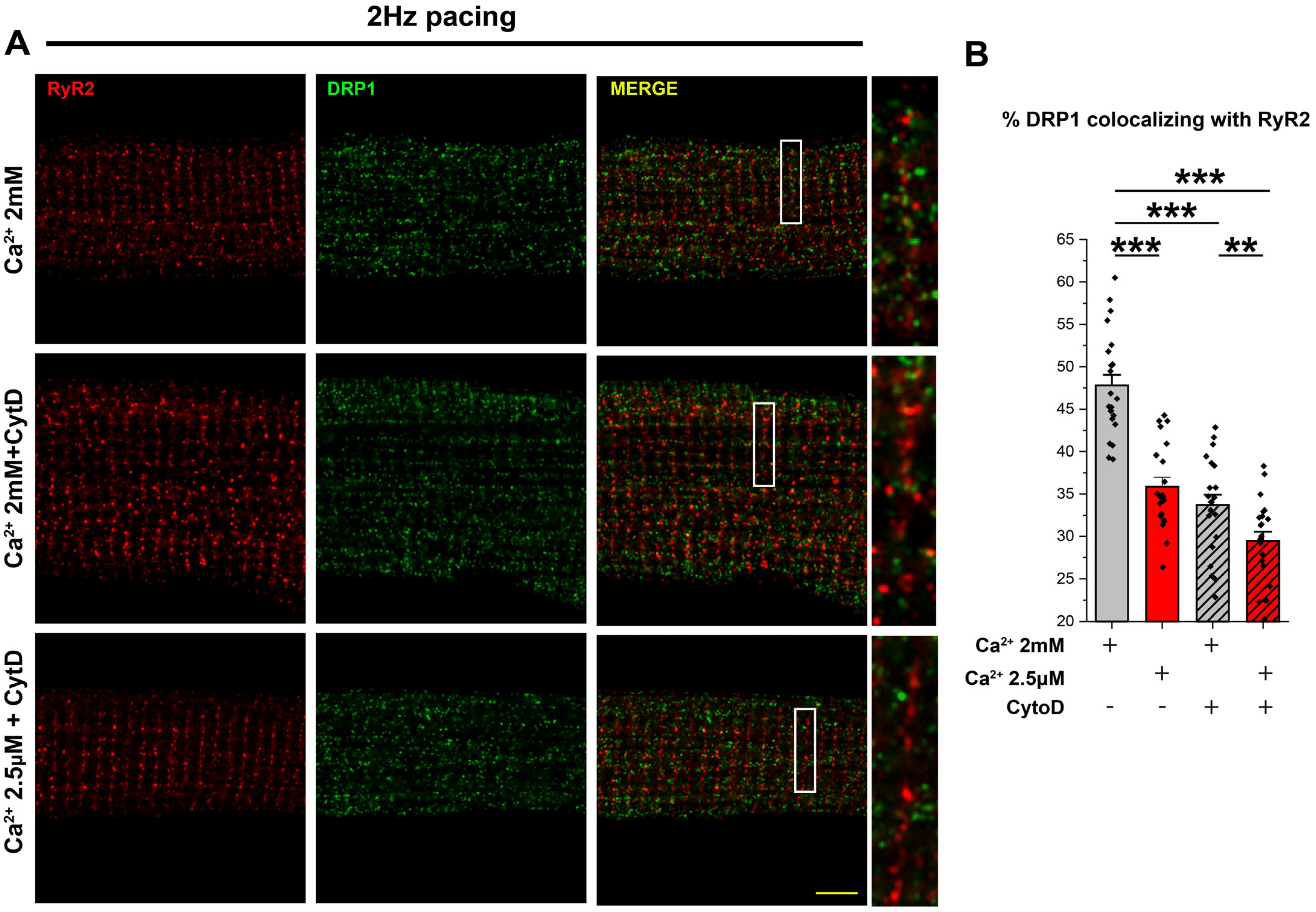
3.4. Ca2+ Transient Activity Is Involved in DRP1 Positioning at the Mitochondria–SR Contacts in the Heart
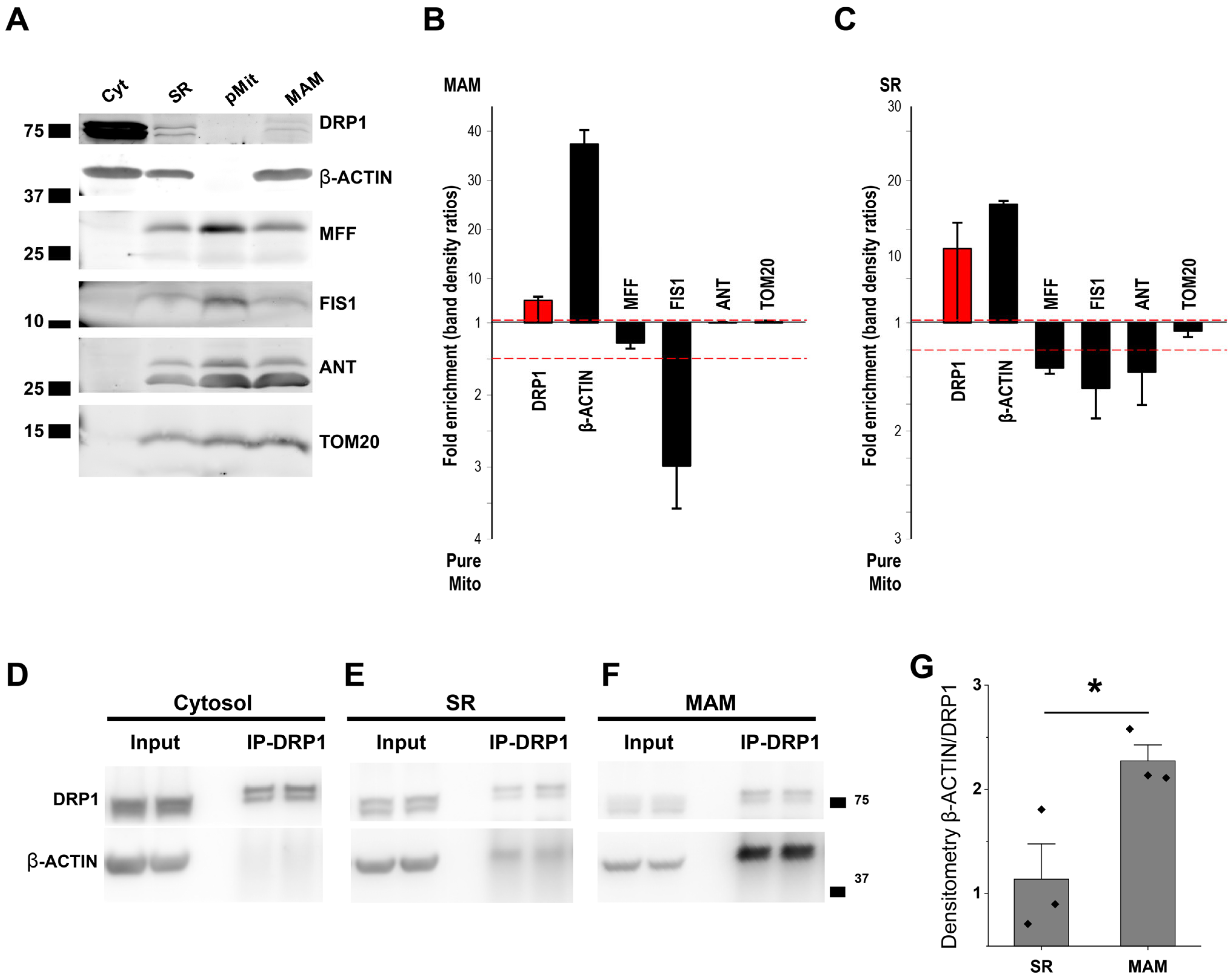
3.5. β-ACTIN Is Responsible for DRP1 Accumulation in MAM
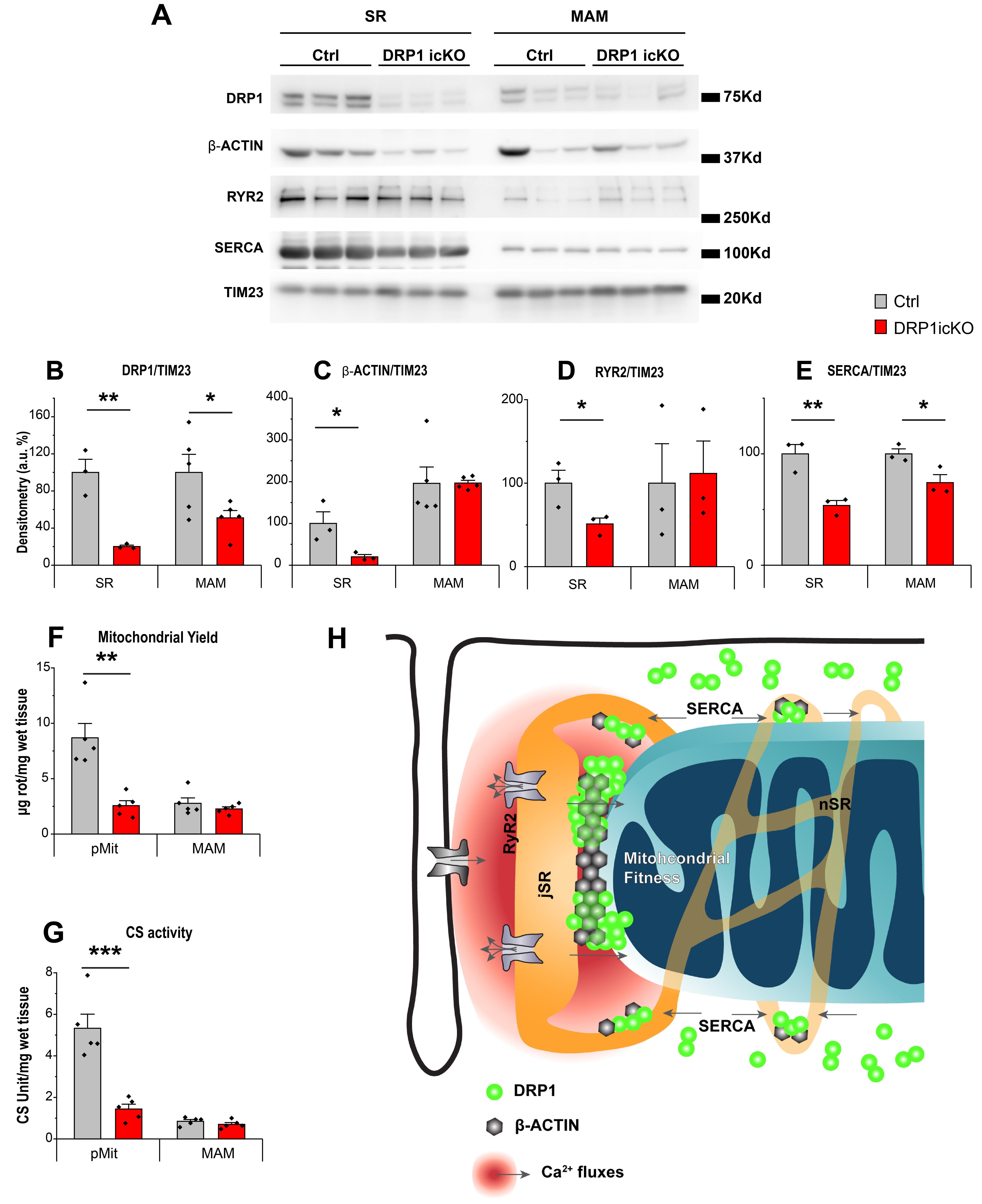
3.6. MAM-Localized DRP1 and β-ACTIN Are Sustained in DRP1-Deficient Hearts
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Friedman, J.R.; Nunnari, J. Mitochondrial Form and Function. Nature 2014, 505, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Kasahara, A.; Scorrano, L. Mitochondria: From Cell Death Executioners to Regulators of Cell Differentiation. Trends Cell Biol. 2014, 24, 761–770. [Google Scholar] [CrossRef] [PubMed]
- Pham, A.H.; Meng, S.; Chu, Q.N.; Chan, D.C. Loss of Mfn2 Results in Progressive, Retrograde Degeneration of Dopaminergic Neurons in the Nigrostriatal Circuit. Hum. Mol. Genet. 2012, 21, 4817–4826. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Sterky, F.H.; Mourier, A.; Terzioglu, M.; Cullheim, S.; Olson, L.; Larsson, N.-G. Mitofusin 2 Is Necessary for Striatal Axonal Projections of Midbrain Dopamine Neurons. Hum. Mol. Genet. 2012, 21, 4827–4835. [Google Scholar] [CrossRef]
- Wasiak, S.; Zunino, R.; McBride, H.M. Bax/Bak Promote Sumoylation of DRP1 and Its Stable Association with Mitochondria during Apoptotic Cell Death. J. Cell Biol. 2007, 177, 439–450. [Google Scholar] [CrossRef]
- Rambold, A.S.; Kostelecky, B.; Elia, N.; Lippincott-Schwartz, J. Tubular Network Formation Protects Mitochondria from Autophagosomal Degradation during Nutrient Starvation. Proc. Natl. Acad. Sci. USA 2011, 108, 10190–10195. [Google Scholar] [CrossRef]
- Burté, F.; Carelli, V.; Chinnery, P.F.; Yu-Wai-Man, P. Disturbed Mitochondrial Dynamics and Neurodegenerative Disorders. Nat. Rev. Neurol. 2015, 11, 11–24. [Google Scholar] [CrossRef]
- Wang, W.; Karamanlidis, G.; Tian, R. Novel Targets for Mitochondrial Medicine. Sci. Transl. Med. 2016, 8, 326rv3. [Google Scholar] [CrossRef]
- Dorn II, G.W. Mitochondrial Dynamism and Heart Disease: Changing Shape and Shaping Change. EMBO Mol. Med. 2015, 7, 865–877. [Google Scholar] [CrossRef]
- Yoon, Y.; Pitts, K.R.; McNiven, M.A. Mammalian Dynamin-like Protein DLP1 Tubulates Membranes. Mol. Biol. Cell 2001, 12, 2894–2905. [Google Scholar] [CrossRef]
- Mishra, P.; Chan, D.C. Metabolic Regulation of Mitochondrial Dynamics. J. Cell Biol. 2016, 212, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Basu, K.; Lajoie, D.; Aumentado-Armstrong, T.; Chen, J.; Koning, R.I.; Bossy, B.; Bostina, M.; Sik, A.; Bossy-Wetzel, E.; Rouiller, I. Molecular Mechanism of DRP1 Assembly Studied in Vitro by Cryo-Electron Microscopy. PLoS ONE 2017, 12, e0179397. [Google Scholar] [CrossRef] [PubMed]
- Imoto, M.; Tachibana, I.; Urrutia, R. Identification and Functional Characterization of a Novel Human Protein Highly Related to the Yeast Dynamin-like GTPase Vps1p. J. Cell. Sci. 1998, 111 Pt 10, 1341–1349. [Google Scholar] [CrossRef] [PubMed]
- Santel, A.; Frank, S.; Gaume, B.; Herrler, M.; Youle, R.J.; Fuller, M.T. Mitofusin-1 Protein Is a Generally Expressed Mediator of Mitochondrial Fusion in Mammalian Cells. J. Cell. Sci. 2003, 116, 2763–2774. [Google Scholar] [CrossRef]
- Dorn, G.W. Evolving Concepts of Mitochondrial Dynamics. Annu. Rev. Physiol. 2019, 81, 1–17. [Google Scholar] [CrossRef]
- Song, M.; Mihara, K.; Chen, Y.; Scorrano, L.; Dorn, G.W. Mitochondrial Fission and Fusion Factors Reciprocally Orchestrate Mitophagic Culling in Mouse Hearts and Cultured Fibroblasts. Cell Metab. 2015, 21, 273–285. [Google Scholar] [CrossRef]
- Korobova, F.; Ramabhadran, V.; Higgs, H.N. An Actin-Dependent Step in Mitochondrial Fission Mediated by the ER-Associated Formin INF2. Science 2013, 339, 464–467. [Google Scholar] [CrossRef]
- Manor, U.; Bartholomew, S.; Golani, G.; Christenson, E.; Kozlov, M.; Higgs, H.; Spudich, J.; Lippincott-Schwartz, J. A Mitochondria-Anchored Isoform of the Actin-Nucleating Spire Protein Regulates Mitochondrial Division. eLife Sci. 2015, 4, e08828. [Google Scholar] [CrossRef]
- Chakrabarti, R.; Ji, W.-K.; Stan, R.V.; de Juan Sanz, J.; Ryan, T.A.; Higgs, H.N. INF2-Mediated Actin Polymerization at the ER Stimulates Mitochondrial Calcium Uptake, Inner Membrane Constriction, and Division. J. Cell Biol. 2018, 217, 251–268. [Google Scholar] [CrossRef]
- Song, M.; Dorn, G.W., 2nd. Mitoconfusion: Noncanonical Functioning of Dynamism Factors in Static Mitochondria of the Heart. Cell Metab. 2015, 21, 195–205. [Google Scholar] [CrossRef]
- Ishihara, T.; Ban-Ishihara, R.; Maeda, M.; Matsunaga, Y.; Ichimura, A.; Kyogoku, S.; Aoki, H.; Katada, S.; Nakada, K.; Nomura, M.; et al. Dynamics of Mitochondrial DNA Nucleoids Regulated by Mitochondrial Fission Is Essential for Maintenance of Homogeneously Active Mitochondria during Neonatal Heart Development. Mol. Cell. Biol. 2015, 35, 211–223. [Google Scholar] [CrossRef]
- Papanicolaou, K.N.; Kikuchi, R.; Ngoh, G.A.; Coughlan, K.A.; Dominguez, I.; Stanley, W.C.; Walsh, K. Mitofusins 1 and 2 Are Essential for Postnatal Metabolic Remodeling in Heart. Circ. Res. 2012, 111, 1012–1026. [Google Scholar] [CrossRef]
- Silva Ramos, E.; Larsson, N.-G.; Mourier, A. Bioenergetic Roles of Mitochondrial Fusion. Biochim. Biophys. Acta 2016, 1857, 1277–1283. [Google Scholar] [CrossRef]
- Shirakabe, A.; Zhai, P.; Ikeda, Y.; Saito, T.; Maejima, Y.; Hsu, C.-P.; Nomura, M.; Egashira, K.; Levine, B.; Sadoshima, J. Drp1-Dependent Mitochondrial Autophagy Plays a Protective Role Against Pressure Overload-Induced Mitochondrial Dysfunction and Heart Failure. Circulation 2016, 133, 1249–1263. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, P.; Bisetto, S.; Yoon, Y.; Chen, Q.; Sheu, S.-S.; Wang, W. A Novel Fission-Independent Role of Dynamin-Related Protein 1 in Cardiac Mitochondrial Respiration. Cardiovasc. Res. 2017, 113, 160–170. [Google Scholar] [CrossRef] [PubMed]
- Ji, W.; Hatch, A.L.; Merrill, R.A.; Strack, S.; Higgs, H.N. Actin Filaments Target the Oligomeric Maturation of the Dynamin GTPase Drp1 to Mitochondrial Fission Sites. eLife 2015, 4, e11553. [Google Scholar] [CrossRef] [PubMed]
- Ji, W.-K.; Chakrabarti, R.; Fan, X.; Schoenfeld, L.; Strack, S.; Higgs, H.N. Receptor-Mediated Drp1 Oligomerization on Endoplasmic Reticulum. J. Cell Biol. 2017, 216, 4123–4139. [Google Scholar] [CrossRef]
- Dorn, G.W. Mitochondrial Dynamics in Heart Disease. Biochim. Biophys. Acta 2013, 1833, 233–241. [Google Scholar] [CrossRef]
- Sharma, V.K.; Ramesh, V.; Franzini-Armstrong, C.; Sheu, S.S. Transport of Ca2+ from Sarcoplasmic Reticulum to Mitochondria in Rat Ventricular Myocytes. J. Bioenerg. Biomembr. 2000, 32, 97–104. [Google Scholar] [CrossRef]
- Glancy, B.; Balaban, R.S. Role of Mitochondrial Ca2+ in the Regulation of Cellular Energetics. Biochemistry 2012, 51, 2959–2973. [Google Scholar] [CrossRef]
- Luongo, T.S.; Lambert, J.P.; Yuan, A.; Zhang, X.; Gross, P.; Song, J.; Shanmughapriya, S.; Gao, E.; Jain, M.; Houser, S.R.; et al. The Mitochondrial Calcium Uniporter Matches Energetic Supply with Cardiac Workload during Stress and Modulates Permeability Transition. Cell Rep. 2015, 12, 23–34. [Google Scholar] [CrossRef]
- Rasmussen, T.P.; Wu, Y.; Joiner, M.A.; Koval, O.M.; Wilson, N.R.; Luczak, E.D.; Wang, Q.; Chen, B.; Gao, Z.; Zhu, Z.; et al. Inhibition of MCU Forces Extramitochondrial Adaptations Governing Physiological and Pathological Stress Responses in Heart. Proc. Natl. Acad. Sci. USA 2015, 112, 9129–9134. [Google Scholar] [CrossRef]
- Wang, P.; Fernandez-Sanz, C.; Wang, W.; Sheu, S.-S. Why Don’t Mice Lacking the Mitochondrial Ca2+ Uniporter Experience an Energy Crisis? J. Physiol. 2018, 598, 1307–1326. [Google Scholar] [CrossRef]
- Hom, J.; Yu, T.; Yoon, Y.; Porter, G.; Sheu, S.-S. Regulation of Mitochondrial Fission by Intracellular Ca2+ in Rat Ventricular Myocytes. Biochim. Biophys. Acta 2010, 1797, 913–921. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, Y.; Shirakabe, A.; Maejima, Y.; Zhai, P.; Sciarretta, S.; Toli, J.; Nomura, M.; Mihara, K.; Egashira, K.; Ohishi, M.; et al. Endogenous Drp1 Mediates Mitochondrial Autophagy and Protects the Heart against Energy Stress. Circ. Res. 2015, 116, 264–278. [Google Scholar] [CrossRef] [PubMed]
- Ishihara, N.; Nomura, M.; Jofuku, A.; Kato, H.; Suzuki, S.O.; Masuda, K.; Otera, H.; Nakanishi, Y.; Nonaka, I.; Goto, Y.-I.; et al. Mitochondrial Fission Factor Drp1 Is Essential for Embryonic Development and Synapse Formation in Mice. Nat. Cell Biol. 2009, 11, 958–966. [Google Scholar] [CrossRef] [PubMed]
- De La Fuente, S.; Fernandez-Sanz, C.; Vail, C.; Agra, E.J.; Holmstrom, K.; Sun, J.; Mishra, J.; Williams, D.; Finkel, T.; Murphy, E.; et al. Strategic Positioning and Biased Activity of the Mitochondrial Calcium Uniporter in Cardiac Muscle. J. Biol. Chem. 2016, 291, 23343–23362. [Google Scholar] [CrossRef]
- O’Connell, T.D.; Rodrigo, M.C.; Simpson, P.C. Isolation and Culture of Adult Mouse Cardiac Myocytes. Methods Mol. Biol. 2007, 357, 271–296. [Google Scholar] [CrossRef]
- Fernandez-Sanz, C.; Ruiz-Meana, M.; Miro-Casas, E.; Nuñez, E.; Castellano, J.; Loureiro, M.; Barba, I.; Poncelas, M.; Rodriguez-Sinovas, A.; Vázquez, J.; et al. Defective Sarcoplasmic Reticulum-Mitochondria Calcium Exchange in Aged Mouse Myocardium. Cell Death Dis. 2014, 5, e1573. [Google Scholar] [CrossRef]
- De La Fuente, S.; Lambert, J.P.; Nichtova, Z.; Fernandez Sanz, C.; Elrod, J.W.; Sheu, S.-S.; Csordás, G. Spatial Separation of Mitochondrial Calcium Uptake and Extrusion for Energy-Efficient Mitochondrial Calcium Signaling in the Heart. Cell Rep. 2018, 24, 3099–3107.e4. [Google Scholar] [CrossRef]
- Sánchez, J.A. Role of Connexin 43 in Ischemia-Reperfusion Injury; Universitat Autònoma de Barcelona: Barcelona, Spain, 2013; ISBN 978-84-490-3678-1. [Google Scholar]
- Matsuoka, Y.; Srere, P.A. Kinetic Studies of Citrate Synthase from Rat Kidney and Rat Brain. J. Biol. Chem. 1973, 248, 8022–8030. [Google Scholar] [CrossRef] [PubMed]
- Segretain, D.; Rambourg, A.; Clermont, Y. Three Dimensional Arrangement of Mitochondria and Endoplasmic Reticulum in the Heart Muscle Fiber of the Rat. Anat. Rec. 1981, 200, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Csordás, G.; Jowdy, C.; Schneider, T.G.; Csordás, N.; Wang, W.; Liu, Y.; Kohlhaas, M.; Meiser, M.; Bergem, S.; et al. Mitofusin 2-Containing Mitochondrial-Reticular Microdomains Direct Rapid Cardiomyocyte Bioenergetic Responses via Interorganelle Ca2+ Crosstalk. Circ. Res. 2012, 111, 863–875. [Google Scholar] [CrossRef] [PubMed]
- Dorn, G.W. Mitofusins as Mitochondrial Anchors and Tethers. J. Mol. Cell Cardiol. 2020, 142, 146–153. [Google Scholar] [CrossRef]
- Naon, D.; Scorrano, L. At the Right Distance: ER-Mitochondria Juxtaposition in Cell Life and Death. Biochim. Biophys. Acta 2014, 1843, 2184–2194. [Google Scholar] [CrossRef]
- Friedman, J.R.; Lackner, L.L.; West, M.; DiBenedetto, J.R.; Nunnari, J.; Voeltz, G.K. ER Tubules Mark Sites of Mitochondrial Division. Science 2011, 334, 358–362. [Google Scholar] [CrossRef]
- Wieckowski, M.R.; Giorgi, C.; Lebiedzinska, M.; Duszynski, J.; Pinton, P. Isolation of Mitochondria-Associated Membranes and Mitochondria from Animal Tissues and Cells. Nat. Protoc. 2009, 4, 1582–1590. [Google Scholar] [CrossRef]
- Macdonald, P.J.; Stepanyants, N.; Mehrotra, N.; Mears, J.A.; Qi, X.; Sesaki, H.; Ramachandran, R. A Dimeric Equilibrium Intermediate Nucleates Drp1 Reassembly on Mitochondrial Membranes for Fission. Mol. Biol. Cell 2014, 25, 1905–1915. [Google Scholar] [CrossRef]
- Kalia, R.; Wang, R.Y.-R.; Yusuf, A.; Thomas, P.V.; Agard, D.A.; Shaw, J.M.; Frost, A. Structural Basis of Mitochondrial Receptor Binding and Constriction by DRP1. Nature 2018, 558, 401–405. [Google Scholar] [CrossRef]
- Fekkes, P.; Shepard, K.A.; Yaffe, M.P. Gag3p, an Outer Membrane Protein Required for Fission of Mitochondrial Tubules. J. Cell Biol. 2000, 151, 333–340. [Google Scholar] [CrossRef]
- Mozdy, A.D.; McCaffery, J.M.; Shaw, J.M. Dnm1p GTPase-Mediated Mitochondrial Fission Is a Multi-Step Process Requiring the Novel Integral Membrane Component Fis1p. J. Cell Biol. 2000, 151, 367–380. [Google Scholar] [CrossRef]
- Tieu, Q.; Nunnari, J. Mdv1p Is a WD Repeat Protein That Interacts with the Dynamin-Related GTPase, Dnm1p, to Trigger Mitochondrial Division. J. Cell Biol. 2000, 151, 353–366. [Google Scholar] [CrossRef]
- van der Bliek, A.M. A Mitochondrial Division Apparatus Takes Shape. J. Cell Biol. 2000, 151, f1–f4. [Google Scholar] [CrossRef]
- Hatch, A.L.; Ji, W.-K.; Merrill, R.A.; Strack, S.; Higgs, H.N. Actin Filaments as Dynamic Reservoirs for Drp1 Recruitment. Mol. Biol. Cell 2016, 27, 3109–3121. [Google Scholar] [CrossRef] [PubMed]
- Wales, P.; Schuberth, C.E.; Aufschnaiter, R.; Fels, J.; García-Aguilar, I.; Janning, A.; Dlugos, C.P.; Schäfer-Herte, M.; Klingner, C.; Wälte, M.; et al. Calcium-Mediated Actin Reset (CaAR) Mediates Acute Cell Adaptations. eLife 2016, 5, e19850. [Google Scholar] [CrossRef] [PubMed]
- Davidson, S.M.; Duchen, M.R. Calcium Microdomains and Oxidative Stress. Cell Calcium 2006, 40, 561–574. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Meana, M.; Fernandez-Sanz, C.; Garcia-Dorado, D. The SR-Mitochondria Interaction: A New Player in Cardiac Pathophysiology. Cardiovasc. Res. 2010, 88, 30–39. [Google Scholar] [CrossRef]
- Kaludercic, N.; Deshwal, S.; Di Lisa, F. Reactive Oxygen Species and Redox Compartmentalization. Front. Physiol. 2014, 5, 285. [Google Scholar] [CrossRef]
- Vance, J.E. Phospholipid Synthesis and Transport in Mammalian Cells. Traffic 2015, 16, 1–18. [Google Scholar] [CrossRef]
- Boyman, L.; Karbowski, M.; Lederer, W.J. Regulation of Mitochondrial ATP Production: Ca2+ Signaling and Quality Control. Trends Mol. Med. 2020, 26, 21–39. [Google Scholar] [CrossRef]
- Wang, W.; Fernandez-Sanz, C.; Sheu, S.-S. Regulation of Mitochondrial Bioenergetics by the Non-Canonical Roles of Mitochondrial Dynamics Proteins in the Heart. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 1991–2001. [Google Scholar] [CrossRef]
- Yoon, Y.; Pitts, K.R.; Dahan, S.; McNiven, M.A. A Novel Dynamin-like Protein Associates with Cytoplasmic Vesicles and Tubules of the Endoplasmic Reticulum in Mammalian Cells. J. Cell Biol. 1998, 140, 779–793. [Google Scholar] [CrossRef] [PubMed]
- Prudent, J.; McBride, H.M. Mitochondrial Dynamics: ER Actin Tightens the Drp1 Noose. Curr. Biol. 2016, 26, R207–R209. [Google Scholar] [CrossRef] [PubMed]
- Adachi, Y.; Kato, T.; Yamada, T.; Murata, D.; Arai, K.; Stahelin, R.V.; Chan, D.C.; Iijima, M.; Sesaki, H. Drp1 Tubulates the ER in a GTPase-Independent Manner. Mol. Cell 2020, 80, 624–632.e6. [Google Scholar] [CrossRef] [PubMed]
- Francy, C.A.; Clinton, R.W.; Fröhlich, C.; Murphy, C.; Mears, J.A. Cryo-EM Studies of Drp1 Reveal Cardiolipin Interactions That Activate the Helical Oligomer. Sci. Rep. 2017, 7, 10744. [Google Scholar] [CrossRef]
- Takeshita, N.; Evangelinos, M.; Zhou, L.; Serizawa, T.; Somera-Fajardo, R.A.; Lu, L.; Takaya, N.; Nienhaus, G.U.; Fischer, R. Pulses of Ca2+ Coordinate Actin Assembly and Exocytosis for Stepwise Cell Extension. Proc. Natl. Acad. Sci. USA 2017, 114, 5701–5706. [Google Scholar] [CrossRef]
- Song, M.; Franco, A.; Fleischer, J.A.; Zhang, L.; Dorn, G.W. Abrogating Mitochondrial Dynamics in Mouse Hearts Accelerates Mitochondrial Senescence. Cell Metab. 2017, 26, 872–883.e5. [Google Scholar] [CrossRef]
- Piao, L.; Fang, Y.-H.; Fisher, M.; Hamanaka, R.B.; Ousta, A.; Wu, R.; Mutlu, G.M.; Garcia, A.J., III; Archer, S.L.; Sharp, W.W. Dynamin-Related Protein 1 Is a Critical Regulator of Mitochondrial Calcium Homeostasis during Myocardial Ischemia/Reperfusion Injury. FASEB J. 2024, 38, e23379. [Google Scholar] [CrossRef]
- Wu, D.; Hu, Q.; Li, H.; Yin, Y.; Wang, P.; Wang, W. Drp1 Knockdown Aggravates Obesity-Induced Cardiac Dysfunction and Remodeling. Mitochondrion 2025, 83, 102023. [Google Scholar] [CrossRef]
- Wittig, I.; Malacarne, P.F. Complexome Profiling: Assembly and Remodeling of Protein Complexes. Int. J. Mol. Sci. 2021, 22, 7809. [Google Scholar] [CrossRef]
- Kohlhaas, M.; Maack, C. Calcium Release Microdomains and Mitochondria. Cardiovasc. Res. 2013, 98, 259–268. [Google Scholar] [CrossRef]
- Vance, J.E. MAM (Mitochondria-Associated Membranes) in Mammalian Cells: Lipids and Beyond. Biochim. Biophys. Acta 2014, 1841, 595–609. [Google Scholar] [CrossRef]
- Maack, C.; O’Rourke, B. Excitation-Contraction Coupling and Mitochondrial Energetics. Basic Res. Cardiol. 2007, 102, 369–392. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernandez-Sanz, C.; De la Fuente, S.; Nichtova, Z.; Federico, M.; Duvezin-Caubet, S.; Lanvermann, S.; Tsai, H.-Y.; Xin, Y.; Csordas, G.; Wang, W.; et al. Highly Oligomeric DRP1 Strategic Positioning at Mitochondria–Sarcoplasmic Reticulum Contacts in Adult Murine Heart Through ACTIN Anchoring. Cells 2025, 14, 1259. https://doi.org/10.3390/cells14161259
Fernandez-Sanz C, De la Fuente S, Nichtova Z, Federico M, Duvezin-Caubet S, Lanvermann S, Tsai H-Y, Xin Y, Csordas G, Wang W, et al. Highly Oligomeric DRP1 Strategic Positioning at Mitochondria–Sarcoplasmic Reticulum Contacts in Adult Murine Heart Through ACTIN Anchoring. Cells. 2025; 14(16):1259. https://doi.org/10.3390/cells14161259
Chicago/Turabian StyleFernandez-Sanz, Celia, Sergio De la Fuente, Zuzana Nichtova, Marilen Federico, Stephane Duvezin-Caubet, Sebastian Lanvermann, Hui-Ying Tsai, Yanguo Xin, Gyorgy Csordas, Wang Wang, and et al. 2025. "Highly Oligomeric DRP1 Strategic Positioning at Mitochondria–Sarcoplasmic Reticulum Contacts in Adult Murine Heart Through ACTIN Anchoring" Cells 14, no. 16: 1259. https://doi.org/10.3390/cells14161259
APA StyleFernandez-Sanz, C., De la Fuente, S., Nichtova, Z., Federico, M., Duvezin-Caubet, S., Lanvermann, S., Tsai, H.-Y., Xin, Y., Csordas, G., Wang, W., Mourier, A., & Sheu, S.-S. (2025). Highly Oligomeric DRP1 Strategic Positioning at Mitochondria–Sarcoplasmic Reticulum Contacts in Adult Murine Heart Through ACTIN Anchoring. Cells, 14(16), 1259. https://doi.org/10.3390/cells14161259







