Whole RNA-Seq Analysis Reveals Longitudinal Proteostasis Network Responses to Photoreceptor Outer Segment Trafficking and Degradation in RPE Cells
Abstract
1. Introduction
2. Materials and Methods
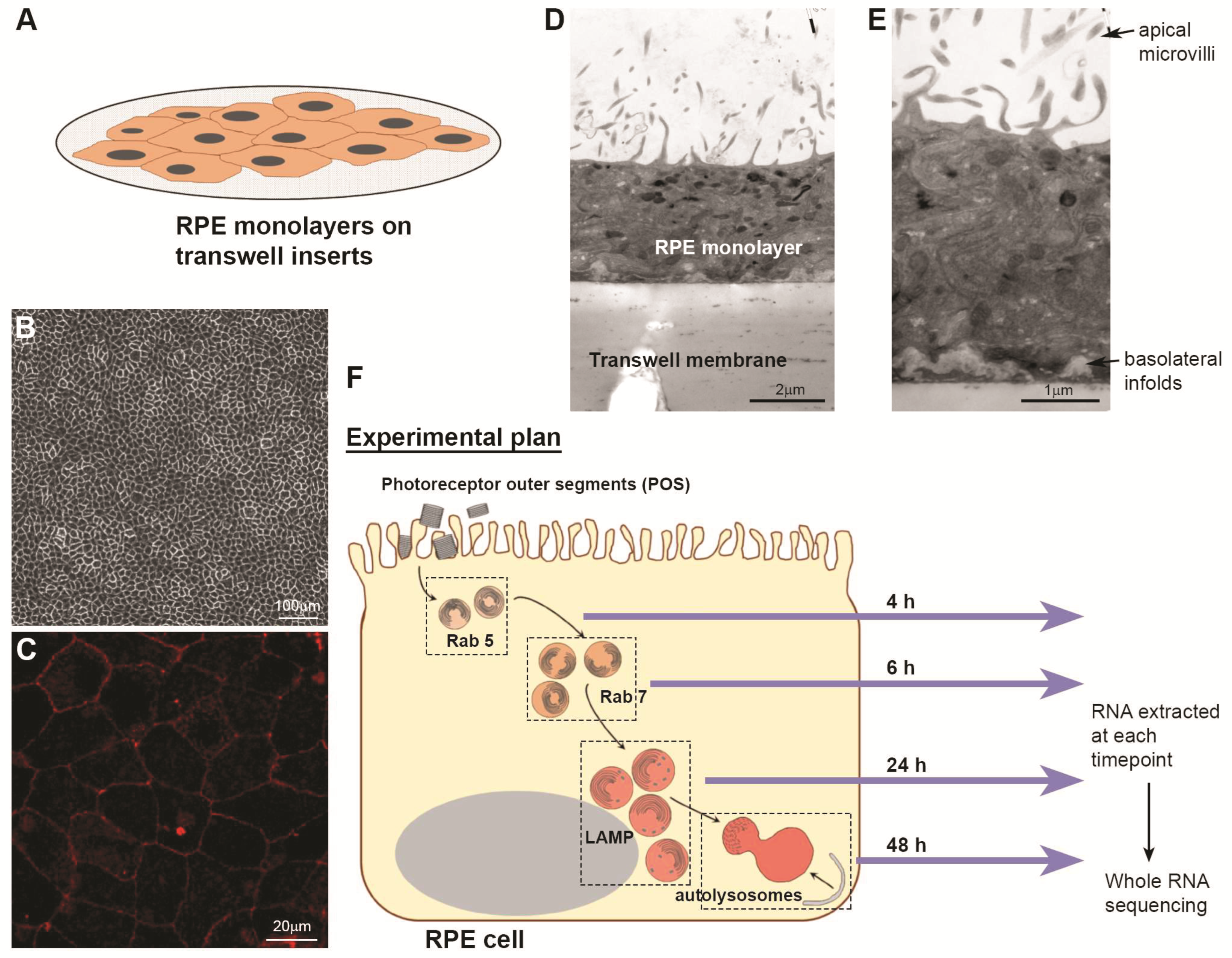
2.1. Culture of RPE Cells
2.2. Isolation and Quantification of Photoreceptor Outer Segments (POS)
2.3. Synchronised POS Feeding Assay
2.4. RNA Extraction and Quantification
2.5. RNA-Seq
2.6. Post Sequencing Analysis of Whole mRNA
2.7. Pathway Formation Using Cytoscape and Analysis Using DAVID
2.8. Statistical Analyses
3. Results
3.1. Whole RNA-Seq Analyses of Highly Differentiated Monolayers of the Human RPE Cell Line (ARPE-19) Showing the Expression of RPE-Specific Genes and Genes Linked with Retinopathies
3.2. Whole RNA-Seq Analyses of Genes in the RPE Proteostasis Network
3.3. Whole RNA Sequencing Reveals Transcriptomic Changes in the Proteostasis Network in Response to Photoreceptor Outer Segment Trafficking by RPE Cells
3.4. A Targeted Scrutiny of Genes Related to the Autophagy–Lysosomal Pathway Provides Additional Insights into POS-Induced Transcriptomic Changes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wong, W.L.; Su, X.; Li, X.; Cheung, C.M.; Klein, R.; Cheng, C.Y.; Wong, T.Y. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: A systematic review and meta-analysis. Lancet Glob. Health 2014, 2, e106–e116. [Google Scholar] [CrossRef]
- Watson, A.; Lako, M. Retinal organoids provide unique insights into molecular signatures of inherited retinal disease throughout retinogenesis. J. Anat. 2023, 243, 186–203. [Google Scholar] [CrossRef]
- Mustafi, D.; Bharathan, S.P.; Calderon, R.; Nagiel, A. Human Cellular Models for Retinal Disease: From Induced Pluripotent Stem Cells to Organoids. Retina 2022, 42, 1829–1835. [Google Scholar] [CrossRef] [PubMed]
- Christensen, D.R.G.; Brown, F.E.; Cree, A.J.; Ratnayaka, J.A.; Lotery, A.J. Sorsby fundus dystrophy—A review of pathology and disease mechanisms. Exp. Eye Res. 2017, 165, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Keeling, E.; Chatelet, D.S.; Tan, N.Y.T.; Khan, F.; Richards, R.; Thisainathan, T.; Goggin, P.; Page, A.; Tumbarello, D.A.; Lotery, A.J.; et al. 3D-Reconstructed Retinal Pigment Epithelial Cells Provide Insights into the Anatomy of the Outer Retina. Int. J. Mol. Sci. 2020, 21, 8408. [Google Scholar] [CrossRef]
- Volland, S.; Esteve-Rudd, J.; Hoo, J.; Yee, C.; Williams, D.S. A comparison of some organizational characteristics of the mouse central retina and the human macula. PLoS ONE 2015, 10, e0125631. [Google Scholar] [CrossRef] [PubMed]
- Keeling, E.; Lotery, A.J.; Tumbarello, D.A.; Ratnayaka, J.A. Impaired Cargo Clearance in the Retinal Pigment Epithelium (RPE) Underlies Irreversible Blinding Diseases. Cells 2018, 7, 16. [Google Scholar] [CrossRef]
- Lieffrig, S.A.; Gyimesi, G.; Mao, Y.; Finnemann, S.C. Clearance phagocytosis by the retinal pigment epithelial during photoreceptor outer segment renewal: Molecular mechanisms and relation to retinal inflammation. Immunol. Rev. 2023, 319, 81–99. [Google Scholar] [CrossRef]
- Kim, H.J.; Sparrow, J.R. Bisretinoid phospholipid and vitamin A aldehyde: Shining a light. J. Lipid Res. 2021, 62, 100042. [Google Scholar] [CrossRef]
- Gliem, M.; Muller, P.L.; Finger, R.P.; McGuinness, M.B.; Holz, F.G.; Charbel Issa, P. Quantitative Fundus Autofluorescence in Early and Intermediate Age-Related Macular Degeneration. JAMA Ophthalmol. 2016, 134, 817–824. [Google Scholar] [CrossRef]
- Holz, F.G.; Sadda, S.R.; Staurenghi, G.; Lindner, M.; Bird, A.C.; Blodi, B.A.; Bottoni, F.; Chakravarthy, U.; Chew, E.Y.; Csaky, K.; et al. Imaging Protocols in Clinical Studies in Advanced Age-Related Macular Degeneration: Recommendations from Classification of Atrophy Consensus Meetings. Ophthalmology 2017, 124, 464–478. [Google Scholar] [CrossRef] [PubMed]
- Keeling, E.; Chatelet, D.S.; Johnston, D.A.; Page, A.; Tumbarello, D.A.; Lotery, A.J.; Ratnayaka, J.A. Oxidative Stress and Dysfunctional Intracellular Traffic Linked to an Unhealthy Diet Results in Impaired Cargo Transport in the Retinal Pigment Epithelium (RPE). Mol. Nutr. Food Res. 2019, 63, e1800951. [Google Scholar] [CrossRef] [PubMed]
- Keeling, E.; Culling, A.J.; Johnston, D.A.; Chatelet, D.S.; Page, A.; Tumbarello, D.A.; Lotery, A.J.; Ratnayaka, J.A. An In-Vitro Cell Model of Intracellular Protein Aggregation Provides Insights into RPE Stress Associated with Retinopathy. Int. J. Mol. Sci. 2020, 21, 6647. [Google Scholar] [CrossRef] [PubMed]
- Lynn, S.A.; Johnston, D.A.; Scott, J.A.; Munday, R.; Desai, R.S.; Keeling, E.; Weaterton, R.; Simpson, A.; Davis, D.; Freeman, T.; et al. Oligomeric Aβ(1–42) Induces an AMD-Like Phenotype and Accumulates in Lysosomes to Impair RPE Function. Cells 2021, 10, 413. [Google Scholar] [CrossRef]
- Dunn, K.C.; Aotaki-Keen, A.E.; Putkey, F.R.; Hjelmeland, L.M. ARPE-19, a human retinal pigment epithelial cell line with differentiated properties. Exp. Eye Res. 1996, 62, 155–169. [Google Scholar] [CrossRef]
- Lynn, S.A.; Keeling, E.; Dewing, J.M.; Johnston, D.A.; Page, A.; Cree, A.J.; Tumbarello, D.A.; Newman, T.A.; Lotery, A.J.; Ratnayaka, J.A. A convenient protocol for establishing a human cell culture model of the outer retina. F1000Research 2018, 7, 1107. [Google Scholar] [CrossRef]
- Schraermeyer, U.; Enzmann, V.; Kohen, L.; Addicks, K.; Wiedemann, P.; Heimann, K. Porcine iris pigment epithelial cells can take up retinal outer segments. Exp. Eye Res. 1997, 65, 277–287. [Google Scholar] [CrossRef]
- Krohne, T.U.; Stratmann, N.K.; Kopitz, J.; Holz, F.G. Effects of lipid peroxidation products on lipofuscinogenesis and autophagy in human retinal pigment epithelial cells. Exp. Eye Res. 2010, 90, 465–471. [Google Scholar] [CrossRef]
- Hall, M.O.; Abrams, T. Kinetic studies of rod outer segment binding and ingestion by cultured rat RPE cells. Exp. Eye Res. 1987, 45, 907–922. [Google Scholar] [CrossRef]
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Chen, S. Ultrafast one-pass FASTQ data preprocessing, quality control, and deduplication using fastp. iMETA 2023, 2, e107. [Google Scholar] [CrossRef] [PubMed]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Tzioutziou, N.A.; Stephen, G.; Milne, I.; Calixto, C.P.; Waugh, R.; Brown, J.W.S.; Zhang, R. 3D RNA-seq: A powerful and flexible tool for rapid and accurate differential expression and alternative splicing analysis of RNA-seq data for biologists. RNA Biol. 2021, 18, 1574–1587. [Google Scholar] [CrossRef] [PubMed]
- Soneson, C.; Love, M.I.; Robinson, M.D. Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Research 2015, 4, 1521. [Google Scholar] [CrossRef]
- Deyneko, I.V.; Mustafaev, O.N.; Tyurin, A.; Zhukova, K.V.; Varzari, A.; Goldenkova-Pavlova, I.V. Modeling and cleaning RNA-seq data significantly improve detection of differentially expressed genes. BMC Bioinform. 2022, 23, 488. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Damgaard, R.B. The ubiquitin system: From cell signalling to disease biology and new therapeutic opportunities. Cell Death Differ. 2021, 28, 423–426. [Google Scholar] [CrossRef]
- Potjewyd, F.M.; Axtman, A.D. Exploration of Aberrant E3 Ligases Implicated in Alzheimer’s Disease and Development of Chemical Tools to Modulate Their Function. Front. Cell. Neurosci. 2021, 15, 768655. [Google Scholar] [CrossRef]
- Rosenzweig, R.; Nillegoda, N.B.; Mayer, M.P.; Bukau, B. The Hsp70 chaperone network. Nat. Rev. Mol. Cell Biol. 2019, 20, 665–680. [Google Scholar] [CrossRef]
- Zhan, S.; Wang, T.; Ge, W. Multiple functions of the E3 ubiquitin ligase CHIP in immunity. Int. Rev. Immunol. 2017, 36, 300–312. [Google Scholar] [CrossRef] [PubMed]
- Schopf, F.H.; Biebl, M.M.; Buchner, J. The HSP90 chaperone machinery. Nat. Rev. Mol. Cell Biol. 2017, 18, 345–360. [Google Scholar] [CrossRef] [PubMed]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef] [PubMed]
- Lynn, S.A.; Ward, G.; Keeling, E.; Scott, J.A.; Cree, A.J.; Johnston, D.A.; Page, A.; Cuan-Urquizo, E.; Bhaskar, A.; Grossel, M.C.; et al. Ex-vivo models of the Retinal Pigment Epithelium (RPE) in long-term culture faithfully recapitulate key structural and physiological features of native RPE. Tissue Cell 2017, 49, 447–460. [Google Scholar] [CrossRef]
- Ahmado, A.; Carr, A.J.; Vugler, A.A.; Semo, M.; Gias, C.; Lawrence, J.M.; Chen, L.L.; Chen, F.K.; Turowski, P.; da Cruz, L.; et al. Induction of differentiation by pyruvate and DMEM in the human retinal pigment epithelium cell line ARPE-19. Investig. Ophthalmol. Vis. Sci. 2011, 52, 7148–7159. [Google Scholar] [CrossRef]
- Amin, S.; Chong, N.H.; Bailey, T.A.; Zhang, J.; Knupp, C.; Cheetham, M.E.; Greenwood, J.; Luthert, P.J. Modulation of Sub-RPE deposits in vitro: A potential model for age-related macular degeneration. Investig. Ophthalmol. Vis. Sci. 2004, 45, 1281–1288. [Google Scholar] [CrossRef]
- Johnson, L.V.; Forest, D.L.; Banna, C.D.; Radeke, C.M.; Maloney, M.A.; Hu, J.; Spencer, C.N.; Walker, A.M.; Tsie, M.S.; Bok, D.; et al. Cell culture model that mimics drusen formation and triggers complement activation associated with age-related macular degeneration. Proc. Natl. Acad. Sci. USA 2011, 108, 18277–18282. [Google Scholar] [CrossRef]
- Shaw, E.M.; Tate, A.J.; Periasamy, R.; Lipinski, D.M. Characterization of drusen formation in a primary porcine tissue culture model of dry AMD. Mol. Ther. Methods Clin. Dev. 2024, 32, 101331. [Google Scholar] [CrossRef]
- Samuel, W.; Jaworski, C.; Postnikova, O.A.; Kutty, R.K.; Duncan, T.; Tan, L.X.; Poliakov, E.; Lakkaraju, A.; Redmond, T.M. Appropriately differentiated ARPE-19 cells regain phenotype and gene expression profiles similar to those of native RPE cells. Mol. Vis. 2017, 23, 60–89. [Google Scholar]
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition). Autophagy 2021, 17, 1–382. [Google Scholar] [CrossRef]
- Feeney-Burns, L.; Hilderbrand, E.S.; Eldridge, S. Aging human RPE: Morphometric analysis of macular, equatorial, and peripheral cells. Investig. Ophthalmol. Vis. Sci. 1984, 25, 195–200. [Google Scholar]
- Feeney-Burns, L.; Gao, C.L.; Tidwell, M. Lysosomal enzyme cytochemistry of human RPE, Bruch’s membrane and drusen. Investig. Ophthalmol. Vis. Sci. 1987, 28, 1138–1147. [Google Scholar]
- Mazzoni, F.; Safa, H.; Finnemann, S.C. Understanding photoreceptor outer segment phagocytosis: Use and utility of RPE cells in culture. Exp. Eye Res. 2014, 126, 51–60. [Google Scholar] [CrossRef]
- Bird, A.C.; Phillips, R.L.; Hageman, G.S. Geographic atrophy: A histopathological assessment. JAMA Ophthalmol. 2014, 132, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Sohn, E.H.; Flamme-Wiese, M.J.; Whitmore, S.S.; Workalemahu, G.; Marneros, A.G.; Boese, E.A.; Kwon, Y.H.; Wang, K.; Abramoff, M.D.; Tucker, B.A.; et al. Choriocapillaris Degeneration in Geographic Atrophy. Am. J. Pathol. 2019, 189, 1473–1480. [Google Scholar] [CrossRef] [PubMed]
- Sanie-Jahromi, F.; Nowroozzadeh, M.H. RPE based gene and cell therapy for inherited retinal diseases: A review. Exp. Eye Res. 2022, 217, 108961. [Google Scholar] [CrossRef]
- Al-Hussaini, H.; Kam, J.H.; Vugler, A.; Semo, M.; Jeffery, G. Mature retinal pigment epithelium cells are retained in the cell cycle and proliferate in vivo. Mol. Vis. 2008, 14, 1784–1791. [Google Scholar]
- Zhou, M.; Geathers, J.S.; Grillo, S.L.; Weber, S.R.; Wang, W.; Zhao, Y.; Sundstrom, J.M. Role of Epithelial-Mesenchymal Transition in Retinal Pigment Epithelium Dysfunction. Front. Cell Dev. Biol. 2020, 8, 501. [Google Scholar] [CrossRef]
- Sarks, J.P.; Sarks, S.H.; Killingsworth, M.C. Evolution of geographic atrophy of the retinal pigment epithelium. Eye 1988, 2 Pt 5, 552–577. [Google Scholar] [CrossRef]
- Kaushik, S.; Cuervo, A.M. Proteostasis and aging. Nat. Med. 2015, 21, 1406–1415. [Google Scholar] [CrossRef]
- Hipp, M.S.; Kasturi, P.; Hartl, F.U. The proteostasis network and its decline in ageing. Nat. Rev. Mol. Cell Biol. 2019, 20, 421–435. [Google Scholar] [CrossRef]
- Hyttinen, J.M.; Amadio, M.; Viiri, J.; Pascale, A.; Salminen, A.; Kaarniranta, K. Clearance of misfolded and aggregated proteins by aggrephagy and implications for aggregation diseases. Ageing Res. Rev. 2014, 18, 16–28. [Google Scholar] [CrossRef] [PubMed]
- Ferrington, D.A.; Sinha, D.; Kaarniranta, K. Defects in retinal pigment epithelial cell proteolysis and the pathology associated with age-related macular degeneration. Prog. Retin. Eye Res. 2016, 51, 69–89. [Google Scholar] [CrossRef] [PubMed]
- Paraoan, L.; Sharif, U.; Carlsson, E.; Supharattanasitthi, W.; Mahmud, N.M.; Kamalden, T.A.; Hiscott, P.; Jackson, M.; Grierson, I. Secretory proteostasis of the retinal pigmented epithelium: Impairment links to age-related macular degeneration. Prog. Retin. Eye Res. 2020, 79, 100859. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.T.; Yoon, B.W.; Seo, J.H. Comparison of risk allele frequencies of single nucleotide polymorphisms associated with age-related macular degeneration in different ethnic groups. BMC Ophthalmol. 2021, 21, 97. [Google Scholar] [CrossRef]
- Zhou, X.; Zhang, J.; Ding, Y.; Huang, H.; Li, Y.; Chen, W. Predicting late-stage age-related macular degeneration by integrating marginally weak SNPs in GWA studies. Front. Genet. 2023, 14, 1075824. [Google Scholar] [CrossRef]
- Li, W.; Wang, Y.; Zhu, L.; Du, S.; Mao, J.; Wang, Y.; Wang, S.; Bo, Q.; Tu, Y.; Yi, Q. The P300/XBP1s/Herpud1 axis promotes macrophage M2 polarization and the development of choroidal neovascularization. J. Cell Mol. Med. 2021, 25, 6709–6720. [Google Scholar] [CrossRef]
- Kaye, R.A.; Patasova, K.; Patel, P.J.; Hysi, P.; Lotery, A.J. Macular thickness varies with age-related macular degeneration genetic risk variants in the UK Biobank cohort. Sci. Rep. 2021, 11, 23255. [Google Scholar] [CrossRef]
- Altay, L.; Scholz, P.; Schick, T.; Felsch, M.; Hoyng, C.B.; den Hollander, A.I.; Langmann, T.; Fauser, S. Association of Hyperreflective Foci Present in Early Forms of Age-Related Macular Degeneration With Known Age-Related Macular Degeneration Risk Polymorphisms. Investig. Ophthalmol. Vis. Sci. 2016, 57, 4315–4320. [Google Scholar] [CrossRef]
- Holtes, L.K.; de Bruijn, S.E.; Cremers, F.P.M.; Roosing, S. Dual inheritance patterns: A spectrum of non-syndromic inherited retinal disease phenotypes with varying molecular mechanisms. Prog. Retin. Eye Res. 2025, 104, 101308. [Google Scholar] [CrossRef]
- Priglinger, C.S.; Gerhardt, M.J.; Priglinger, S.G.; Schaumberger, M.; Neuhann, T.M.; Bolz, H.J.; Mehraein, Y.; Rudolph, G. Phenotypic and Genetic Spectrum in 309 Consecutive Pediatric Patients with Inherited Retinal Disease. Int. J. Mol. Sci. 2024, 25, 12259. [Google Scholar] [CrossRef]
- Singh, S.R.; Meyer-Jens, M.; Alizoti, E.; Bacon, W.C.; Davis, G.; Osinska, H.; Gulick, J.; Reischmann-Düsener, S.; Orthey, E.; McLendon, P.M.; et al. A high-throughput screening identifies ZNF418 as a novel regulator of the ubiquitin-proteasome system and autophagy-lysosomal pathway. Autophagy 2021, 17, 3124–3139. [Google Scholar] [CrossRef] [PubMed]
- Castellano, E.; Santos, E. Functional specificity of ras isoforms: So similar but so different. Genes. Cancer 2011, 2, 216–231. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.Y.; Zoncu, R. The lysosome as a command-and-control center for cellular metabolism. J. Cell Biol. 2016, 214, 653–664. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.N.; Yu, C.G.; Li, R.; Xi, Y.; Jian, Y.J.; Xu, N.; Chen, S. The nonautophagic functions of autophagy-related proteins. Autophagy 2024, 20, 720–734. [Google Scholar] [CrossRef]
- Noda, N.N.; Mizushima, N. Atg101: Not Just an Accessory Subunit in the Autophagy-initiation Complex. Cell Struct. Funct. 2016, 41, 13–20. [Google Scholar] [CrossRef]
- Zhu, X.; Wang, K.; Zhou, F.; Zhu, L. Paeoniflorin attenuates atRAL-induced oxidative stress, mitochondrial dysfunction and endoplasmic reticulum stress in retinal pigment epithelial cells via triggering Ca(2+)/CaMKII-dependent activation of AMPK. Arch. Pharm. Res. 2018, 41, 1009–1018. [Google Scholar] [CrossRef]
- Yang, C.; Zhao, X.; Zhou, W.; Li, Q.; Lazarovici, P.; Zheng, W. Artemisinin conferred cytoprotection to human retinal pigment epithelial cells exposed to amiodarone-induced oxidative insult by activating the CaMKK2/AMPK/Nrf2 pathway. J. Transl. Med. 2024, 22, 844. [Google Scholar] [CrossRef]
- Proikas-Cezanne, T.; Takacs, Z.; Dönnes, P.; Kohlbacher, O. WIPI proteins: Essential PtdIns3P effectors at the nascent autophagosome. J. Cell Sci. 2015, 128, 207–217. [Google Scholar] [CrossRef]
- Sun, Q.; Westphal, W.; Wong, K.N.; Tan, I.; Zhong, Q. Rubicon controls endosome maturation as a Rab7 effector. Proc. Natl. Acad. Sci. USA 2010, 107, 19338–19343. [Google Scholar] [CrossRef]
- Sun, Q.; Zhang, J.; Fan, W.; Wong, K.N.; Ding, X.; Chen, S.; Zhong, Q. The RUN domain of rubicon is important for hVps34 binding, lipid kinase inhibition, and autophagy suppression. J. Biol. Chem. 2011, 286, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Muniz-Feliciano, L.; Doggett, T.A.; Zhou, Z.; Ferguson, T.A. RUBCN/rubicon and EGFR regulate lysosomal degradative processes in the retinal pigment epithelium (RPE) of the eye. Autophagy 2017, 13, 2072–2085. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, T.; Han, C.; He, D.; Liu, H.; An, H.; Cai, Z.; Cao, X. Lysosome-associated small Rab GTPase Rab7b negatively regulates TLR4 signaling in macrophages by promoting lysosomal degradation of TLR4. Blood 2007, 110, 962–971. [Google Scholar] [CrossRef] [PubMed]
- Gong, Q.; Luo, D.; Wang, H.; Xu, X.; Fan, Y.; Zheng, Z.; Qian, T. Inhibiting autophagy by miR-19a-3p/PTEN regulation protected retinal pigment epithelial cells from hyperglycemic damage. Biochim. Biophys. Acta Mol. Cell Res. 2023, 1870, 119530. [Google Scholar] [CrossRef]
- Nishimura, T.; Tooze, S.A. Emerging roles of ATG proteins and membrane lipids in autophagosome formation. Cell Discov. 2020, 6, 32. [Google Scholar] [CrossRef]
- Diao, J.; Liu, R.; Rong, Y.; Zhao, M.; Zhang, J.; Lai, Y.; Zhou, Q.; Wilz, L.M.; Li, J.; Vivona, S.; et al. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature 2015, 520, 563–566. [Google Scholar] [CrossRef]
- Ragupathi, A.; Kim, C.; Jacinto, E. The mTORC2 signaling network: Targets and cross-talks. Biochem. J. 2024, 481, 45–91. [Google Scholar] [CrossRef]
- Conus, S.; Pop, C.; Snipas, S.J.; Salvesen, G.S.; Simon, H.U. Cathepsin D primes caspase-8 activation by multiple intra-chain proteolysis. J. Biol. Chem. 2012, 287, 21142–21151. [Google Scholar] [CrossRef]
- Yadati, T.; Houben, T.; Bitorina, A.; Shiri-Sverdlov, R. The Ins and Outs of Cathepsins: Physiological Function and Role in Disease Management. Cells 2020, 9, 1679. [Google Scholar] [CrossRef]
- Rakoczy, P.E.; Lai, C.M.; Baines, M.; Di Grandi, S.; Fitton, J.H.; Constable, I.J. Modulation of cathepsin D activity in retinal pigment epithelial cells. Biochem. J. 1997, 324 Pt 3, 935–940. [Google Scholar] [CrossRef]
- Rakoczy, P.E.; Sarks, S.H.; Daw, N.; Constable, I.J. Distribution of cathepsin D in human eyes with or without age-related maculopathy. Exp. Eye Res. 1999, 69, 367–374. [Google Scholar] [CrossRef]
- Boulton, M.; Moriarty, P.; Jarvis-Evans, J.; Marcyniuk, B. Regional variation and age-related changes of lysosomal enzymes in the human retinal pigment epithelium. Br. J. Ophthalmol. 1994, 78, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Ng, E.S.Y.; Hu, J.; Jiang, Z.; Radu, R.A. Impaired cathepsin D in retinal pigment epithelium cells mediates Stargardt disease pathogenesis. FASEB J. 2024, 38, e23720. [Google Scholar] [CrossRef] [PubMed]
- Pareek, G.; Kundu, M. Physiological functions of ULK1/2. J. Mol. Biol. 2024, 436, 168472. [Google Scholar] [CrossRef] [PubMed]
- Strappazzon, F.; Vietri-Rudan, M.; Campello, S.; Nazio, F.; Florenzano, F.; Fimia, G.M.; Piacentini, M.; Levine, B.; Cecconi, F. Mitochondrial BCL-2 inhibits AMBRA1-induced autophagy. EMBO J. 2011, 30, 1195–1208. [Google Scholar] [CrossRef]
- Li, M.; Hou, Y.; Wang, J.; Chen, X.; Shao, Z.M.; Yin, X.M. Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 substrates. J. Biol. Chem. 2011, 286, 7327–7338. [Google Scholar] [CrossRef]
- Di Bartolomeo, S.; Corazzari, M.; Nazio, F.; Oliverio, S.; Lisi, G.; Antonioli, M.; Pagliarini, V.; Matteoni, S.; Fuoco, C.; Giunta, L.; et al. The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J. Cell Biol. 2010, 191, 155–168. [Google Scholar] [CrossRef]
- Blasiak, J.; Pawlowska, E.; Szczepanska, J.; Kaarniranta, K. Interplay between Autophagy and the Ubiquitin-Proteasome System and Its Role in the Pathogenesis of Age-Related Macular Degeneration. Int. J. Mol. Sci. 2019, 20, 210. [Google Scholar] [CrossRef]
- Park, C.; Cuervo, A.M. Selective autophagy: Talking with the UPS. Cell Biochem. Biophys. 2013, 67, 3–13. [Google Scholar] [CrossRef]
- Ryhanen, T.; Hyttinen, J.M.; Kopitz, J.; Rilla, K.; Kuusisto, E.; Mannermaa, E.; Viiri, J.; Holmberg, C.I.; Immonen, I.; Meri, S.; et al. Crosstalk between Hsp70 molecular chaperone, lysosomes and proteasomes in autophagy-mediated proteolysis in human retinal pigment epithelial cells. J. Cell. Mol. Med. 2009, 13, 3616–3631. [Google Scholar] [CrossRef]
- Perl, K.; Ushakov, K.; Pozniak, Y.; Yizhar-Barnea, O.; Bhonker, Y.; Shivatzki, S.; Geiger, T.; Avraham, K.B.; Shamir, R. Reduced changes in protein compared to mRNA levels across non-proliferating tissues. BMC Genom. 2017, 18, 305. [Google Scholar] [CrossRef]
- Koussounadis, A.; Langdon, S.P.; Um, I.H.; Harrison, D.J.; Smith, V.A. Relationship between differentially expressed mRNA and mRNA-protein correlations in a xenograft model system. Sci. Rep. 2015, 5, 10775. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miller, R.D.; Mondon, I.; Ellis, C.; Muir, A.-M.; Turner, S.; Keeling, E.; Wai, H.A.; Chatelet, D.S.; Johnson, D.A.; Tumbarello, D.A.; et al. Whole RNA-Seq Analysis Reveals Longitudinal Proteostasis Network Responses to Photoreceptor Outer Segment Trafficking and Degradation in RPE Cells. Cells 2025, 14, 1166. https://doi.org/10.3390/cells14151166
Miller RD, Mondon I, Ellis C, Muir A-M, Turner S, Keeling E, Wai HA, Chatelet DS, Johnson DA, Tumbarello DA, et al. Whole RNA-Seq Analysis Reveals Longitudinal Proteostasis Network Responses to Photoreceptor Outer Segment Trafficking and Degradation in RPE Cells. Cells. 2025; 14(15):1166. https://doi.org/10.3390/cells14151166
Chicago/Turabian StyleMiller, Rebecca D., Isaac Mondon, Charles Ellis, Anna-Marie Muir, Stephanie Turner, Eloise Keeling, Htoo A. Wai, David S. Chatelet, David A. Johnson, David A. Tumbarello, and et al. 2025. "Whole RNA-Seq Analysis Reveals Longitudinal Proteostasis Network Responses to Photoreceptor Outer Segment Trafficking and Degradation in RPE Cells" Cells 14, no. 15: 1166. https://doi.org/10.3390/cells14151166
APA StyleMiller, R. D., Mondon, I., Ellis, C., Muir, A.-M., Turner, S., Keeling, E., Wai, H. A., Chatelet, D. S., Johnson, D. A., Tumbarello, D. A., Lotery, A. J., Baralle, D., & Ratnayaka, J. A. (2025). Whole RNA-Seq Analysis Reveals Longitudinal Proteostasis Network Responses to Photoreceptor Outer Segment Trafficking and Degradation in RPE Cells. Cells, 14(15), 1166. https://doi.org/10.3390/cells14151166






