Microproteins in Metabolism
Abstract
1. Introduction
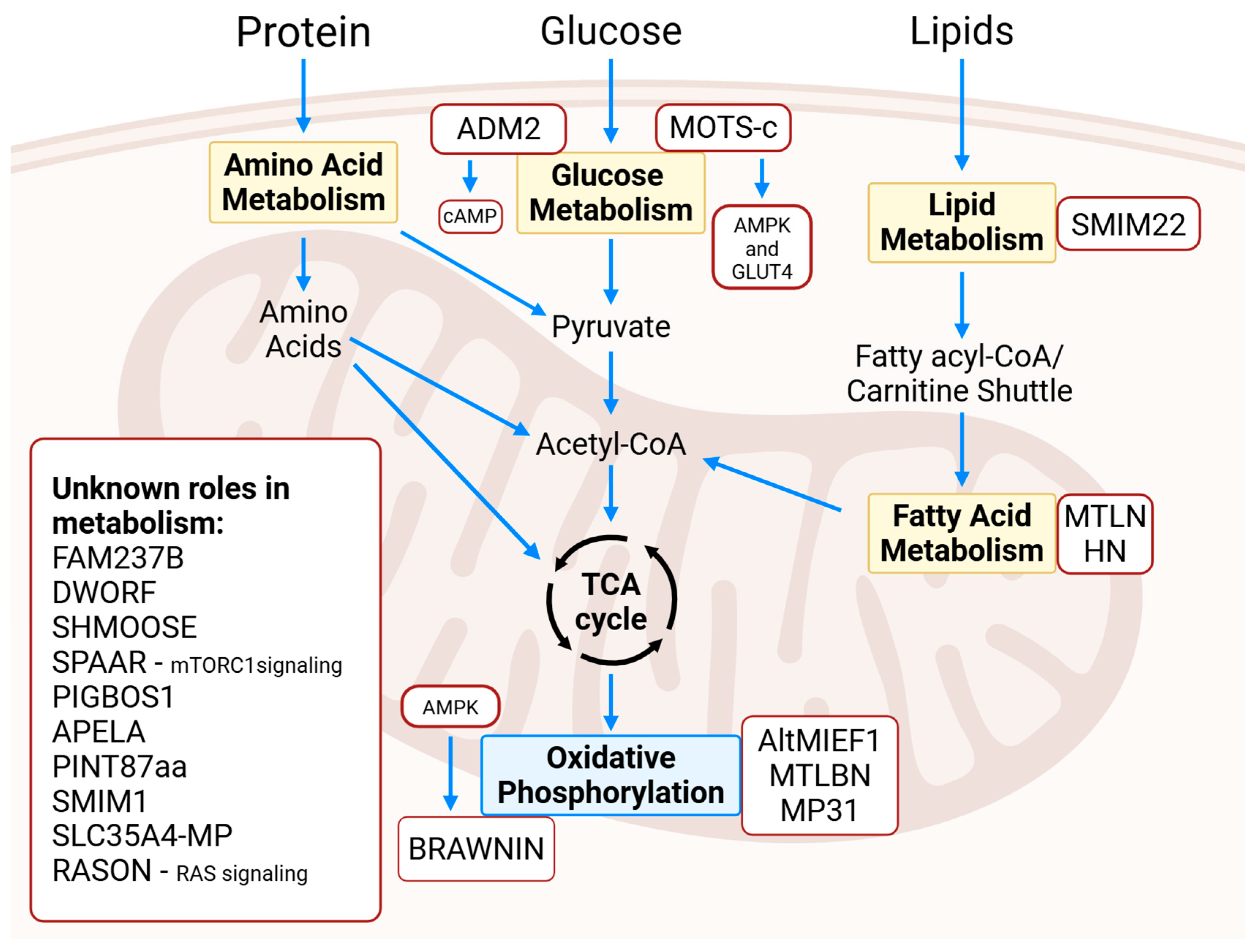
2. Microproteins in Metabolism
2.1. Glucose Metabolism
2.1.1. Mitochondrial Opening Reading Frame of the 12S Ribosomal RNA Type-c
2.1.2. Adrenomedullin-2
2.2. Fatty Acid Metabolism
2.2.1. Mitoregulin
2.2.2. Humanin
2.3. Lipid Metabolism
Small Integral Membrane Protein 22
2.4. Oxidative Phosphorylation
2.4.1. Mitochondrial Ribosome and Complex I Assembly Factor
2.4.2. Ubiquinol–cytochrome c Reductase Complex Assembly Factor 6
2.4.3. Mitolamban
2.4.4. Micropeptide 31
2.5. Erythrocyte Metabolism
Small Integral Membrane Protein 1
2.6. General Metabolism
2.6.1. Small Regulatory Polypeptide of Amino Acid Sequences
2.6.2. PIGB Opposite Protein 1
2.6.3. Small Human Mitochondrial ORF over Serine tRNA
2.6.4. Dwarf Open Reading Frame
2.6.5. Apelin Receptor Early Endogenous Ligand
2.6.6. PINT87aa
2.6.7. SLC35A4 Upstream Open Reading Frame Protein
2.6.8. Family with Sequencing Similarity 237 Member B
2.6.9. RAS-ON
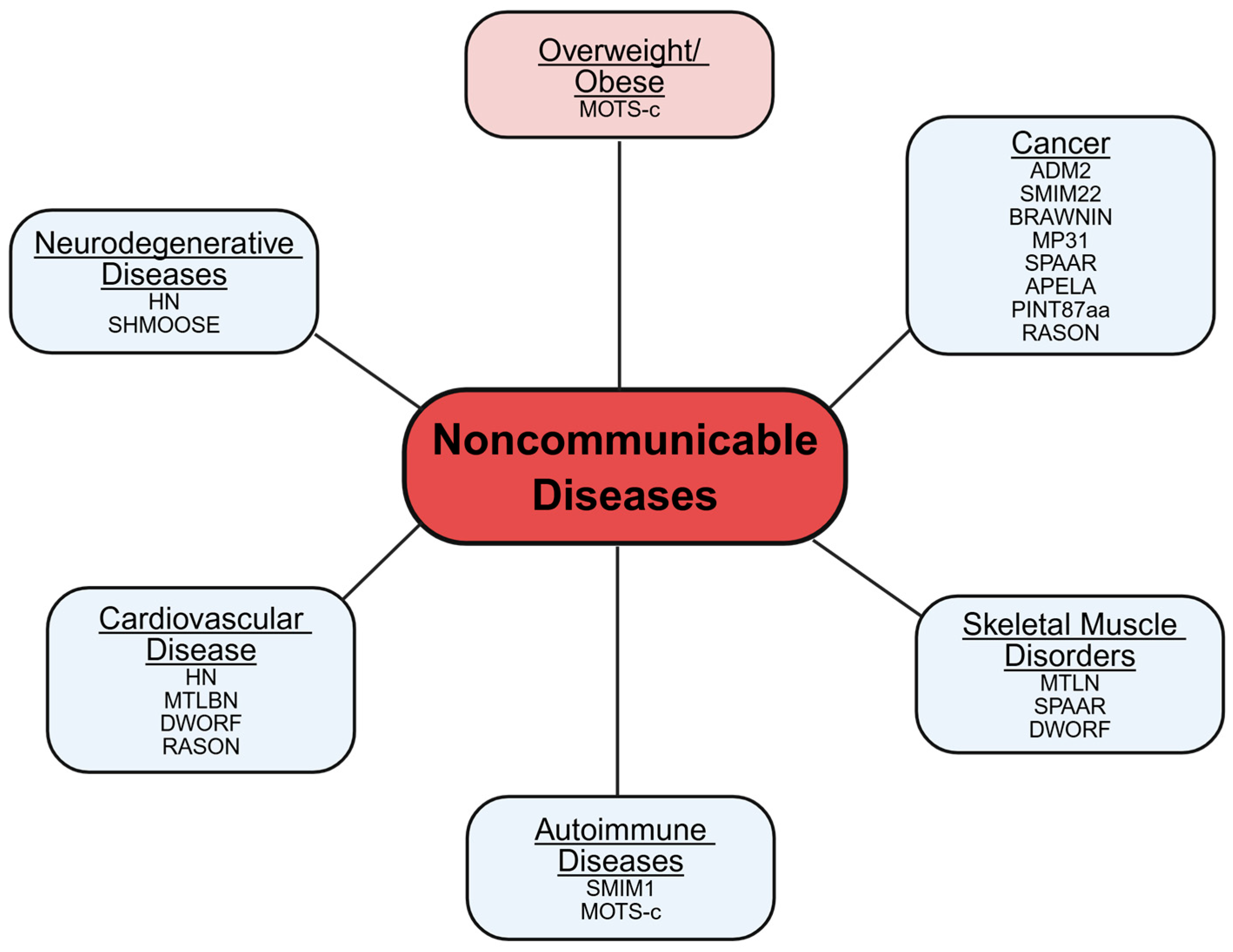
3. Conclusions
Funding
Data Availability Statement
Conflicts of Interest
References
- Judge, A.; Dodd, M.S. Metabolism. Essays Biochem. 2020, 64, 607–647. [Google Scholar] [CrossRef] [PubMed]
- Hotamisligil, G.S. Inflammation and metabolic disorders. Nature 2006, 444, 860–867. [Google Scholar] [CrossRef]
- Carney, T.J.; Wiltz, J.L.; Davis, K.; Briss, P.A.; Hacker, K. Advancing Chronic Disease Practice Through the CDC Data Modernization Initiative. Prev. Chronic Dis. 2023, 20, E110. [Google Scholar] [CrossRef]
- Catherman, A.D.; Li, M.; Tran, J.C.; Durbin, K.R.; Compton, P.D.; Early, B.P.; Thomas, P.M.; Kelleher, N.L. Top down proteomics of human membrane proteins from enriched mitochondrial fractions. Anal. Chem. 2013, 85, 1880–1888. [Google Scholar] [CrossRef]
- Bazzini, A.A.; Johnstone, T.G.; Christiano, R.; Mackowiak, S.D.; Obermayer, B.; Fleming, E.S.; Vejnar, C.E.; Lee, M.T.; Rajewsky, N.; Walther, T.C.; et al. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 2014, 33, 981–993. [Google Scholar] [CrossRef] [PubMed]
- Makarewich, C.A. The hidden world of membrane microproteins. Exp. Cell Res. 2020, 388, 111853. [Google Scholar] [CrossRef] [PubMed]
- Hassel, K.R.; Brito-Estrada, O.; Makarewich, C.A. Microproteins: Overlooked regulators of physiology and disease. iScience 2023, 26, 106781. [Google Scholar] [CrossRef]
- Miller, B.; Kim, S.J.; Kumagai, H.; Mehta, H.H.; Xiang, W.; Liu, J.; Yen, K.; Cohen, P. Peptides derived from small mitochondrial open reading frames: Genomic, biological, and therapeutic implications. Exp. Cell Res. 2020, 393, 112056. [Google Scholar] [CrossRef]
- Chen, Y.; Ho, L.; Tergaonkar, V. sORF-Encoded MicroPeptides: New players in inflammation, metabolism, and precision medicine. Cancer Lett. 2021, 500, 263–270. [Google Scholar] [CrossRef]
- Hassel, K.R.; Gibson, A.M.; Seflova, J.; Cho, E.E.; Blair, N.S.; Van Raamsdonk, C.D.; Anderson, D.M.; Robia, S.L.; Makarewich, C.A. Another-regulin regulates cardiomyocyte calcium handling via integration of neuroendocrine signaling with SERCA2a activity. J. Mol. Cell Cardiol. 2024, 197, 45–58. [Google Scholar] [CrossRef]
- Makarewich, C.A.; Munir, A.Z.; Schiattarella, G.G.; Bezprozvannaya, S.; Raguimova, O.N.; Cho, E.E.; Vidal, A.H.; Robia, S.L.; Bassel-Duby, R.; Olson, E.N. The DWORF micropeptide enhances contractility and prevents heart failure in a mouse model of dilated cardiomyopathy. eLife 2018, 7, e38319. [Google Scholar] [CrossRef] [PubMed]
- Makarewich, C.A.; Bezprozvannaya, S.; Gibson, A.M.; Bassel-Duby, R.; Olson, E.N. Gene Therapy With the DWORF Micropeptide Attenuates Cardiomyopathy in Mice. Circ. Res. 2020, 127, 1340–1342. [Google Scholar] [CrossRef] [PubMed]
- Morales, E.D.; Yue, Y.; Watkins, T.B.; Han, J.; Pan, X.; Gibson, A.M.; Hu, B.; Brito-Estrada, O.; Yao, G.; Makarewich, C.A.; et al. Dwarf Open Reading Frame (DWORF) Gene Therapy Ameliorated Duchenne Muscular Dystrophy Cardiomyopathy in Aged mdx Mice. J. Am. Heart Assoc. 2023, 12, e027480. [Google Scholar] [CrossRef]
- Nakrani, M.N.; Wineland, R.H.; Anjum, F. Physiology, Glucose Metabolism. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Esmailnasab, N.; Moradi, G.; Delaveri, A. Risk factors of non-communicable diseases and metabolic syndrome. Iran. J. Public Health 2012, 41, 77–85. [Google Scholar] [PubMed]
- Reynolds, J.C.; Lai, R.W.; Woodhead, J.S.T.; Joly, J.H.; Mitchell, C.J.; Cameron-Smith, D.; Lu, R.; Cohen, P.; Graham, N.A.; Benayoun, B.A.; et al. MOTS-c is an exercise-induced mitochondrial-encoded regulator of age-dependent physical decline and muscle homeostasis. Nat. Commun. 2021, 12, 470. [Google Scholar] [CrossRef]
- Kong, B.S.; Min, S.H.; Lee, C.; Cho, Y.M. Mitochondrial-encoded MOTS-c prevents pancreatic islet destruction in autoimmune diabetes. Cell Rep. 2021, 36, 109447. [Google Scholar] [CrossRef]
- Lee, C.; Zeng, J.; Drew, B.G.; Sallam, T.; Martin-Montalvo, A.; Wan, J.; Kim, S.J.; Mehta, H.; Hevener, A.L.; de Cabo, R.; et al. The mitochondrial-derived peptide MOTS-c promotes metabolic homeostasis and reduces obesity and insulin resistance. Cell Metab. 2015, 21, 443–454. [Google Scholar] [CrossRef]
- Kong, B.S.; Lee, C.; Cho, Y.M. Mitochondrial-Encoded Peptide MOTS-c, Diabetes, and Aging-Related Diseases. Diabetes Metab. J. 2023, 47, 315–324. [Google Scholar] [CrossRef]
- Kitamura, K.; Kangawa, K.; Kawamoto, M.; Ichiki, Y.; Nakamura, S.; Matsuo, H.; Eto, T. Adrenomedullin: A novel hypotensive peptide isolated from human pheochromocytoma. Biochem. Biophys. Res. Commun. 1993, 192, 553–560. [Google Scholar] [CrossRef]
- Liang, Y.L.; Belousoff, M.J.; Fletcher, M.M.; Zhang, X.; Khoshouei, M.; Deganutti, G.; Koole, C.; Furness, S.G.B.; Miller, L.J.; Hay, D.L.; et al. Structure and Dynamics of Adrenomedullin Receptors AM(1) and AM(2) Reveal Key Mechanisms in the Control of Receptor Phenotype by Receptor Activity-Modifying Proteins. ACS Pharmacol. Transl. Sci. 2020, 3, 263–284. [Google Scholar] [CrossRef]
- Kim, J.T.; Lim, M.A.; Lee, S.E.; Kim, H.J.; Koh, H.Y.; Lee, J.H.; Jun, S.M.; Kim, J.M.; Kim, K.H.; Shin, H.S.; et al. Adrenomedullin2 stimulates progression of thyroid cancer in mice and humans under nutrient excess conditions. J. Pathol. 2022, 258, 264–277. [Google Scholar] [CrossRef] [PubMed]
- Talley, J.T.; Mohiuddin, S.S. Biochemistry, Fatty Acid Oxidation. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Billingsley, H.E.; Carbone, S.; Lavie, C.J. Dietary Fats and Chronic Noncommunicable Diseases. Nutrients 2018, 10, 1385. [Google Scholar] [CrossRef] [PubMed]
- Koundouros, N.; Poulogiannis, G. Reprogramming of fatty acid metabolism in cancer. Br. J. Cancer 2020, 122, 4–22. [Google Scholar] [CrossRef]
- Stein, C.S.; Jadiya, P.; Zhang, X.; McLendon, J.M.; Abouassaly, G.M.; Witmer, N.H.; Anderson, E.J.; Elrod, J.W.; Boudreau, R.L. Mitoregulin: A lncRNA-Encoded Microprotein that Supports Mitochondrial Supercomplexes and Respiratory Efficiency. Cell Rep. 2018, 23, 3710–3720.e8. [Google Scholar] [CrossRef]
- Makarewich, C.A.; Baskin, K.K.; Munir, A.Z.; Bezprozvannaya, S.; Sharma, G.; Khemtong, C.; Shah, A.M.; McAnally, J.R.; Malloy, C.R.; Szweda, L.I.; et al. MOXI Is a Mitochondrial Micropeptide That Enhances Fatty Acid beta-Oxidation. Cell Rep. 2018, 23, 3701–3709. [Google Scholar] [CrossRef]
- Chugunova, A.; Loseva, E.; Mazin, P.; Mitina, A.; Navalayeu, T.; Bilan, D.; Vishnyakova, P.; Marey, M.; Golovina, A.; Serebryakova, M.; et al. LINC00116 codes for a mitochondrial peptide linking respiration and lipid metabolism. Proc. Natl. Acad. Sci. USA 2019, 116, 4940–4945. [Google Scholar] [CrossRef]
- Xiao, M.H.; Lin, Y.F.; Xie, P.P.; Chen, H.X.; Deng, J.W.; Zhang, W.; Zhao, N.; Xie, C.; Meng, Y.; Liu, X.; et al. Downregulation of a mitochondrial micropeptide, MPM, promotes hepatoma metastasis by enhancing mitochondrial complex I activity. Mol. Ther. 2022, 30, 714–725. [Google Scholar] [CrossRef]
- Friesen, M.; Warren, C.R.; Yu, H.; Toyohara, T.; Ding, Q.; Florido, M.H.C.; Sayre, C.; Pope, B.D.; Goff, L.A.; Rinn, J.L.; et al. Mitoregulin Controls beta-Oxidation in Human and Mouse Adipocytes. Stem Cell Rep. 2020, 14, 590–602. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Fan, J.; Han, L.; Qi, H.; Wang, Y.; Wang, H.; Chen, S.; Du, L.; Li, S.; Zhang, Y.; et al. The micropeptide LEMP plays an evolutionarily conserved role in myogenesis. Cell Death Dis. 2020, 11, 357. [Google Scholar] [CrossRef]
- Zhang, S.; Guo, Y.; Fidelito, G.; Robinson, D.R.L.; Liang, C.; Lim, R.; Bichler, Z.; Guo, R.; Wu, G.; Xu, H.; et al. LINC00116-encoded microprotein mitoregulin regulates fatty acid metabolism at the mitochondrial outer membrane. iScience 2023, 26, 107558. [Google Scholar] [CrossRef]
- Hashimoto, Y.; Ito, Y.; Niikura, T.; Shao, Z.; Hata, M.; Oyama, F.; Nishimoto, I. Mechanisms of neuroprotection by a novel rescue factor humanin from Swedish mutant amyloid precursor protein. Biochem. Biophys. Res. Commun. 2001, 283, 460–468. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhang, D.; Yuan, W.; Wang, C.; Huang, Q.; Luo, J. Humanin Ameliorates Free Fatty Acid-Induced Endothelial Inflammation by Suppressing the NLRP3 Inflammasome. ACS Omega 2020, 5, 22039–22045. [Google Scholar] [CrossRef] [PubMed]
- Chandel, N.S. Lipid Metabolism. Cold Spring Harb. Perspect. Biol. 2021, 13, a040576. [Google Scholar] [CrossRef]
- Xu, L.; Yang, Q.; Zhou, J. Mechanisms of Abnormal Lipid Metabolism in the Pathogenesis of Disease. Int. J. Mol. Sci. 2024, 25, 8465. [Google Scholar] [CrossRef]
- Zhao, Q.; Li, B.; Zhang, X.; Zhao, H.; Xue, W.; Yuan, Z.; Xu, S.; Duan, G. M2 macrophage-derived lncRNA NORAD in EVs promotes NSCLC progression via miR-520g-3p/SMIM22/GALE axis. NPJ Precis. Oncol. 2024, 8, 185. [Google Scholar] [CrossRef]
- Polycarpou-Schwarz, M.; Gross, M.; Mestdagh, P.; Schott, J.; Grund, S.E.; Hildenbrand, C.; Rom, J.; Aulmann, S.; Sinn, H.P.; Vandesompele, J.; et al. The cancer-associated microprotein CASIMO1 controls cell proliferation and interacts with squalene epoxidase modulating lipid droplet formation. Oncogene 2018, 37, 4750–4768. [Google Scholar] [CrossRef] [PubMed]
- Seyama, Y.; Sudo, K.; Hirose, S.; Hamano, Y.; Yamada, T.; Hiroyama, T.; Sasaki, R.; Hirai, M.Y.; Hyodo, I.; Tsuchiya, K.; et al. Identification of a gene set that maintains tumorigenicity of the hepatocellular carcinoma cell line Li-7. Hum. Cell 2023, 36, 2074–2086. [Google Scholar] [CrossRef]
- Samandi, S.; Roy, A.V.; Delcourt, V.; Lucier, J.F.; Gagnon, J.; Beaudoin, M.C.; Vanderperre, B.; Breton, M.A.; Motard, J.; Jacques, J.F.; et al. Deep transcriptome annotation enables the discovery and functional characterization of cryptic small proteins. eLlife 2017, 6, e27860. [Google Scholar] [CrossRef]
- Dibley, M.G.; Formosa, L.E.; Lyu, B.; Reljic, B.; McGann, D.; Muellner-Wong, L.; Kraus, F.; Sharpe, A.J.; Stroud, D.A.; Ryan, M.T. The Mitochondrial Acyl-carrier Protein Interaction Network Highlights Important Roles for LYRM Family Members in Complex I and Mitoribosome Assembly. Mol. Cell Proteom. 2020, 19, 65–77. [Google Scholar] [CrossRef]
- Zhang, S.; Reljic, B.; Liang, C.; Kerouanton, B.; Francisco, J.C.; Peh, J.H.; Mary, C.; Jagannathan, N.S.; Olexiouk, V.; Tang, C.; et al. Mitochondrial peptide BRAWNIN is essential for vertebrate respiratory complex III assembly. Nat. Commun. 2020, 11, 1312. [Google Scholar] [CrossRef]
- Makarewich, C.A.; Munir, A.Z.; Bezprozvannaya, S.; Gibson, A.M.; Young Kim, S.; Martin-Sandoval, M.S.; Mathews, T.P.; Szweda, L.I.; Bassel-Duby, R.; Olson, E.N. The cardiac-enriched microprotein mitolamban regulates mitochondrial respiratory complex assembly and function in mice. Proc. Natl. Acad. Sci. USA 2022, 119, e2120476119. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Wang, F.Y.; Sang, Y.; Chen, B.; Huang, J.H.; He, F.J.; Li, H.; Zhu, Y.; Liu, X.; Zhuang, S.M.; et al. Mitochondrial Micropeptide STMP1 Enhances Mitochondrial Fission to Promote Tumor Metastasis. Cancer Res. 2022, 82, 2431–2443. [Google Scholar] [CrossRef]
- Zhang, D.; Xi, Y.; Coccimiglio, M.L.; Mennigen, J.A.; Jonz, M.G.; Ekker, M.; Trudeau, V.L. Functional prediction and physiological characterization of a novel short trans-membrane protein 1 as a subunit of mitochondrial respiratory complexes. Physiol. Genom. 2012, 44, 1133–1140. [Google Scholar] [CrossRef]
- Sang, Y.; Liu, J.Y.; Wang, F.Y.; Luo, X.Y.; Chen, Z.Q.; Zhuang, S.M.; Zhu, Y. Mitochondrial micropeptide STMP1 promotes G1/S transition by enhancing mitochondrial complex IV activity. Mol. Ther. 2022, 30, 2844–2855. [Google Scholar] [CrossRef]
- See, K.; Tan, W.L.W.; Lim, E.H.; Tiang, Z.; Lee, L.T.; Li, P.Y.Q.; Luu, T.D.A.; Ackers-Johnson, M.; Foo, R.S. Single cardiomyocyte nuclear transcriptomes reveal a lincRNA-regulated de-differentiation and cell cycle stress-response in vivo. Nat. Commun. 2017, 8, 225. [Google Scholar] [CrossRef] [PubMed]
- Hofman, D.A.; Prensner, J.R.; van Heesch, S. Microproteins in cancer: Identification, biological functions, and clinical implications. Trends Genet. 2025, 41, 146–161. [Google Scholar] [CrossRef]
- Bhatta, A.; Atianand, M.; Jiang, Z.; Crabtree, J.; Blin, J.; Fitzgerald, K.A. A Mitochondrial Micropeptide Is Required for Activation of the Nlrp3 Inflammasome. J. Immunol. 2020, 204, 428–437. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Wang, M.; Liu, S.; Chen, H.; Li, Y.; Yuan, F.; Yang, L.; Qiu, S.; Wang, H.; Xie, Z.; et al. A lncRNA-encoded mitochondrial micropeptide exacerbates microglia-mediated neuroinflammation in retinal ischemia/reperfusion injury. Cell Death Dis. 2023, 14, 126. [Google Scholar] [CrossRef]
- Huang, N.; Li, F.; Zhang, M.; Zhou, H.; Chen, Z.; Ma, X.; Yang, L.; Wu, X.; Zhong, J.; Xiao, F.; et al. An Upstream Open Reading Frame in Phosphatase and Tensin Homolog Encodes a Circuit Breaker of Lactate Metabolism. Cell Metab. 2021, 33, 128–144.e129. [Google Scholar] [CrossRef]
- Chatzinikolaou, P.N.; Margaritelis, N.V.; Paschalis, V.; Theodorou, A.A.; Vrabas, I.S.; Kyparos, A.; D’Alessandro, A.; Nikolaidis, M.G. Erythrocyte metabolism. Acta Physiol 2024, 240, e14081. [Google Scholar] [CrossRef]
- Ballif, B.A.; Helias, V.; Peyrard, T.; Menanteau, C.; Saison, C.; Lucien, N.; Bourgouin, S.; Le Gall, M.; Cartron, J.P.; Arnaud, L. Disruption of SMIM1 causes the Vel- blood type. EMBO Mol. Med. 2013, 5, 751–761. [Google Scholar] [CrossRef]
- Storry, J.R.; Joud, M.; Christophersen, M.K.; Thuresson, B.; Akerstrom, B.; Sojka, B.N.; Nilsson, B.; Olsson, M.L. Homozygosity for a null allele of SMIM1 defines the Vel-negative blood group phenotype. Nat. Genet. 2013, 45, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, A.; Pasut, A.; Matsumoto, M.; Yamashita, R.; Fung, J.; Monteleone, E.; Saghatelian, A.; Nakayama, K.I.; Clohessy, J.G.; Pandolfi, P.P. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature 2017, 541, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Wacholder, A.; Carvunis, A.R. Evolutionary Characterization of the Short Protein SPAAR. Genes 2021, 12, 1864. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Liu, J.; Zhang, Y.H.; Shen, D.; Liu, S.; Lin, F.; Su, J.; Lin, Q.F.; Yan, S.; Li, Y.; et al. Long noncoding RNA LINC00961 inhibits cell invasion and metastasis in human non-small cell lung cancer. Biomed. Pharmacother. 2018, 97, 1311–1318. [Google Scholar] [CrossRef]
- Chu, Q.; Martinez, T.F.; Novak, S.W.; Donaldson, C.J.; Tan, D.; Vaughan, J.M.; Chang, T.; Diedrich, J.K.; Andrade, L.; Kim, A.; et al. Regulation of the ER stress response by a mitochondrial microprotein. Nat. Commun. 2019, 10, 4883. [Google Scholar] [CrossRef]
- Sak, F.; Sengul, F.; Vatansev, H. The Role of Endoplasmic Reticulum Stress in Metabolic Diseases. Metab. Syndr. Relat. Disord. 2024, 22, 487–493. [Google Scholar] [CrossRef]
- Ghemrawi, R.; Battaglia-Hsu, S.F.; Arnold, C. Endoplasmic Reticulum Stress in Metabolic Disorders. Cells 2018, 7, 63. [Google Scholar] [CrossRef]
- John, G.B.; Shang, Y.; Li, L.; Renken, C.; Mannella, C.A.; Selker, J.M.; Rangell, L.; Bennett, M.J.; Zha, J. The mitochondrial inner membrane protein mitofilin controls cristae morphology. Mol. Biol. Cell 2005, 16, 1543–1554. [Google Scholar] [CrossRef]
- Miller, B.; Kim, S.J.; Mehta, H.H.; Cao, K.; Kumagai, H.; Thumaty, N.; Leelaprachakul, N.; Braniff, R.G.; Jiao, H.; Vaughan, J.; et al. Mitochondrial DNA variation in Alzheimer’s disease reveals a unique microprotein called SHMOOSE. Mol. Psychiatry 2023, 28, 1813–1826. [Google Scholar] [CrossRef]
- Nelson, B.R.; Makarewich, C.A.; Anderson, D.M.; Winders, B.R.; Troupes, C.D.; Wu, F.; Reese, A.L.; McAnally, J.R.; Chen, X.; Kavalali, E.T.; et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 2016, 351, 271–275. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Read, C.; Kuc, R.E.; Buonincontri, G.; Southwood, M.; Torella, R.; Upton, P.D.; Crosby, A.; Sawiak, S.J.; Carpenter, T.A.; et al. Elabela/Toddler Is an Endogenous Agonist of the Apelin APJ Receptor in the Adult Cardiovascular System, and Exogenous Administration of the Peptide Compensates for the Downregulation of Its Expression in Pulmonary Arterial Hypertension. Circulation 2017, 135, 1160–1173. [Google Scholar] [CrossRef]
- Wang, Z.; Yu, D.; Wang, M.; Wang, Q.; Kouznetsova, J.; Yang, R.; Qian, K.; Wu, W.; Shuldiner, A.; Sztalryd, C.; et al. Elabela-apelin receptor signaling pathway is functional in mammalian systems. Sci. Rep. 2015, 5, 8170. [Google Scholar] [CrossRef]
- Liu, L.; Yi, X.; Lu, C.; Wang, Y.; Xiao, Q.; Zhang, L.; Pang, Y.; Guan, X. Study Progression of Apelin/APJ Signaling and Apela in Different Types of Cancer. Front. Oncol. 2021, 11, 658253. [Google Scholar] [CrossRef] [PubMed]
- Ganguly, D.; Cai, C.; Sims, M.M.; Yang, C.H.; Thomas, M.; Cheng, J.; Saad, A.G.; Pfeffer, L.M. APELA Expression in Glioma, and Its Association with Patient Survival and Tumor Grade. Pharmaceuticals 2019, 12, 45. [Google Scholar] [CrossRef] [PubMed]
- Soulet, F.; Bodineau, C.; Hooks, K.B.; Descarpentrie, J.; Alves, I.; Dubreuil, M.; Mouchard, A.; Eugenie, M.; Hoepffner, J.L.; Lopez, J.J.; et al. ELA/APELA precursor cleaved by furin displays tumor suppressor function in renal cell carcinoma through mTORC1 activation. JCI Insight 2020, 5, e129070. [Google Scholar] [CrossRef]
- Liet, B.; Nys, N.; Siegfried, G. Elabela/toddler: New peptide with a promising future in cancer diagnostic and therapy. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 119065. [Google Scholar] [CrossRef]
- Zhang, M.; Zhao, K.; Xu, X.; Yang, Y.; Yan, S.; Wei, P.; Liu, H.; Xu, J.; Xiao, F.; Zhou, H.; et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat. Commun. 2018, 9, 4475. [Google Scholar] [CrossRef]
- Rocha, A.L.; Pai, V.; Perkins, G.; Chang, T.; Ma, J.; De Souza, E.V.; Chu, Q.; Vaughan, J.M.; Diedrich, J.K.; Ellisman, M.H.; et al. An Inner Mitochondrial Membrane Microprotein from the SLC35A4 Upstream ORF Regulates Cellular Metabolism. J. Mol. Biol. 2024, 436, 168559. [Google Scholar] [CrossRef]
- Martinez, T.F.; Lyons-Abbott, S.; Bookout, A.L.; De Souza, E.V.; Donaldson, C.; Vaughan, J.M.; Lau, C.; Abramov, A.; Baquero, A.F.; Baquero, K.; et al. Profiling mouse brown and white adipocytes to identify metabolically relevant small ORFs and functional microproteins. Cell Metab. 2023, 35, 166–183.e11. [Google Scholar] [CrossRef]
- Cheng, R.; Li, F.; Zhang, M.; Xia, X.; Wu, J.; Gao, X.; Zhou, H.; Zhang, Z.; Huang, N.; Yang, X.; et al. A novel protein RASON encoded by a lncRNA controls oncogenic RAS signaling in KRAS mutant cancers. Cell Res. 2023, 33, 30–45. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, S.; Vander Heiden, M.G.; McCormick, F. The Metabolic Landscape of RAS-Driven Cancers from biology to therapy. Nat. Cancer 2021, 2, 271–283. [Google Scholar] [CrossRef] [PubMed]
| Human Gene | Microprotein Size (Amino Acids) | Tissue Expression | Mouse Homolog Gene | Reference |
|---|---|---|---|---|
| Adrenomedullin-2 (ADM2, AM2) | 148 | Pancreas, thyroid grand, kidney, liver | Adm2, Am2 | [20,21,22] |
| Apelin receptor early endogenous ligand (APELA, ELABELA, TODDLER) | 54 | Kidney, male reproductive tissues, pluripotent stem cells, blood vessels | Apela, Ela, Ende, Gm10664, Tdl | [64,65,66,67,68,69] |
| Dwarf opening reading frame (DWORF, STRIT1) | 35 | Skeletal and cardiac muscle | Dworf, Strit1 | [11,12,63] |
| Family with sequence similarity 237 member B (FAM237B) | 139 | Brain, eye, adipose tissue, reproductive organs | Fam237b, Gm8773, EG667705 | [72] |
| Humanin (HN) | 24 | Male reproductive tissues, eye, cardiac and skeletal muscle, liver, kidney | Mtrnr2l7, Gm20594 | [33,34] |
| Microprotein 31 (MP31) | 31 | Brain, specifically studied in glioblastomas | MP35 | [51] |
| Mitochondrial ribosome and complex I assembly factor (AltMIEF1, AltMiD51) | 70 | N/a | N/a | [40,41] |
| Mitochondrial opening reading frame of the 12S ribosomal RNA type-c (MOTS-c) | 16 | Plasma and skeletal muscle | N/a | [16,17,18,19] |
| Mitolamban (MTLBN, STMP1, C7orf73) | 47 | Ubiquitous, highest in skeletal and cardiac muscle | Mtlbn, Stmp1, Mm47 | [43,44,45,46,49] |
| Mitoregulin (MTLN, LEMP, MOXI, MPM, SMIM37) | 56 | Ubiquitous, highest in skeletal and cardiac muscle | Mtln, Lemp, Moxi, Mpm, Smim37 | [26,27,28,29,30,31,32] |
| PIGB opposite protein 1 (PIGBOS1) | 54 | Ubiquitous, highest in pancreas, thyroid, and spleen | N/a | [58] |
| PINT87aa | 87 | Brain, liver, kidney, stomach, breast tissue, intestine, and thyroid | N/a | [70] |
| RAS-ON (RASON) | 108 | Tumors | N/a | [73] |
| SLC35A4 upstream open reading frame protein (SLC35A4-MP) | 103 | Ubiquitous, highest in skeletal muscle | N/a | [71] |
| Small human mitochondrial ORF over serine tRNA (SHMOOSE) | 58 | Brain | N/a | [62] |
| Small integral membrane protein 1 (SMIM1) | 78 | Male reproductive tissues, skeletal muscle, bone marrow, red blood cells | Smim1 | [53,54] |
| Small integral membrane protein 22 (SMIM22, CASIMO1) | 83 | Stomach, digestive tract | Smim22, Gm5480 | [37,38,39] |
| Small regulatory polypeptide of amino acid sequences (SPAAR, SPAR) | 90 | Ubiquitously expressed, most highly expressed in adipose tissue, placenta, lung, and skeletal and cardiac muscle | Spaar, Spar | [55,56,57] |
| Ubiquinol–cytochrome c reductase complex assembly factor 6 (UQCC6, BR, BRAWNIN, C12orf73) | 71 | Ubiquitous, highest in skeletal muscle and brown adipose tissue | Uqcc6, Br, Brawnin | [42] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wadding-Lee, C.A.; Makarewich, C.A. Microproteins in Metabolism. Cells 2025, 14, 859. https://doi.org/10.3390/cells14120859
Wadding-Lee CA, Makarewich CA. Microproteins in Metabolism. Cells. 2025; 14(12):859. https://doi.org/10.3390/cells14120859
Chicago/Turabian StyleWadding-Lee, Caris A., and Catherine A. Makarewich. 2025. "Microproteins in Metabolism" Cells 14, no. 12: 859. https://doi.org/10.3390/cells14120859
APA StyleWadding-Lee, C. A., & Makarewich, C. A. (2025). Microproteins in Metabolism. Cells, 14(12), 859. https://doi.org/10.3390/cells14120859





