Modified Methylation Following Electrostimulation in a Standardized Setting—Complementing a Transcriptomic Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Methylomics
2.3. Differential Analysis
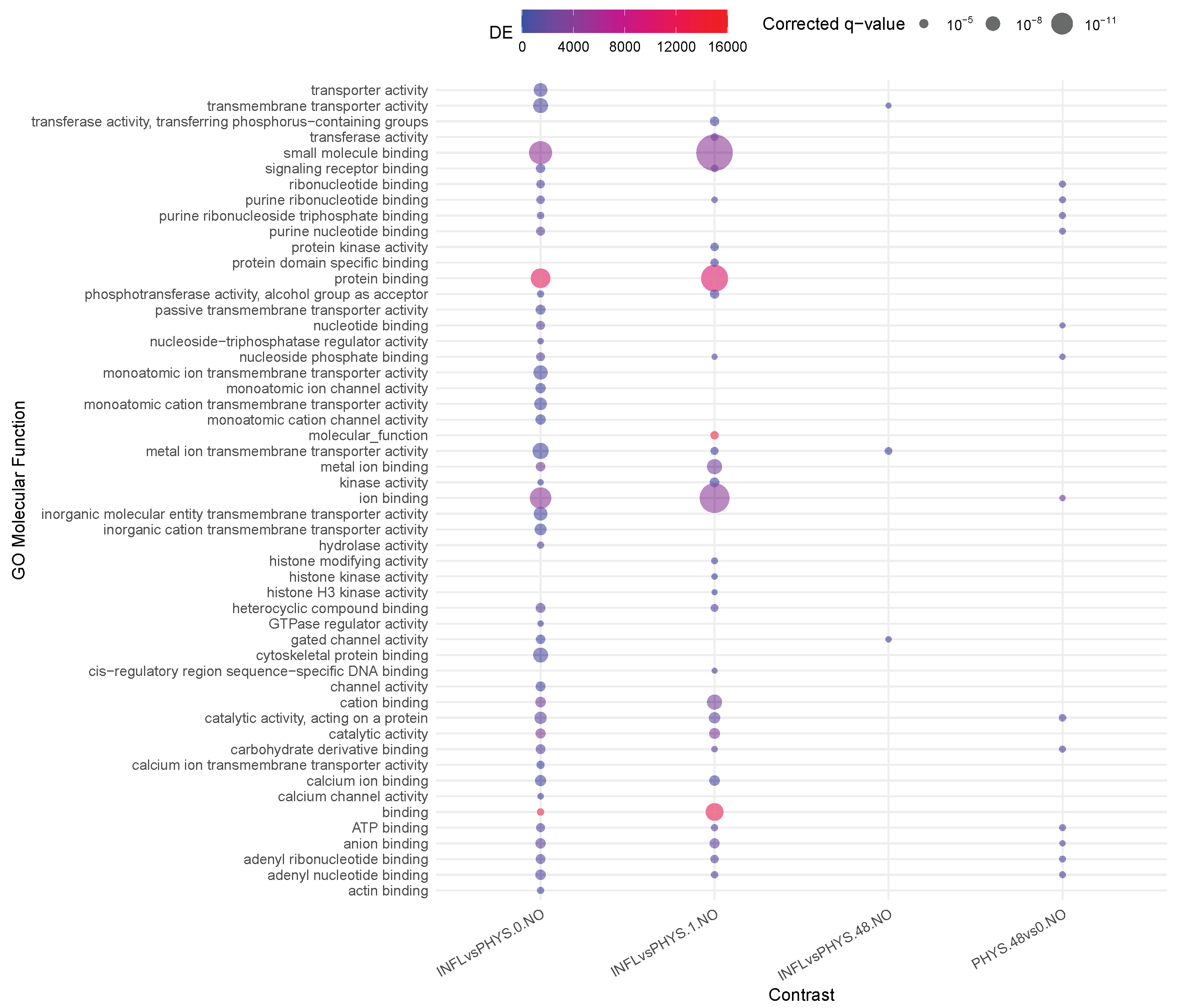
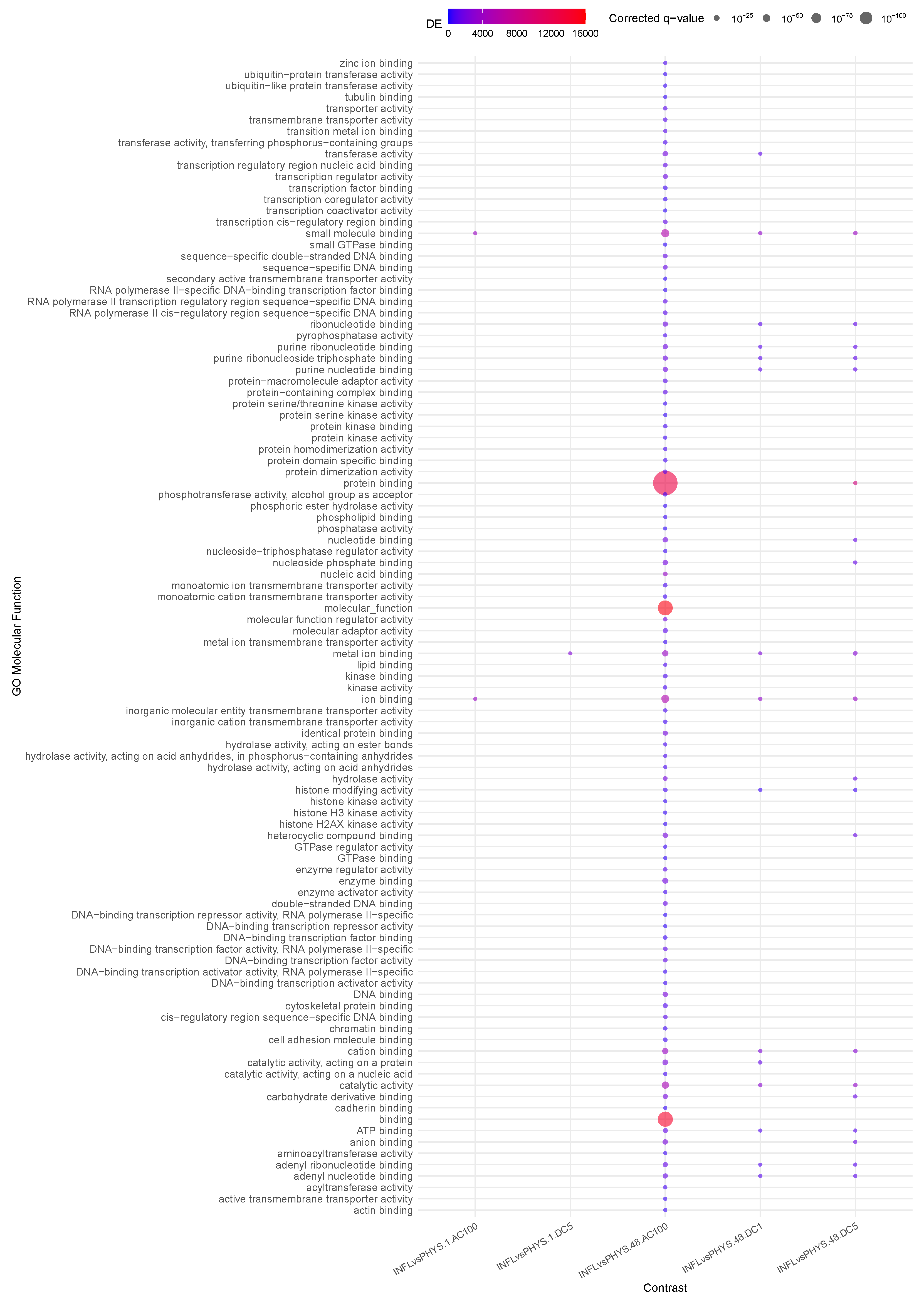
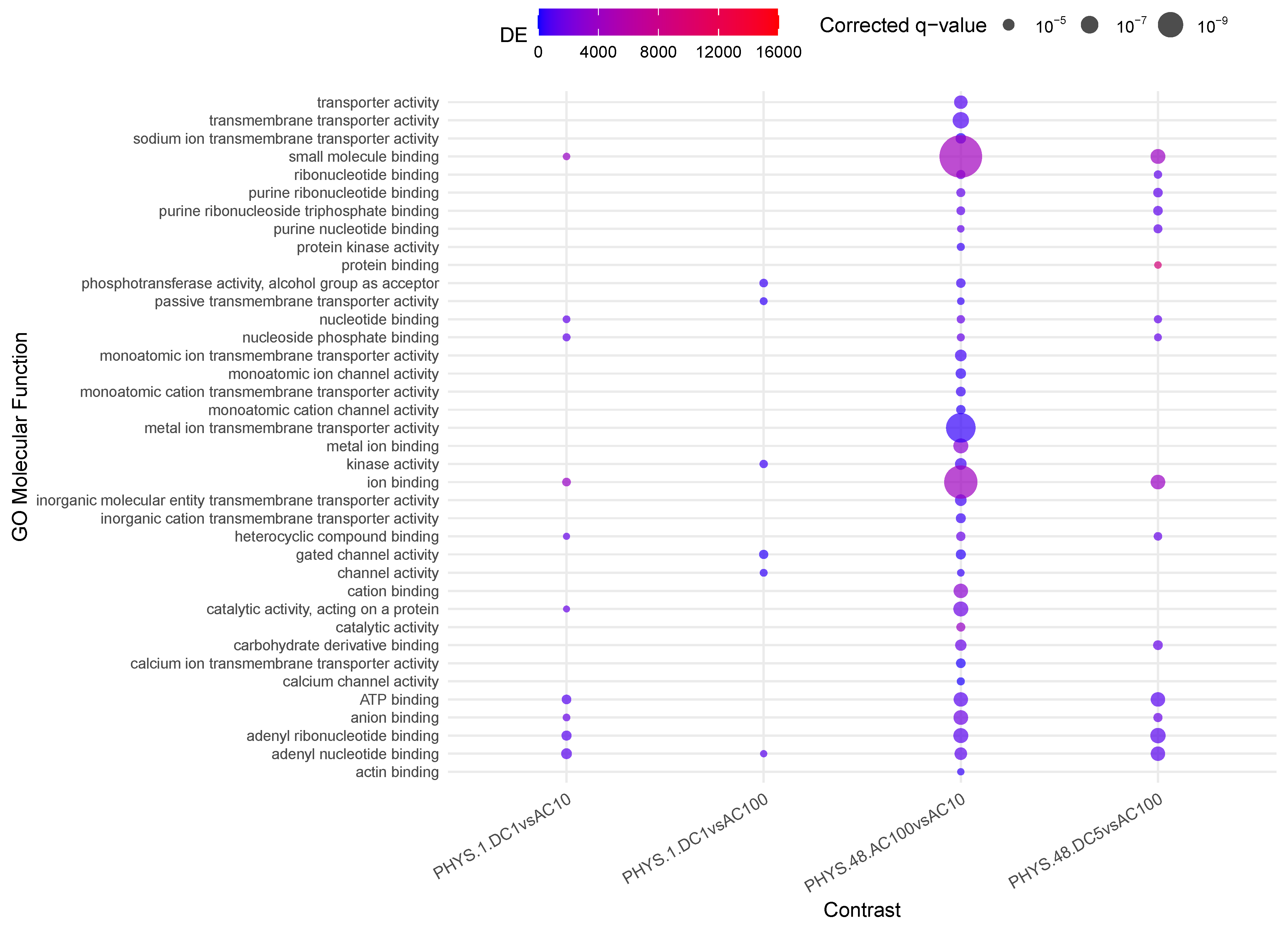
2.4. Enrichment
2.5. Methylage
3. Results and Discussion
3.1. Question 1: Impact of Time
3.2. Question 2: Impact of Stimuli on Physiological Samples
3.3. Question 3: Effects of Stimuli on Inflamed Samples
3.4. Question 4: Impact of States on Stimulus
3.5. Question 5: Differential Impact of Stimuli on PHYS
3.6. Question 6: Differential Impact of Stimuli on INFL
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rabbani, M.; Rahman, E.; Powner, M.B.; Triantis, I.F. Making sense of electrical stimulation: A meta-analysis for wound healing. Ann. Biomed. Eng. 2024, 52, 153–177. [Google Scholar] [CrossRef] [PubMed]
- Tracey, K.J. The inflammatory reflex. Nature 2002, 420, 853–859. [Google Scholar] [CrossRef] [PubMed]
- Martelli, D.; Farmer, D.G.; Yao, S.T. The splanchnic anti-inflammatory pathway: Could it be the efferent arm of the inflammatory reflex? Exp. Physiol. 2016, 101, 1245–1252. [Google Scholar] [CrossRef] [PubMed]
- Saha, A.; Alleyne, G. Recognizing noncommunicable diseases as a global health security threat. Bull World Health Organ. 2018, 96, 792–793. [Google Scholar] [CrossRef] [PubMed]
- Di Pietro, B.; Villata, S.; Dal MOnego, S.; Degasperi, M.; Ghini, V.; Guarnieri, T.; Plaksienko, A.; Liu, Y.; Pecchioli, V.; Manni, L.; et al. Differential Anti-Inflammatory Effects of Electrostimulation in a Standardized Setting. Int. J. Mol. Sci. 2024, 25, 9808. [Google Scholar] [CrossRef] [PubMed]
- Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 2013, 14, R115. [Google Scholar] [CrossRef] [PubMed]
- Di Lena, P.; Sala, C.; Nardini, C. Evaluation of different computational methods for DNA methylation-based biological age. Brief Bioinform. 2022, 23, bbac274. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wong, A.; Kuh, D.; Paul, D.S.; Rakyan, V.K.; Leslie, R.D.; Zheng, S.C.; Widschwendter, M.; Beck, S.; Teschendorff, A.E. Correlation of an epigenetic mitotic clock with cancer risk. Genome Biol. 2016, 17, 205. [Google Scholar] [CrossRef] [PubMed]
- Bott, K.; Upton, Z.; Schrobback, K.; Ehrbar, M.; Hubbell, J.A.; Lutolf, M.P.; Rizzi, S.C. The effect of matrix characteristics on fibroblast proliferation in 3D gels. Biomaterials 2010, 31, 8454–8464. [Google Scholar] [CrossRef] [PubMed]
- Maas-Szabowski, N.; Shimotoyodome, A.; Fusenig, N.E. Keratinocyte growth regulation in fibroblast cocultures via a double paracrine mechanism. J. Cell Sci. 1999, 112, 1843–1853. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, Y.; Farhangfar, F.; Zimmer, M.; Zhang, Y. Enhanced keratinocyte proliferation and migration in co-culture with fibroblasts. PLoS ONE 2012, 7, e40951. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Morris, T.J.; Webster, A.P.; Yang, Z.; Beck, S.; Feber, A.; Teschendorff, A.E. ChAMP: Updated methylation analysis pipeline for Illumina BeadChips. Bioinformatics 2017, 33, 3982–3984. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Phipson, B.; Maksimovic, J. missMethyl: Analysing Illumina HumanMethylation BeadChip Data. Bioinformatics 2016, 32, 286–288. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef] [PubMed]



| INFL | PHYS | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NO | DC | AC | NO | DC | AC | ||||||||||||||||
| 1 V | 5 V | 10 Hz | 100 Hz | 1 V | 5 V | 10 Hz | 100 Hz | ||||||||||||||
| t0 | t1 | t48 | t1 | t48 | t1 | t48 | t1 | t48 | t1 | t48 | t0 | t1 | t48 | t1 | t48 | t1 | t48 | t1 | t48 | t1 | t48 |
| Question | Contrasts | RNA & Metabolites | mDNA & Methylage |
|---|---|---|---|
| 1. What is the impact of time? | PHYS.1vs0.NO, PHYS.48vs1.NO, PHYS.48vs0.NO, INFL.1vs0.NO, INFL.48vs1.NO, INFL.48vs0.NO, INFLvsPHYS.0.NO, INFLvsPHYS.1.NO, INFLvsPHYS.48.NO |
|
|
| 2. What is the impact of stimuli on the physiological state? | PHYS.1.DC1vsNO, PHYS.1.DC5vsNO, PHYS.1.AC10vsNO, PHYS.1.AC100vsNO, PHYS.48.DC1vsNO, PHYS.48.DC5vsNO, PHYS.48.AC10vsNO, PHYS.48.AC100vsNO |
|
|
| 3. What is the impact of stimuli on the inflamed state? | INFL.1.DC1vsNO, INFL.1.DC5vsNO, INFL.1.AC10vsNO, INFL.1.AC100vsNO, INFL.48.DC1vsNO, INFL.48.DC5vsNO, INFL.48.AC10vsNO, INFL.48.AC100vsNO |
|
|
| 4. What is the impact of states on stimulus? | INFLvsPHYS.1.DC1, INFLvsPHYS.1.DC5, INFLvsPHYS.1.AC10, INFLvsPHYS.1.AC100, INFLvsPHYS.48.DC1, INFLvsPHYS.48.DC5, INFLvsPHYS.48.AC10, INFLvsPHYS.48.AC100 |
|
|
| 5. What is the differential impact of stimuli on PHYS? | PHYS.1.DC5vsDC1, PHYS.48.DC5vsDC1, PHYS.1.AC100vsAC10, PHYS.48.AC100vsAC10, PHYS.1.DC5vsAC10, PHYS.1.DC5vsAC100, PHYS.48.DC5vsAC10, PHYS.48.DC5vsAC100, PHYS.1.DC1vsAC10, PHYS.1.DC1vsAC100, PHYS.48.DC1vsAC10, PHYS.48.DC1vsAC100 |
|
|
| 6. What is the differential impact of stimuli on INFL? | INFL.1.DC5vsDC1, INFL.48.DC5vsDC1, INFL.1.AC100vsAC10, INFL.48.AC100vsAC10, INFL.1.DC5vsAC10, INFL.1.DC5vsAC100, INFL.48.DC5vsAC10, INFL.48.DC5vsAC100, INFL.1.DC1vsAC10, INFL.1.DC1vsAC100, INFL.48.DC1vsAC10, INFL.48.DC1vsAC100 |
|
|
| Contrast | RNA | DNAm | t | p-Value | BH p-Value | |
|---|---|---|---|---|---|---|
| INFLvsPHYS.48.NO | ✓ | ✓ | 0.0181 | 10.385 | 0.000883 | 0.0503 |
| PHYS.48.DC1vsNO | ✓ | - | 0.0081 | 3.1856 | 0.0577 | 0.4111 |
| PHYS.48.AC10vsNO | ✓ | - | 0.00565 | 2.6269 | 0.0784 | 0.4965 |
| PHYS.48.DC5vsNO | - | ✓ | 0.00658 | 4.0624 | 0.01736 | 0.9895 |
| INFLvsPHYS.48.DC1 | - | ✓ | −0.012 | −4.1344 | 0.0041 | 0.2341 |
| INFL.48.DC1vsNO | - | ✓ | −0.01356 | −9.429 | 0.01106 | 0.6305 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pietro, B.D.; Villata, S.; Plaksienko, A.; Guarnieri, T.; Monego, S.D.; Degasperi, M.; Lena, P.D.; Licastro, D.; Angelini, C.; Frascella, F.; et al. Modified Methylation Following Electrostimulation in a Standardized Setting—Complementing a Transcriptomic Analysis. Cells 2025, 14, 838. https://doi.org/10.3390/cells14110838
Pietro BD, Villata S, Plaksienko A, Guarnieri T, Monego SD, Degasperi M, Lena PD, Licastro D, Angelini C, Frascella F, et al. Modified Methylation Following Electrostimulation in a Standardized Setting—Complementing a Transcriptomic Analysis. Cells. 2025; 14(11):838. https://doi.org/10.3390/cells14110838
Chicago/Turabian StylePietro, Biagio Di, Simona Villata, Anna Plaksienko, Tiziana Guarnieri, Simeone Dal Monego, Margherita Degasperi, Pietro Di Lena, Danilo Licastro, Claudia Angelini, Francesca Frascella, and et al. 2025. "Modified Methylation Following Electrostimulation in a Standardized Setting—Complementing a Transcriptomic Analysis" Cells 14, no. 11: 838. https://doi.org/10.3390/cells14110838
APA StylePietro, B. D., Villata, S., Plaksienko, A., Guarnieri, T., Monego, S. D., Degasperi, M., Lena, P. D., Licastro, D., Angelini, C., Frascella, F., Napione, L., & Nardini, C. (2025). Modified Methylation Following Electrostimulation in a Standardized Setting—Complementing a Transcriptomic Analysis. Cells, 14(11), 838. https://doi.org/10.3390/cells14110838












