Development of SNAP-Tag Based Nanobodies as Secondary Antibody Mimics for Indirect Immunofluorescence Assays
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Anti-IgG-SNAP Nanobody Fusion Proteins Expression and Purification
2.3. Conjugation of Nbs Anti-IgG-SNAP Fusion Proteins with Benzylguanine-Modified Fluorescent Substrates
2.4. Fluorescence Western Blot
2.5. Flow Cytometry
2.6. Fluorescence Microscopy
2.7. Preparation of Cell Block
2.8. Immunohistochemistry (IHC) of the Cell Blocks
2.9. Multicolor Staining of the Cell Block
2.10. Statistical Analysis
3. Results
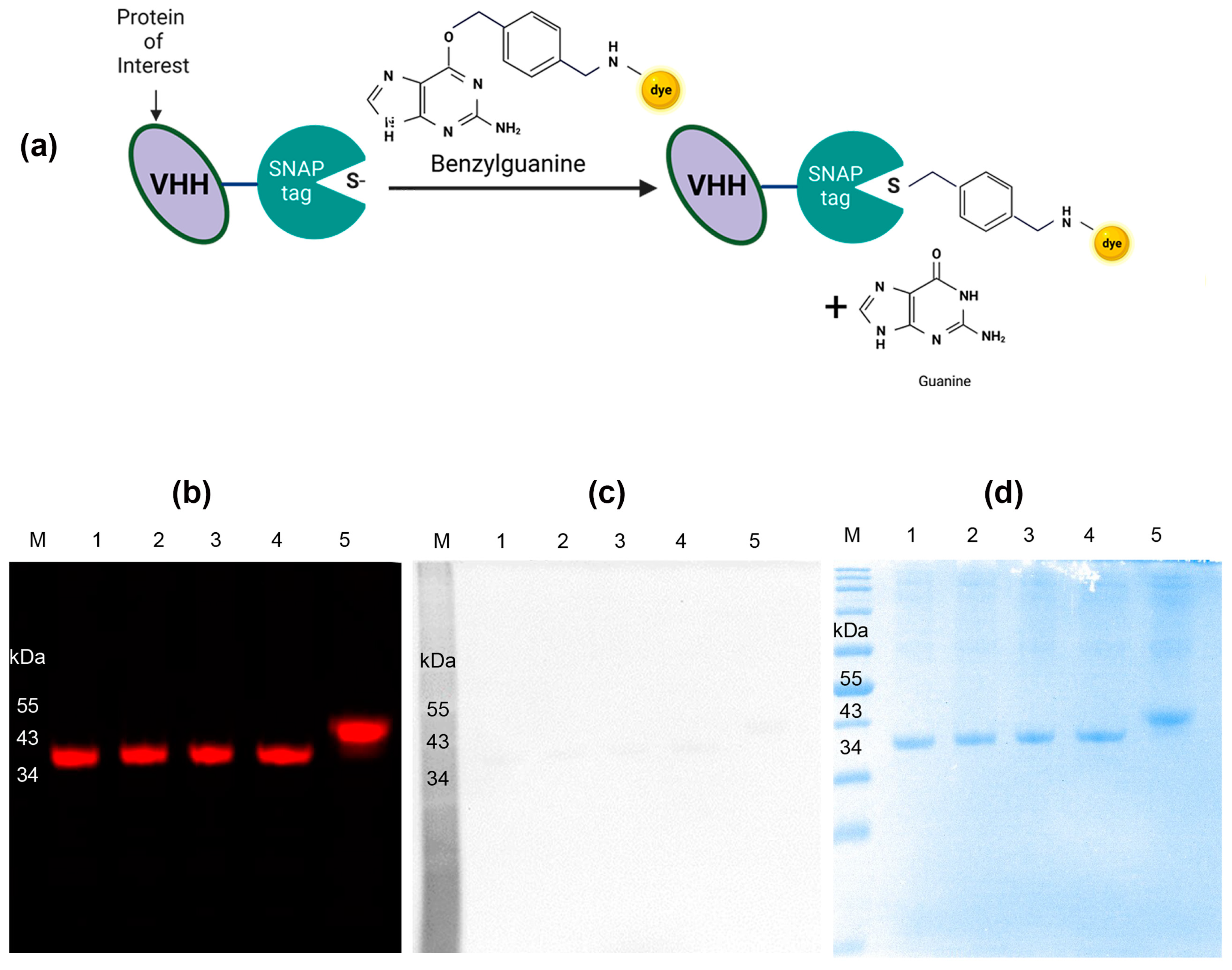
3.1. Production of Nb Anti-IgG-SNAP Fusion Protein
3.2. Conjugation of Nb Anti-IgG-SNAP Fusion Proteins with Benzylguanine-Modified Fluorescent Substrates
3.3. The Specific Binding of Nb Anti-IgG-SNAP Fusion Proteins in Fluorescence Western Blot
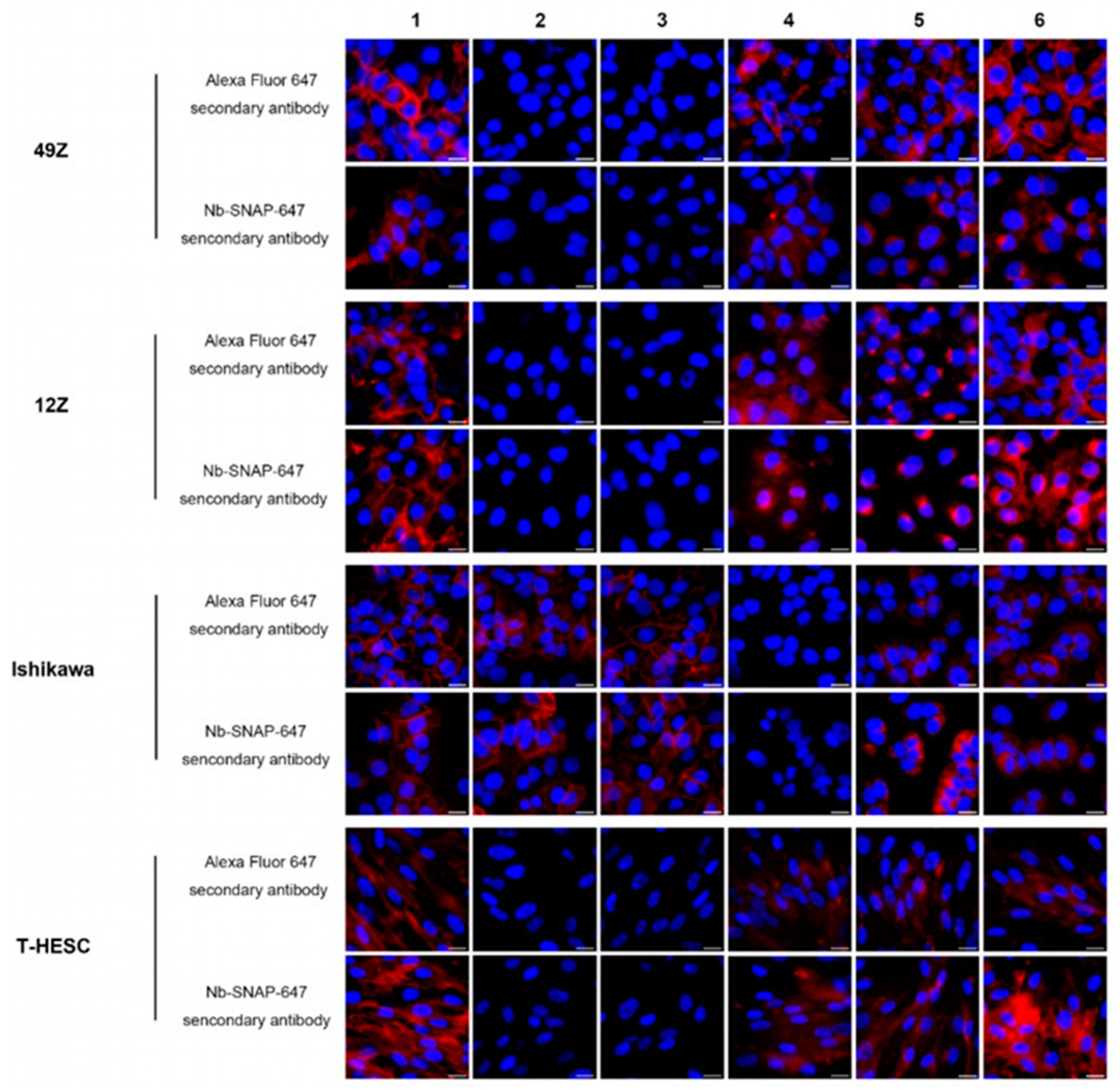
3.4. The Application of Nb Anti-IgG-SNAP Fusion Proteins as Secondary Antibodies for Flow Cytometry and Fluorescence Microscopy for Fixed Cells
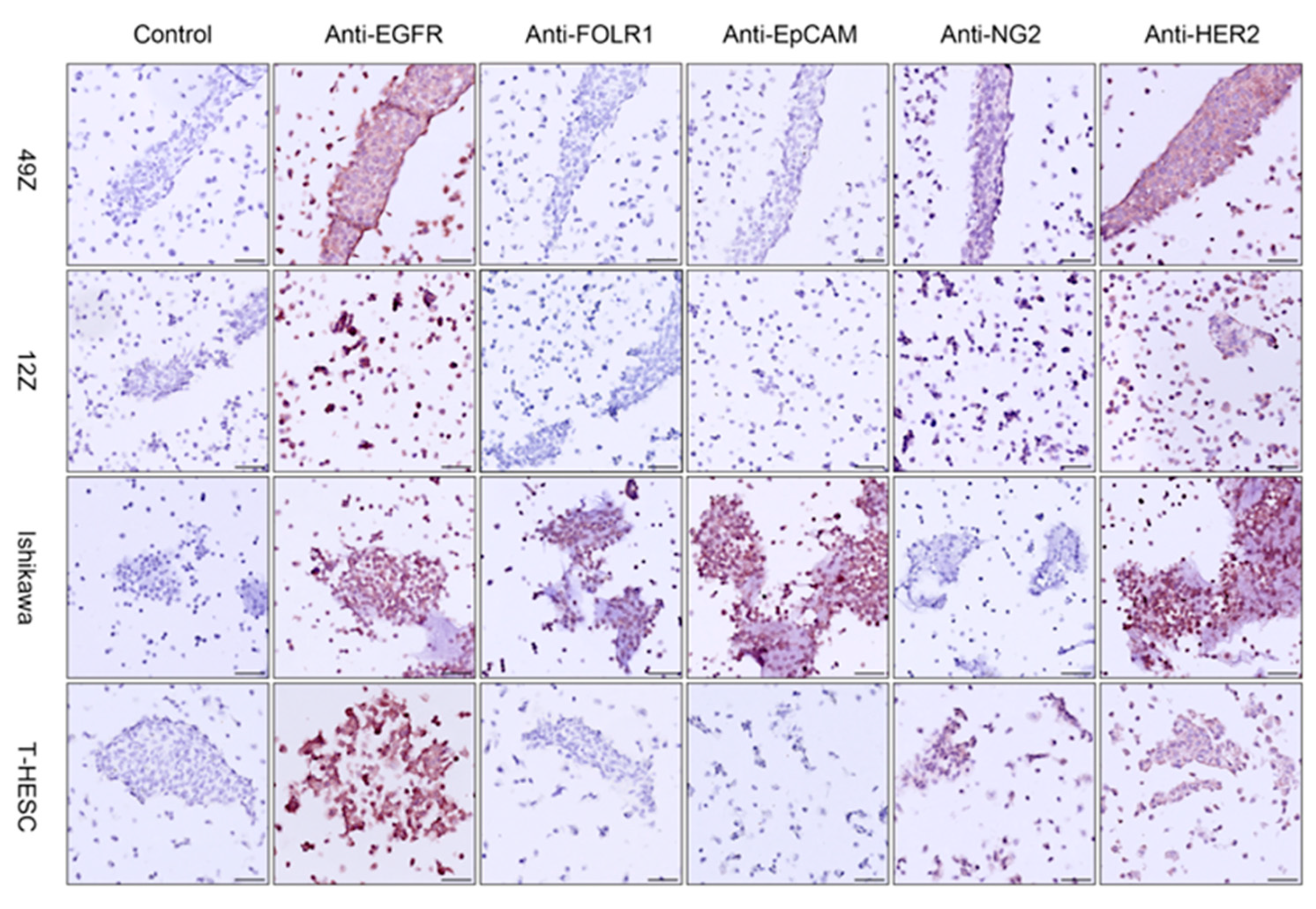
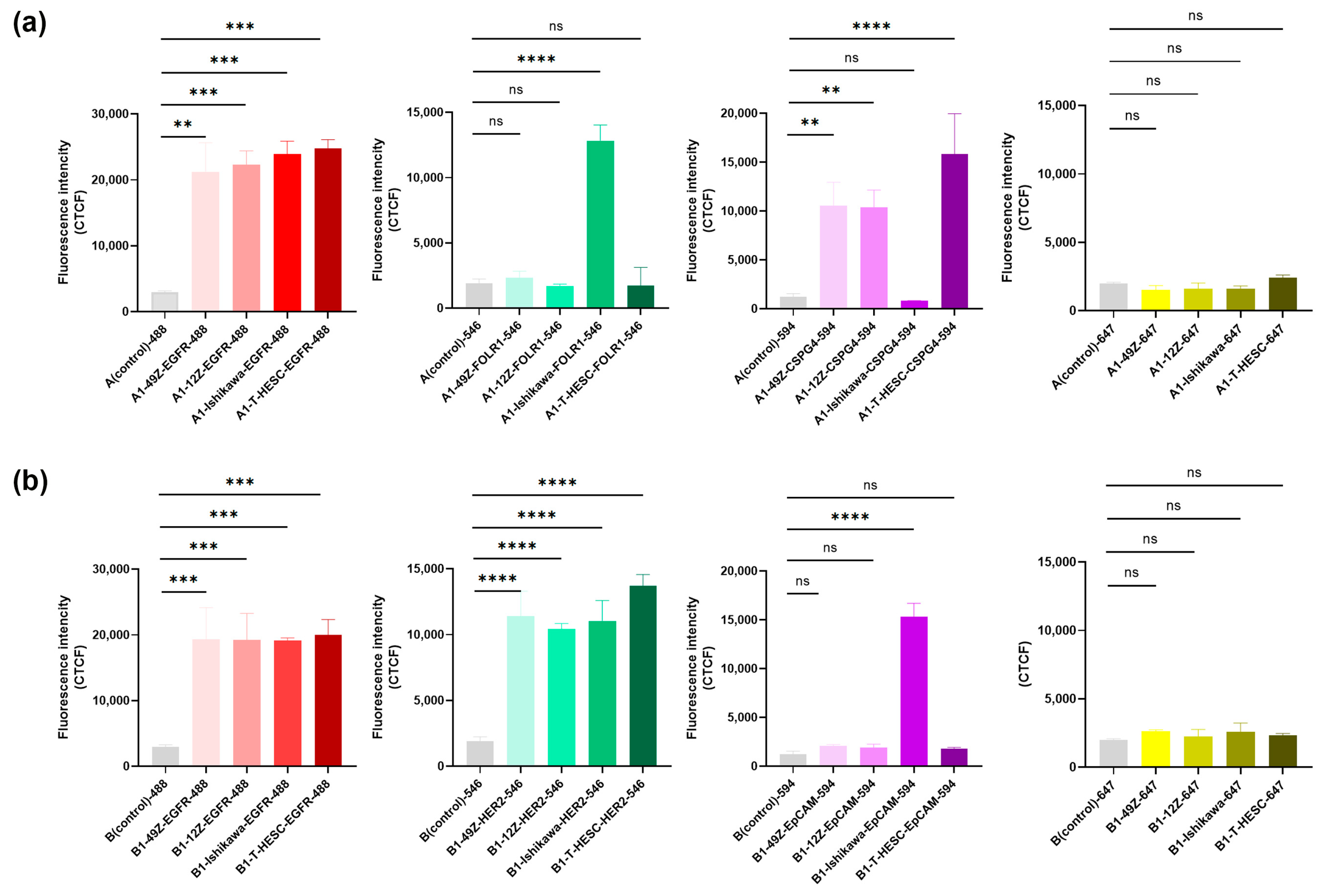
3.5. Immunohistochemistry and Multicolor Immunofluorescence in Cell Blocks
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| FPs | Fluorescent proteins |
| GFP | Green fluorescent protein |
| POI | Protein of interest |
| hAGT | O6-alkylguanine-DNA alkyltransferase |
| BG | Benzylguanine |
| Nb | Nanobody |
| VHH | Variable heavy chain |
| CDRs | Complementarity determining regions |
| IFA | Indirect immunofluorescence assay |
| FBS | Fetal calf serum |
| ITS-X | Insulin-transferrin-selenium-ethanolamine |
| PVDF | Polyvinylidene difluoride |
| IHC | Immunohistochemistry |
| ICC | Instant computational clearing |
| CTCF | Corrected total cell fluorescence |
| PTMs | Post-translational modifications |
| NHS | N-hydroxysuccinimide |
| scFv | Single-chain antibody fragment |
| EGFR | Epidermal growth factor receptor |
| EpCAM | Epithelial cell adhesion molecule |
| NG2 | Nerve/glial-antigen 2 |
| FOLR1 | Folate receptor alpha |
| CTNNB1 | beta Catenin 1 |
| ANK3 | ankyrin 3 |
References
- Tanenbaum, M.E.; Gilbert, L.A.; Qi, L.S.; Weissman, J.S.; Vale, R.D. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 2014, 159, 635–646. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, R.M. The multiple uses of fluorescent proteins to visualize cancer in vivo. Nat. Rev. Cancer 2005, 5, 796–806. [Google Scholar] [CrossRef]
- Giepmans, B.N.; Adams, S.R.; Ellisman, M.H.; Tsien, R.Y. The fluorescent toolbox for assessing protein location and function. Science 2006, 312, 217–224. [Google Scholar] [CrossRef]
- Tsien, R.Y. The green fluorescent protein. Annu. Rev. Biochem. 1998, 67, 509–544. [Google Scholar] [CrossRef]
- Chalfie, M.; Tu, Y.; Euskirchen, G.; Ward, W.W.; Prasher, D.C. Green fluorescent protein as a marker for gene expression. Science 1994, 263, 802–805. [Google Scholar] [CrossRef]
- Shaner, N.C.; Steinbach, P.A.; Tsien, R.Y. A guide to choosing fluorescent proteins. Nat. Methods 2005, 2, 905–909. [Google Scholar] [CrossRef]
- Xia, T.; Li, N.; Fang, X. Single-molecule fluorescence imaging in living cells. Annu. Rev. Phys. Chem. 2013, 64, 459–480. [Google Scholar] [CrossRef]
- Pegg, A.E. Repair of O6-alkylguanine by alkyltransferases. Mutat. Res. /Rev. Mutat. Res. 2000, 462, 83–100. [Google Scholar] [CrossRef]
- Keppler, A.; Kindermann, M.; Gendreizig, S.; Pick, H.; Vogel, H.; Johnsson, K. Labeling of fusion proteins of O6-alkylguanine-DNA alkyltransferase with small molecules in vivo and in vitro. Methods 2004, 32, 437–444. [Google Scholar] [CrossRef]
- Keppler, A.; Pick, H.; Arrivoli, C.; Vogel, H.; Johnsson, K. Labeling of fusion proteins with synthetic fluorophores in live cells. Proc. Natl. Acad. Sci. USA 2004, 101, 9955–9959. [Google Scholar] [CrossRef]
- Srikun, D.; Albers, A.E.; Nam, C.I.; Iavarone, A.T.; Chang, C.J. Organelle-targetable fluorescent probes for imaging hydrogen peroxide in living cells via SNAP-Tag protein labeling. J. Am. Chem. Soc. 2010, 132, 4455–4465. [Google Scholar] [CrossRef] [PubMed]
- Cole, N.B. Site-specific protein labeling with SNAP-tags. Curr. Protoc. Protein Sci. 2013, 73, 30.1.1–30.1.16. [Google Scholar] [CrossRef] [PubMed]
- Kampmeier, F.; Ribbert, M.; Nachreiner, T.; Dembski, S.; Beaufils, F.; Brecht, A.; Barth, S.J.B.c. Site-specific, covalent labeling of recombinant antibody fragments via fusion to an engineered version of 6-O-alkylguanine DNA alkyltransferase. Bioconjugate Chem. 2009, 20, 1010–1015. [Google Scholar] [CrossRef] [PubMed]
- Cortez-Retamozo, V.; Backmann, N.; Senter, P.D.; Wernery, U.; De Baetselier, P.; Muyldermans, S.; Revets, H. Efficient cancer therapy with a nanobody-based conjugate. Cancer Res. 2004, 64, 2853–2857. [Google Scholar] [CrossRef]
- Decanniere, K.; Desmyter, A.; Lauwereys, M.; Ghahroudi, M.A.; Muyldermans, S.; Wyns, L. A single-domain antibody fragment in complex with RNase A: Non-canonical loop structures and nanomolar affinity using two CDR loops. Structure 1999, 7, 361–370. [Google Scholar] [CrossRef]
- Wesolowski, J.; Alzogaray, V.; Reyelt, J.; Unger, M.; Juarez, K.; Urrutia, M.; Cauerhff, A.; Danquah, W.; Rissiek, B.; Scheuplein, F.; et al. Single domain antibodies: Promising experimental and therapeutic tools in infection and immunity. Med. Microbiol. Immunol. 2009, 198, 157–174. [Google Scholar] [CrossRef]
- De Genst, E.; Silence, K.; Decanniere, K.; Conrath, K.; Loris, R.; Kinne, J.; Muyldermans, S.; Wyns, L. Molecular basis for the preferential cleft recognition by dromedary heavy-chain antibodies. Proc. Natl. Acad. Sci. USA 2006, 103, 4586–4591. [Google Scholar] [CrossRef]
- Decanniere, K.; Muyldermans, S.; Wyns, L. Canonical antigen-binding loop structures in immunoglobulins: More structures, more canonical classes? J. Mol. Biol. 2000, 300, 83–91. [Google Scholar] [CrossRef]
- Herce, H.D.; Deng, W.; Helma, J.; Leonhardt, H.; Cardoso, M.C. Visualization and targeted disruption of protein interactions in living cells. Nat. Commun. 2013, 4, 2660. [Google Scholar] [CrossRef]
- Mujic-Delic, A.; de Wit, R.H.; Verkaar, F.; Smit, M.J. GPCR-targeting nanobodies: Attractive research tools, diagnostics, and therapeutics. Trends Pharmacol. Sci. 2014, 35, 247–255. [Google Scholar] [CrossRef]
- Gray, A.; Bradbury, A.R.M.; Knappik, A.; Pluckthun, A.; Borrebaeck, C.A.K.; Dubel, S. Animal-free alternatives and the antibody iceberg. Nat. Biotechnol. 2020, 38, 1234–1239. [Google Scholar] [CrossRef] [PubMed]
- Gorovits, B.; Hays, A.; Jani, D.; Jones, C.; King, C.; Lundequist, A.; Mora, J.; Partridge, M.; Pathania, D.; Ramaswamy, S.S.; et al. AAPS Perspective on the EURL Recommendation on the use of Non-Animal-Derived Antibodies. AAPS J. 2021, 23, 34. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.J.N.N. Discovery of goat facility adds to antibody provider’s woes. Nature 2013. [Google Scholar] [CrossRef]
- Pleiner, T.; Bates, M.; Gorlich, D. A toolbox of anti-mouse and anti-rabbit IgG secondary nanobodies. J. Cell Biol. 2018, 217, 1143–1154. [Google Scholar] [CrossRef]
- Hussain, A.F.; Heppenstall, P.A.; Kampmeier, F.; Meinhold-Heerlein, I.; Barth, S. One-step site-specific antibody fragment auto-conjugation using SNAP-tag technology. Nat. Protoc. 2019, 14, 3101–3125. [Google Scholar] [CrossRef]
- Zhang, C.; Sheng, W.; Al-Rawe, M.; Mohiuddin, T.M.; Niebert, M.; Zeppernick, F.; Meihold-Heerlein, I.; Hussain, A.F. EpCAM- and EGFR-Specific Antibody Drug Conjugates for Triple-Negative Breast Cancer Treatment. Int. J. Mol. Sci. 2022, 23, 6122. [Google Scholar] [CrossRef]
- Varghese, F.; Bukhari, A.B.; Malhotra, R.; De, A. IHC Profiler: An open source plugin for the quantitative evaluation and automated scoring of immunohistochemistry images of human tissue samples. PLoS ONE 2014, 9, e96801. [Google Scholar] [CrossRef]
- EC. Directive 2010/63/EU of the European Parliament and of the Council of 22 September 2010 on the protection of animals used for scientific purposes. Off. J. Eur. Union 2010, 50, 33–79. [Google Scholar]
- Kunert, R.; Reinhart, D. Advances in recombinant antibody manufacturing. Appl. Microbiol. Biotechnol. 2016, 100, 3451–3461. [Google Scholar] [CrossRef]
- Liu, Y.; Huang, H. Expression of single-domain antibody in different systems. Appl. Microbiol. Biotechnol. 2018, 102, 539–551. [Google Scholar] [CrossRef]
- Muyldermans, S. Single domain camel antibodies: Current status. J. Biotechnol. 2001, 74, 277–302. [Google Scholar] [CrossRef] [PubMed]
- Virdi, V.; Coddens, A.; De Buck, S.; Millet, S.; Goddeeris, B.M.; Cox, E.; De Greve, H.; Depicker, A. Orally fed seeds producing designer IgAs protect weaned piglets against enterotoxigenic Escherichia coli infection. Proc. Natl. Acad. Sci. USA 2013, 110, 11809–11814. [Google Scholar] [CrossRef] [PubMed]
- Lentz, E.M.; Garaicoechea, L.; Alfano, E.F.; Parreno, V.; Wigdorovitz, A.; Bravo-Almonacid, F.F. Translational fusion and redirection to thylakoid lumen as strategies to improve the accumulation of a camelid antibody fragment in transplastomic tobacco. Planta 2012, 236, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Kooijmans, S.A.A.; Gitz-Francois, J.; Schiffelers, R.M.; Vader, P. Recombinant phosphatidylserine-binding nanobodies for targeting of extracellular vesicles to tumor cells: A plug-and-play approach. Nanoscale 2018, 10, 2413–2426. [Google Scholar] [CrossRef]
- Dhara, V.G.; Naik, H.M.; Majewska, N.I.; Betenbaugh, M.J. Recombinant Antibody Production in CHO and NS0 Cells: Differences and Similarities. BioDrugs 2018, 32, 571–584. [Google Scholar] [CrossRef]
- Kelley, B.; Kiss, R.; Laird, M. A Different Perspective: How Much Innovation Is Really Needed for Monoclonal Antibody Production Using Mammalian Cell Technology? Adv. Biochem. Eng. Biotechnol. 2018, 165, 443–462. [Google Scholar] [CrossRef]
- Aricescu, A.R.; Lu, W.; Jones, E.Y. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr. D Biol. Crystallogr. 2006, 62, 1243–1250. [Google Scholar] [CrossRef]
- Tan, E.; Chin, C.S.H.; Lim, Z.F.S.; Ng, S.K. HEK293 Cell Line as a Platform to Produce Recombinant Proteins and Viral Vectors. Front. Bioeng. Biotechnol. 2021, 9, 796991. [Google Scholar] [CrossRef]
- Alexander, E.; Leong, K.W. Discovery of nanobodies: A comprehensive review of their applications and potential over the past five years. J. Nanobiotechnol. 2024, 22, 661. [Google Scholar] [CrossRef]
- Fridy, P.C.; Li, Y.; Keegan, S.; Thompson, M.K.; Nudelman, I.; Scheid, J.F.; Oeffinger, M.; Nussenzweig, M.C.; Fenyö, D.; Chait, B.T.; et al. A robust pipeline for rapid production of versatile nanobody repertoires. Nature Methods 2014, 11, 1253–1260. [Google Scholar] [CrossRef]
- Massa, S.; Vikani, N.; Betti, C.; Ballet, S.; Vanderhaegen, S.; Steyaert, J.; Descamps, B.; Vanhove, C.; Bunschoten, A.; van Leeuwen, F.W. Sortase A-mediated site-specific labeling of camelid single-domain antibody-fragments: A versatile strategy for multiple molecular imaging modalities. Contrast Media Mol. Imaging 2016, 11, 328–339. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, D.; Helma, J.; Schneider, A.F.L.; Leonhardt, H.; Hackenberger, C.P.R. Nanobodies: Chemical Functionalization Strategies and Intracellular Applications. Angew. Chem. Int. Ed. Engl. 2018, 57, 2314–2333. [Google Scholar] [CrossRef] [PubMed]
- Pleiner, T.; Bates, M.; Trakhanov, S.; Lee, C.T.; Schliep, J.E.; Chug, H.; Bohning, M.; Stark, H.; Urlaub, H.; Gorlich, D. Correction: Nanobodies: Site-specific labeling for super-resolution imaging, rapid epitope-mapping and native protein complex isolation. Elife 2016, 5, 2314–2333. [Google Scholar] [CrossRef] [PubMed]
- Fung, H.Y.J.; McKibben, K.M.; Ramirez, J.; Gupta, K.; Rhoades, E. Structural Characterization of Tau in Fuzzy Tau:Tubulin Complexes. Structure 2020, 28, 378–384.e4. [Google Scholar] [CrossRef]
- Morgan, H.E.; Turnbull, W.B.; Webb, M.E. Challenges in the use of sortase and other peptide ligases for site-specific protein modification. Chem. Soc. Rev. 2022, 51, 4121–4145. [Google Scholar] [CrossRef]
- Wilhelm, J.; Kuhn, S.; Tarnawski, M.; Gotthard, G.; Tunnermann, J.; Tanzer, T.; Karpenko, J.; Mertes, N.; Xue, L.; Uhrig, U.; et al. Kinetic and Structural Characterization of the Self-Labeling Protein Tags HaloTag7, SNAP-tag, and CLIP-tag. Biochemistry 2021, 60, 2560–2575. [Google Scholar] [CrossRef]
- Gostner, J.M.; Fong, D.; Wrulich, O.A.; Lehne, F.; Zitt, M.; Hermann, M.; Krobitsch, S.; Martowicz, A.; Gastl, G.; Spizzo, G. Effects of EpCAM overexpression on human breast cancer cell lines. BMC Cancer 2011, 11, 45. [Google Scholar] [CrossRef]
- Wang, X.; Osada, T.; Wang, Y.; Yu, L.; Sakakura, K.; Katayama, A.; McCarthy, J.B.; Brufsky, A.; Chivukula, M.; Khoury, T.; et al. CSPG4 protein as a new target for the antibody-based immunotherapy of triple-negative breast cancer. J. Natl. Cancer Inst. 2010, 102, 1496–1512. [Google Scholar] [CrossRef]
- Jin, L.L.; Lu, H.J.; Shao, J.K.; Wang, Y.; Lu, S.P.; Huang, B.F.; Hu, G.N.; Jin, H.C.; Wang, C.Q. Relevance and mechanism of STAT3/miR-221-3p/Fascin-1 axis in EGFR TKI resistance of triple-negative breast cancer. Mol. Cell Biochem. 2024, 479, 3037–3047. [Google Scholar] [CrossRef]
- McKnight, B.N.; Kim, S.; Boerner, J.L.; Viola, N.T. Cetuximab PET delineated changes in cellular distribution of EGFR upon dasatinib treatment in triple negative breast cancer. Breast Cancer Res. 2020, 22, 37. [Google Scholar] [CrossRef]
- (n.d.), The Human Protein Atlas. ANK3 in Cancer. Available online: https://www.proteinatlas.org/ENSG00000151150-ANK3/cancer (accessed on 1 February 2025).
- Kurozumi, S.; Joseph, C.; Raafat, S.; Sonbul, S.; Kariri, Y.; Alsaeed, S.; Pigera, M.; Alsaleem, M.; Nolan, C.C.; Johnston, S.J.; et al. Utility of ankyrin 3 as a prognostic marker in androgen-receptor-positive breast cancer. Breast Cancer Res. Treat. 2019, 176, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Alkaraki, A.; Alshaer, W.; Wehaibi, S.; Gharaibeh, L.; Abuarqoub, D.; Alqudah, D.A.; Al-Azzawi, H.; Zureigat, H.; Souleiman, M.; Awidi, A. Enhancing chemosensitivity of wild-type and drug-resistant MDA-MB-231 triple-negative breast cancer cell line to doxorubicin by silencing of STAT 3, Notch-1, and beta-catenin genes. Breast Cancer 2020, 27, 989–998. [Google Scholar] [CrossRef] [PubMed]
- Jia, T.; Zhang, L.; Duan, Y.; Zhang, M.; Wang, G.; Zhang, J.; Zhao, Z. The differential susceptibilities of MCF-7 and MDA-MB-231 cells to the cytotoxic effects of curcumin are associated with the PI3K/Akt-SKP2-Cip/Kips pathway. Cancer Cell Int. 2014, 14, 126. [Google Scholar] [CrossRef]
- Vess, A.; Blache, U.; Leitner, L.; Kurz, A.R.M.; Ehrenpfordt, A.; Sixt, M.; Posern, G. A dual phenotype of MDA-MB-468 cancer cells reveals mutual regulation of tensin3 and adhesion plasticity. J. Cell Sci. 2017, 130, 2172–2184. [Google Scholar] [CrossRef]
- Albitar, L.; Pickett, G.; Morgan, M.; Wilken, J.A.; Maihle, N.J.; Leslie, K.K. EGFR isoforms and gene regulation in human endometrial cancer cells. Mol. Cancer 2010, 9, 166. [Google Scholar] [CrossRef]
- Gellersen, B.; Wolf, A.; Kruse, M.; Schwenke, M.; Bamberger, A.M. Human endometrial stromal cell-trophoblast interactions: Mutual stimulation of chemotactic migration and promigratory roles of cell surface molecules CD82 and CEACAM1. Biol. Reprod. 2013, 88, 80. [Google Scholar] [CrossRef]
- Grimm, A.; Hussain, A.; Sheng, W.; Zhang, C.; Al-Rawe, M.; Meinhold-Heerlein, I. Development and evaluation of antibody-fluorophore-conjugates to detect endometrial tissue. Geburtshilfe Und Frauenheilkd. 2022, 82, 107. [Google Scholar]
- O’Shannessy, D.J.; Somers, E.B.; Smale, R.; Fu, Y.S. Expression of folate receptor-alpha (FRA) in gynecologic malignancies and its relationship to the tumor type. Int. J. Gynecol. Pathol. 2013, 32, 258–268. [Google Scholar] [CrossRef]
- Wen, K.C.; Sung, P.L.; Chou, Y.T.; Pan, C.M.; Wang, P.H.; Lee, O.K.; Wu, C.W. The role of EpCAM in tumor progression and the clinical prognosis of endometrial carcinoma. Gynecol. Oncol. 2018, 148, 383–392. [Google Scholar] [CrossRef]
- Gronemeyer, T.; Godin, G.; Johnsson, K. Adding value to fusion proteins through covalent labelling. Curr. Opin. Biotechnol. 2005, 16, 453–458. [Google Scholar] [CrossRef]
- Sograte-Idrissi, S.; Schlichthaerle, T.; Duque-Afonso, C.J.; Alevra, M.; Strauss, S.; Moser, T.; Jungmann, R.; Rizzoli, S.O.; Opazo, F. Correction: Circumvention of common labelling artefacts using secondary nanobodies. Nanoscale 2020, 12, 24543. [Google Scholar] [CrossRef] [PubMed]
- Bales, C.E. Laboratory techniques. In Koss’s Diagnostic Cytology and Its Histopathologic Bases; Lippincott Williams & Wilkins (LWW): Philadelphia, PA, USA, 2006; pp. 1569–1634. [Google Scholar]
- Maier, J.; Traenkle, B.; Rothbauer, U. Real-time analysis of epithelial-mesenchymal transition using fluorescent single-domain antibodies. Sci. Rep. 2015, 5, 13402. [Google Scholar] [CrossRef] [PubMed]
- Braun, M.B.; Traenkle, B.; Koch, P.A.; Emele, F.; Weiss, F.; Poetz, O.; Stehle, T.; Rothbauer, U. Peptides in headlock--a novel high-affinity and versatile peptide-binding nanobody for proteomics and microscopy. Sci. Rep. 2016, 6, 19211. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.H.; Kim, S.F.; Liu, Y.; Zhang, X. A Fluorogenic AggTag Method Based on Halo- and SNAP-Tags to Simultaneously Detect Aggregation of Two Proteins in Live Cells. Chembiochem 2019, 20, 1078–1087. [Google Scholar] [CrossRef]
- Gautier, A.; Nakata, E.; Lukinavicius, G.; Tan, K.T.; Johnsson, K. Selective cross-linking of interacting proteins using self-labeling tags. J. Am. Chem. Soc. 2009, 131, 17954–17962. [Google Scholar] [CrossRef]
- Pellett, P.A.; Sun, X.; Gould, T.J.; Rothman, J.E.; Xu, M.Q.; Correa, I.R., Jr.; Bewersdorf, J. Two-color STED microscopy in living cells. Biomed. Opt. Express 2011, 2, 2364–2371. [Google Scholar] [CrossRef]



| Plasmid name | Catalog no. | Features |
| pTP943 | 104157 | Anti-mouse IgG1 Fab specific nanobody TP886 |
| pTP1112 | 104158 | Anti-mouse IgG1 Fc specific nanobody TP1107 |
| pTP1005 | 104160 | Anti-mouse IgG2a Fc specific nanobody TP1129 |
| pTP1174 | 104162 | Anti-mouse kappa (κ) chain specific nanobody TP1170 |
| pTP1183 | 104163 | Anti-rabbit IgG Fc specific nanobody TP897 |
| Antibody | Class and Isotype | Dilution | Supplier | Catalog-No |
|---|---|---|---|---|
| Anti-EGFR | Monoclonal mouse/IgG2a | IF: 1:200 Multi-IF: 1:20 | Invitrogen, Schwerte, Germany | MA5-13319 |
| Anti-EGFR | Recombinant monoclonal rabbit/IgG | IF: 1:100 | Invitrogen, Schwerte, Germany | 700308 |
| Anti-EGFR | Polyclonal rabbit/IgG | IHC: 1:200 Multi-IF: 1:200 | Sigma-Aldrich, Taufkirchen, Germany | HPA018530 |
| Anti-EpCAM | Monoclonal mouse/IgG1, kappa | IF: 1:200 IHC: 1:50 Multi-IF: 1:50 | Invitrogen, Schwerte, Germany | 14-9326-82 |
| Anti-NG2 | Monoclonal mouse/IgG1, kappa | IF: 1:100 IHC: 1:100 Multi-IF: 1:100 | Invitrogen, Schwerte, Germany | 37-2700 |
| Anti-HER2 | Monoclonal Mouse/IgG1 | IF: 1:100 IHC: 1:500 Multi-IF: 1:500 | Invitrogen, Schwerte, Germany | MA5-13675 |
| Anti-FOLR1 | Monoclonal mouse/IgG1 | IF: 1:100 IHC: 1:50 Multi-IF: 1:50 | Invitrogen, Schwerte, Germany | MA5-23917 |
| Anti-CTNNB1 | Polyclonal rabbit/IgG | IF: 1:200 | Sigma-Aldrich, Taufkirchen, Germany | HPA029159 |
| Anti-ANK3 | Polyclonal rabbit/IgG | IF: 1:100 | Sigma-Aldrich, Taufkirchen, Germany | HPA038455 |
| Panel A | Panel B | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Staining Sequence | Antibody | A1 | A2 | A3 | A4 | A Control | Antibody | B1 | B2 | B3 | B4 | B Control |
| 1 | Anti-EGFR (mouse) | + 1 | − 2 | + | + | − | Anti-EGFR (rabbit) | + | − | + | + | − |
| 2 | Nb anti-mouse IgG2a Fc-SNAP-488 | + | + | + | + | + | Nb anti-rabbit IgG Fc-488 | + | + | + | + | + |
| 3 | Nb anti-mouse IgG2a Fc-SNAP-647 | + | + | + | + | + | Nb anti-rabbit IgG Fc-647 | + | + | + | + | + |
| 4 | Anti-FOLR1 | + | + | − | + | − | Anti-HER2 | + | + | − | + | − |
| 5 | Nb anti-mouse IgG1 Fab-SNAP-546 | + | + | + | + | + | Nb anti-mouse IgG1 Fab-SNAP-546 | + | + | + | + | + |
| 6 | Nb anti-mouse IgG1 Fab-SNAP-647 | + | + | + | + | + | Nb anti-mouse IgG1 Fab-SNAP-647 | + | + | + | + | + |
| 7 | Anti-NG2 | + | + | + | − | − | Anti-EpCAM | + | + | + | − | − |
| 8 | Nb anti-mouse IgG1 Fc-SNAP-594 | + | + | + | + | + | Nb anti-mouse kappa chain-SNAP-594 | + | + | + | + | + |
| 9 | Nb anti-mouse IgG1 Fc-SNAP-647 | + | + | + | + | + | Nb anti-mouse kappa chain-SNAP-647 | + | + | + | + | + |
| Target | 49Z | 12Z | Ishikawa | T-HESC |
|---|---|---|---|---|
| EGFR | high | high | high | high |
| HER2 | high | high | high | high |
| NG2 | medium | medium | low | medium |
| EpCAM | low | low | high | low |
| FOLR1 | low | low | high | low |
| Panel A | Panel B | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Endometrial cells | Slide | 488 anti-EGFR | 546 anti-FOLR1 | 594 anti-NG2 | 647 channel | Slide | 488 anti-EGFR | 546 anti-HER2 | 594 anti-EpCAM | 647 channel |
| 49Z | A1 | + 1 | − 2 | + | − | B1 | + | + | − | − |
| A2 | − | − | + | − | B2 | − | + | − | − | |
| A3 | + | − | + | − | B3 | + | − | − | − | |
| A4 | + | − | − | − | B4 | + | + | − | − | |
| A control | − | − | − | − | B control | − | − | − | − | |
| 12Z | A1 | + | − | + | − | B1 | + | + | − | − |
| A2 | - | − | + | − | B2 | − | + | − | − | |
| A3 | + | − | + | − | B3 | + | − | − | − | |
| A4 | + | − | − | − | B4 | + | + | − | − | |
| A control | - | − | − | − | B control | − | − | − | − | |
| Ishikawa | A1 | + | + | − | − | B1 | + | + | + | − |
| A2 | − | + | − | − | B2 | − | + | + | − | |
| A3 | + | − | − | − | B3 | + | − | + | − | |
| A4 | + | + | − | − | B4 | + | + | − | − | |
| A control | − | − | − | − | B control | − | − | − | − | |
| T-HESC | A1 | + | − | + | − | B1 | + | + | − | − |
| A2 | − | − | + | − | B2 | − | + | − | − | |
| A3 | + | − | + | − | B3 | + | − | − | − | |
| A4 | + | − | − | − | B4 | + | + | − | − | |
| A control | − | − | − | − | B control | − | − | − | − | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sheng, W.; Zhang, C.; Mohiuddin, T.M.; Al-Rawe, M.; Schmitz, R.; Niebert, M.; Konrad, L.; Wagner, S.; Zeppernick, F.; Meinhold-Heerlein, I.; et al. Development of SNAP-Tag Based Nanobodies as Secondary Antibody Mimics for Indirect Immunofluorescence Assays. Cells 2025, 14, 691. https://doi.org/10.3390/cells14100691
Sheng W, Zhang C, Mohiuddin TM, Al-Rawe M, Schmitz R, Niebert M, Konrad L, Wagner S, Zeppernick F, Meinhold-Heerlein I, et al. Development of SNAP-Tag Based Nanobodies as Secondary Antibody Mimics for Indirect Immunofluorescence Assays. Cells. 2025; 14(10):691. https://doi.org/10.3390/cells14100691
Chicago/Turabian StyleSheng, Wenjie, Chaoyu Zhang, T. M. Mohiuddin, Marwah Al-Rawe, Roland Schmitz, Marcus Niebert, Lutz Konrad, Steffen Wagner, Felix Zeppernick, Ivo Meinhold-Heerlein, and et al. 2025. "Development of SNAP-Tag Based Nanobodies as Secondary Antibody Mimics for Indirect Immunofluorescence Assays" Cells 14, no. 10: 691. https://doi.org/10.3390/cells14100691
APA StyleSheng, W., Zhang, C., Mohiuddin, T. M., Al-Rawe, M., Schmitz, R., Niebert, M., Konrad, L., Wagner, S., Zeppernick, F., Meinhold-Heerlein, I., & Hussain, A. F. (2025). Development of SNAP-Tag Based Nanobodies as Secondary Antibody Mimics for Indirect Immunofluorescence Assays. Cells, 14(10), 691. https://doi.org/10.3390/cells14100691









