Advances and Challenges in Sepsis Management: Modern Tools and Future Directions
Abstract
1. Introduction
2. Sepsis Pathophysiology: A Multi-Level Perspective
2.1. Molecular and Immune Mechanisms in Sepsis
2.2. Sepsis Pathophysiology across Organ Systems
- Central to this process is the cardiovascular system, which undergoes significant changes during the progression of sepsis from localized infection to severe systemic inflammation and septic shock. Despite normal or increased cardiac output, patients with sepsis often experience acute biventricular dysfunction and elevated lactate levels, indicating a critical imbalance in tissue oxygenation and metabolic dysfunction [40,41,42,43,44].
- At the endothelial level, sepsis induces profound alterations, such as increased leukocyte adhesion, a shift to a procoagulant state, and compromised barrier function, leading to tissue oedema and microvascular disturbances [19,45,46]. These changes, collectively described as endothelial dysfunction, coupled with widespread tissue factor expression and impaired anticoagulant mechanisms, can culminate in disseminated intravascular coagulation (DIC), further exacerbating organ dysfunction, and increasing mortality risk [47].
- In the liver, sepsis impairs crucial functions, including the clearance of bilirubin and processing of pathogen lipids, which intensifies systemic inflammation [48]. Septic acute kidney injury (AKI) involves cytokine and immune-mediated microvascular and tubular dysfunction, rather than mere hypoperfusion or tubular necrosis [49,50,51,52,53].
3. Biomarkers in Sepsis
3.1. Innate Immunity
3.1.1. Neutrophils
3.1.2. Monocytes
3.1.3. Natural Killer (NK) Cells
3.1.4. γδ. T Cells and MAIT Cells
3.2. Adaptive Immunity
3.2.1. T Lymphocytes
3.2.2. B Lymphocytes
3.3. Metabolic Shifts in Immune Cells
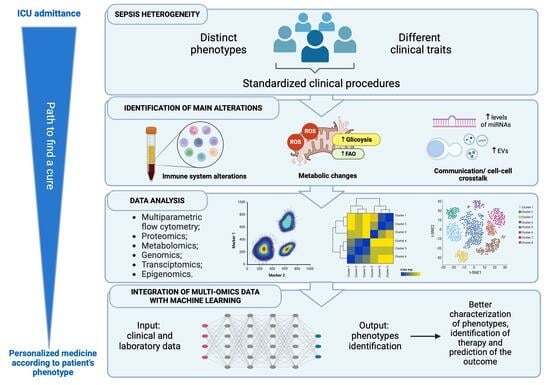
4. Sepsis Therapy: Phenotyping and Personalized Approaches
5. Omics Technologies
6. Future Directions
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Guarino, M.; Perna, B.; Cesaro, A.E.; Maritati, M.; Spampinato, M.D.; Contini, C.; De Giorgio, R. 2023 Update on Sepsis and Septic Shock in Adult Patients: Management in the Emergency Department. J. Clin. Med. 2023, 12, 3188. [Google Scholar] [CrossRef]
- Singer, M.; Deutschman, C.S.; Seymour, C.W.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.-D.; Coopersmith, C.M.; et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016, 315, 801–810. [Google Scholar] [CrossRef]
- Cavaillon, J.-M.; Singer, M.; Skirecki, T. Sepsis Therapies: Learning from 30 Years of Failure of Translational Research to Propose New Leads. EMBO Mol. Med. 2020, 12, e10128. [Google Scholar] [CrossRef]
- de Grooth, H.-J.; Postema, J.; Loer, S.A.; Parienti, J.-J.; Oudemans-van Straaten, H.M.; Girbes, A.R. Unexplained Mortality Differences between Septic Shock Trials: A Systematic Analysis of Population Characteristics and Control-Group Mortality Rates. Intensive Care Med. 2018, 44, 311–322. [Google Scholar] [CrossRef]
- Global Report on the Epidemiology and Burden of Sepsis: Current Evidence, Identifying Gaps and Future Directions—World|ReliefWeb. Available online: https://reliefweb.int/report/world/global-report-epidemiology-and-burden-sepsis-current-evidence-identifying-gaps-and (accessed on 9 January 2024).
- Schlapbach, L.J.; Kissoon, N.; Alhawsawi, A.; Aljuaid, M.H.; Daniels, R.; Gorordo-Delsol, L.A.; Machado, F.; Malik, I.; Nsutebu, E.F.; Finfer, S.; et al. World Sepsis Day: A Global Agenda to Target a Leading Cause of Morbidity and Mortality. Am. J. Physiol. Lung Cell. Mol. Physiol. 2020, 319, L518–L522. [Google Scholar] [CrossRef]
- Seymour, C.W.; Rea, T.D.; Kahn, J.M.; Walkey, A.J.; Yealy, D.M.; Angus, D.C. Severe Sepsis in Pre-Hospital Emergency Care: Analysis of Incidence, Care, and Outcome. Am. J. Respir. Crit. Care Med. 2012, 186, 1264–1271. [Google Scholar] [CrossRef]
- Chiu, C.; Legrand, M. Epidemiology of Sepsis and Septic Shock. Curr. Opin. Anaesthesiol. 2021, 34, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Rudd, K.E.; Johnson, S.C.; Agesa, K.M.; Shackelford, K.A.; Tsoi, D.; Kievlan, D.R.; Colombara, D.V.; Ikuta, K.S.; Kissoon, N.; Finfer, S.; et al. Global, Regional, and National Sepsis Incidence and Mortality, 1990–2017: Analysis for the Global Burden of Disease Study. Lancet 2020, 395, 200–211. [Google Scholar] [CrossRef] [PubMed]
- Brunkhorst, F.M.; Reinhart, K. Diagnostic Approach to Sepsis—State of the Art. In Sepsis and Organ Dysfunction; Baue, A.E., Berlot, G., Gullo, A., Vincent, J.-L., Eds.; Springer: Milano, Italy, 2002; pp. 151–167. [Google Scholar] [CrossRef]
- Suarez-de-la-Rica, A.; Maseda, E. Precision Medicine in Sepsis and Septic Shock. J. Clin. Med. 2022, 11, 5332. [Google Scholar] [CrossRef] [PubMed]
- Fohner, A.E.; Greene, J.D.; Lawson, B.L.; Chen, J.H.; Kipnis, P.; Escobar, G.J.; Liu, V.X. Assessing Clinical Heterogeneity in Sepsis through Treatment Patterns and Machine Learning. J. Am. Med. Inform. Assoc. 2019, 26, 1466–1477. [Google Scholar] [CrossRef] [PubMed]
- Rubio, I.; Osuchowski, M.F.; Shankar-Hari, M.; Skirecki, T.; Winkler, M.S.; Lachmann, G.; La Rosée, P.; Monneret, G.; Venet, F.; Bauer, M.; et al. Current Gaps in Sepsis Immunology: New Opportunities for Translational Research. Lancet Infect. Dis. 2019, 19, e422–e436. [Google Scholar] [CrossRef]
- Vakkalanka, J.P.; Harland, K.K.; Swanson, M.B.; Mohr, N.M. Clinical and Epidemiological Variability in Severe Sepsis: An Ecological Study. J. Epidemiol. Community Health 2018, 72, 741–745. [Google Scholar] [CrossRef]
- Brun-Buisson, C.; Doyon, F.; Carlet, J.; Dellamonica, P.; Gouin, F.; Lepoutre, A.; Mercier, J.C.; Offenstadt, G.; Régnier, B. Incidence, Risk Factors, and Outcome of Severe Sepsis and Septic Shock in Adults. A Multicenter Prospective Study in Intensive Care Units. French ICU Group for Severe Sepsis. JAMA 1995, 274, 968–974. [Google Scholar] [CrossRef] [PubMed]
- Tolsma, V.; Schwebel, C.; Azoulay, E.; Darmon, M.; Souweine, B.; Vesin, A.; Goldgran-Toledano, D.; Lugosi, M.; Jamali, S.; Cheval, C.; et al. Sepsis Severe or Septic Shock: Outcome According to Immune Status and Immunodeficiency Profile. Chest 2014, 146, 1205–1213. [Google Scholar] [CrossRef]
- Williams, M.D.; Braun, L.A.; Cooper, L.M.; Johnston, J.; Weiss, R.V.; Qualy, R.L.; Linde-Zwirble, W. Hospitalized Cancer Patients with Severe Sepsis: Analysis of Incidence, Mortality, and Associated Costs of Care. Crit. Care 2004, 8, R291–R298. [Google Scholar] [CrossRef]
- Sørensen, T.I.; Nielsen, G.G.; Andersen, P.K.; Teasdale, T.W. Genetic and Environmental Influences on Premature Death in Adult Adoptees. N. Engl. J. Med. 1988, 318, 727–732. [Google Scholar] [CrossRef]
- Gotts, J.E.; Matthay, M.A. Sepsis: Pathophysiology and Clinical Management. BMJ 2016, 353, i1585. [Google Scholar] [CrossRef]
- de Haan, K.; Groeneveld, A.B.J.; de Geus, H.R.H.; Egal, M.; Struijs, A. Vitamin D Deficiency as a Risk Factor for Infection, Sepsis and Mortality in the Critically Ill: Systematic Review and Meta-Analysis. Crit. Care 2014, 18, 660. [Google Scholar] [CrossRef] [PubMed]
- Vincent, J.-L.; Rello, J.; Marshall, J.; Silva, E.; Anzueto, A.; Martin, C.D.; Moreno, R.; Lipman, J.; Gomersall, C.; Sakr, Y.; et al. International Study of the Prevalence and Outcomes of Infection in Intensive Care Units. JAMA 2009, 302, 2323–2329. [Google Scholar] [CrossRef] [PubMed]
- Wiersinga, W.J.; van der Poll, T. Immunopathophysiology of Human Sepsis. EBioMedicine 2022, 86, 104363. [Google Scholar] [CrossRef]
- van der Poll, T.; Shankar-Hari, M.; Wiersinga, W.J. The Immunology of Sepsis. Immunity 2021, 54, 2450–2464. [Google Scholar] [CrossRef] [PubMed]
- van der Poll, T.; van de Veerdonk, F.L.; Scicluna, B.P.; Netea, M.G. The Immunopathology of Sepsis and Potential Therapeutic Targets. Nat. Rev. Immunol. 2017, 17, 407–420. [Google Scholar] [CrossRef]
- Kumar, S.; Ingle, H.; Prasad, D.V.R.; Kumar, H. Recognition of Bacterial Infection by Innate Immune Sensors. Crit. Rev. Microbiol. 2013, 39, 229–246. [Google Scholar] [CrossRef]
- Haak, B.W.; Wiersinga, W.J. The Role of the Gut Microbiota in Sepsis. Lancet Gastroenterol. Hepatol. 2017, 2, 135–143. [Google Scholar] [CrossRef]
- Adelman, M.W.; Woodworth, M.H.; Langelier, C.; Busch, L.M.; Kempker, J.A.; Kraft, C.S.; Martin, G.S. The Gut Microbiome’s Role in the Development, Maintenance, and Outcomes of Sepsis. Crit. Care 2020, 24, 278. [Google Scholar] [CrossRef]
- Gentile, L.F.; Cuenca, A.G.; Efron, P.A.; Ang, D.; Bihorac, A.; McKinley, B.A.; Moldawer, L.L.; Moore, F.A. Persistent Inflammation and Immunosuppression: A Common Syndrome and New Horizon for Surgical Intensive Care. J. Trauma Acute Care Surg. 2012, 72, 1491–1501. [Google Scholar] [CrossRef]
- Darden, D.B.; Kelly, L.S.; Fenner, B.P.; Moldawer, L.L.; Mohr, A.M.; Efron, P.A. Dysregulated Immunity and Immunotherapy after Sepsis. J. Clin. Med. 2021, 10, 1742. [Google Scholar] [CrossRef]
- Hotchkiss, R.S.; Monneret, G.; Payen, D. Sepsis-Induced Immunosuppression: From Cellular Dysfunctions to Immunotherapy. Nat. Rev. Immunol. 2013, 13, 862–874. [Google Scholar] [CrossRef]
- Brinkmann, V.; Reichard, U.; Goosmann, C.; Fauler, B.; Uhlemann, Y.; Weiss, D.S.; Weinrauch, Y.; Zychlinsky, A. Neutrophil Extracellular Traps Kill Bacteria. Science 2004, 303, 1532–1535. [Google Scholar] [CrossRef]
- Czaikoski, P.G.; Mota, J.M.S.C.; Nascimento, D.C.; Sônego, F.; Castanheira, F.V.e.S.; Melo, P.H.; Scortegagna, G.T.; Silva, R.L.; Barroso-Sousa, R.; Souto, F.O.; et al. Neutrophil Extracellular Traps Induce Organ Damage during Experimental and Clinical Sepsis. PLoS ONE 2016, 11, e0148142. [Google Scholar] [CrossRef]
- Virgilio, F.D. The Therapeutic Potential of Modifying Inflammasomes and NOD-Like Receptors. Pharmacol. Rev. 2013, 65, 872–905. [Google Scholar] [CrossRef]
- Dinarello, C.A.; Simon, A.; van der Meer, J.W.M. Treating Inflammation by Blocking Interleukin-1 in a Broad Spectrum of Diseases. Nat. Rev. Drug Discov. 2012, 11, 633–652. [Google Scholar] [CrossRef]
- Shankar-Hari, M.; Singer, M.; Spencer, J. Can Concurrent Abnormalities in Free Light Chains and Immunoglobulin Concentrations Identify a Target Population for Immunoglobulin Trials in Sepsis? Crit. Care Med. 2017, 45, 1829–1836. [Google Scholar] [CrossRef]
- Döcke, W.D.; Randow, F.; Syrbe, U.; Krausch, D.; Asadullah, K.; Reinke, P.; Volk, H.D.; Kox, W. Monocyte Deactivation in Septic Patients: Restoration by IFN-γ Treatment. Nat. Med. 1997, 3, 678–681. [Google Scholar] [CrossRef]
- Vachharajani, V.; Liu, T.; McCall, C.E. Epigenetic Coordination of Acute Systemic Inflammation: Potential Therapeutic Targets. Expert Rev. Clin. Immunol. 2014, 10, 1141–1150. [Google Scholar] [CrossRef]
- Chavan, S.S.; Pavlov, V.A.; Tracey, K.J. Mechanisms and Therapeutic Relevance of Neuro-Immune Communication. Immunity 2017, 46, 927–942. [Google Scholar] [CrossRef] [PubMed]
- Yende, S.; Iwashyna, T.J.; Angus, D.C. Interplay between Sepsis and Chronic Health. Trends Mol. Med. 2014, 20, 234–238. [Google Scholar] [CrossRef] [PubMed]
- Rabuel, C.; Mebazaa, A. Septic Shock: A Heart Story since the 1960s. Intensive Care Med. 2006, 32, 799–807. [Google Scholar] [CrossRef] [PubMed]
- Kimchi, A.; Ellrodt, A.G.; Berman, D.S.; Riedinger, M.S.; Swan, H.J.; Murata, G.H. Right Ventricular Performance in Septic Shock: A Combined Radionuclide and Hemodynamic Study. J. Am. Coll. Cardiol. 1984, 4, 945–951. [Google Scholar] [CrossRef] [PubMed]
- Parker, M.M.; Shelhamer, J.H.; Bacharach, S.L.; Green, M.V.; Natanson, C.; Frederick, T.M.; Damske, B.A.; Parrillo, J.E. Profound but Reversible Myocardial Depression in Patients with Septic Shock. Ann. Intern. Med. 1984, 100, 483–490. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, N.I.; Howell, M.D.; Talmor, D.; Nathanson, L.A.; Lisbon, A.; Wolfe, R.E.; Weiss, J.W. Serum Lactate as a Predictor of Mortality in Emergency Department Patients with Infection. Ann. Emerg. Med. 2005, 45, 524–528. [Google Scholar] [CrossRef] [PubMed]
- Kraut, J.A.; Madias, N.E. Lactic Acidosis. N. Engl. J. Med. 2014, 371, 2309–2319. [Google Scholar] [CrossRef] [PubMed]
- Wolinsky, H. A Proposal Linking Clearance of Circulating Lipoproteins to Tissue Metabolic Activity as a Basis for Understanding Atherogenesis. Circ. Res. 1980, 47, 301–311. [Google Scholar] [CrossRef]
- Aird, W.C. The Role of the Endothelium in Severe Sepsis and Multiple Organ Dysfunction Syndrome. Blood 2003, 101, 3765–3777. [Google Scholar] [CrossRef]
- Levi, M. Pathogenesis and Treatment of Disseminated Intravascular Coagulation in the Septic Patient. J. Crit. Care 2001, 16, 167–177. [Google Scholar] [CrossRef]
- Walley, K.R.; Francis, G.A.; Opal, S.M.; Stein, E.A.; Russell, J.A.; Boyd, J.H. The Central Role of Proprotein Convertase Subtilisin/Kexin Type 9 in Septic Pathogen Lipid Transport and Clearance. Am. J. Respir. Crit. Care Med. 2015, 192, 1275–1286. [Google Scholar] [CrossRef] [PubMed]
- Alobaidi, R.; Basu, R.K.; Goldstein, S.L.; Bagshaw, S.M. Sepsis-Associated Acute Kidney Injury. Semin. Nephrol. 2015, 35, 2–11. [Google Scholar] [CrossRef]
- Wan, L.; Bagshaw, S.M.; Langenberg, C.; Saotome, T.; May, C.; Bellomo, R. Pathophysiology of Septic Acute Kidney Injury: What Do We Really Know? Crit. Care Med. 2008, 36 (Suppl. S4), S198–S203. [Google Scholar] [CrossRef]
- Ishikawa, K.; May, C.N.; Gobe, G.; Langenberg, C.; Bellomo, R. Pathophysiology of Septic Acute Kidney Injury: A Different View of Tubular Injury. In Cardiorenal Syndromes in Critical Care; S.Karger AG: Basel, Switzerland, 2010; Volume 165, pp. 18–27. [Google Scholar] [CrossRef]
- Takasu, O.; Gaut, J.P.; Watanabe, E.; To, K.; Fagley, R.E.; Sato, B.; Jarman, S.; Efimov, I.R.; Janks, D.L.; Srivastava, A.; et al. Mechanisms of Cardiac and Renal Dysfunction in Patients Dying of Sepsis. Am. J. Respir. Crit. Care Med. 2013, 187, 509–517. [Google Scholar] [CrossRef]
- Prowle, J.R.; Bellomo, R. Sepsis-Associated Acute Kidney Injury: Macrohemodynamic and Microhemodynamic Alterations in the Renal Circulation. Semin. Nephrol. 2015, 35, 64–74. [Google Scholar] [CrossRef]
- Hotchkiss, R.S.; Monneret, G.; Payen, D. Immunosuppression in Sepsis: A Novel Understanding of the Disorder and a New Therapeutic Approach. Lancet Infect. Dis. 2013, 13, 260–268. [Google Scholar] [CrossRef]
- Martin, G.S.; Mannino, D.M.; Moss, M. The Effect of Age on the Development and Outcome of Adult Sepsis. Crit. Care Med. 2006, 34, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Reber, A.J.; Chirkova, T.; Kim, J.H.; Cao, W.; Biber, R.; Shay, D.K.; Sambhara, S. Immunosenescence and Challenges of Vaccination against Influenza in the Aging Population. Aging Dis. 2012, 3, 68–90. [Google Scholar] [PubMed]
- Baghela, A.; Pena, O.M.; Lee, A.H.; Baquir, B.; Falsafi, R.; An, A.; Farmer, S.W.; Hurlburt, A.; Mondragon-Cardona, A.; Rivera, J.D.; et al. Predicting Sepsis Severity at First Clinical Presentation: The Role of Endotypes and Mechanistic Signatures. EBioMedicine 2022, 75, 103776. [Google Scholar] [CrossRef] [PubMed]
- Rimmelé, T.; Payen, D.; Cantaluppi, V.; Marshall, J.; Gomez, H.; Gomez, A.; Murray, P.; Kellum, J.A.; ADQI XIV Workgroup. Immune Cell Phenotype and Function in Sepsis. Shock 2016, 45, 282–291. [Google Scholar] [CrossRef] [PubMed]
- Delano, M.J.; Ward, P.A. The Immune System’s Role in Sepsis Progression, Resolution, and Long-Term Outcome. Immunol. Rev. 2016, 274, 330–353. [Google Scholar] [CrossRef] [PubMed]
- Nathan, C. Neutrophils and Immunity: Challenges and Opportunities. Nat. Rev. Immunol. 2006, 6, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Tamayo, E.; Gómez, E.; Bustamante, J.; Gómez-Herreras, J.I.; Fonteriz, R.; Bobillo, F.; Bermejo-Martín, J.F.; Castrodeza, J.; Heredia, M.; Fierro, I.; et al. Evolution of Neutrophil Apoptosis in Septic Shock Survivors and Nonsurvivors. J. Crit. Care 2012, 27, 415.e1. [Google Scholar] [CrossRef] [PubMed]
- Drifte, G.; Dunn-Siegrist, I.; Tissières, P.; Pugin, J. Innate Immune Functions of Immature Neutrophils in Patients with Sepsis and Severe Systemic Inflammatory Response Syndrome. Crit. Care Med. 2013, 41, 820–832. [Google Scholar] [CrossRef]
- Hotchkiss, R.S.; Nicholson, D.W. Apoptosis and Caspases Regulate Death and Inflammation in Sepsis. Nat. Rev. Immunol. 2006, 6, 813–822. [Google Scholar] [CrossRef]
- Daix, T.; Guerin, E.; Tavernier, E.; Mercier, E.; Gissot, V.; Hérault, O.; Mira, J.-P.; Dumas, F.; Chapuis, N.; Guitton, C.; et al. Multicentric Standardized Flow Cytometry Routine Assessment of Patients With Sepsis to Predict Clinical Worsening. Chest 2018, 154, 617–627. [Google Scholar] [CrossRef] [PubMed]
- Guérin, E.; Orabona, M.; Raquil, M.-A.; Giraudeau, B.; Bellier, R.; Gibot, S.; Béné, M.-C.; Lacombe, F.; Droin, N.; Solary, E.; et al. Circulating Immature Granulocytes with T-Cell Killing Functions Predict Sepsis Deterioration. Crit. Care Med. 2014, 42, 2007–2018. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.-F.; Sun, L.; Gao, H.; Shi, K.X.; Rittirsch, D.; Sarma, V.J.; Zetoune, F.S.; Ward, P.A. In Vivo Regulation of Neutrophil Apoptosis by C5a during Sepsis. J. Leukoc. Biol. 2006, 80, 1575–1583. [Google Scholar] [CrossRef] [PubMed]
- Perianayagam, M.C.; Balakrishnan, V.S.; Pereira, B.J.G.; Jaber, B.L. C5a Delays Apoptosis of Human Neutrophils via an Extracellular Signal-Regulated Kinase and Bad-Mediated Signalling Pathway. Eur. J. Clin. Investig. 2004, 34, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Simon, H.-U. Neutrophil Apoptosis Pathways and Their Modifications in Inflammation. Immunol. Rev. 2003, 193, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Alves-Filho, J.C.; Spiller, F.; Cunha, F.Q. Neutrophil Paralysis in Sepsis. Shock 2010, 34 (Suppl. S1), 15–21. [Google Scholar] [CrossRef]
- Kovach, M.A.; Standiford, T.J. The Function of Neutrophils in Sepsis. Curr. Opin. Infect. Dis. 2012, 25, 321–327. [Google Scholar] [CrossRef]
- Morris, A.C.; Brittan, M.; Wilkinson, T.S.; McAuley, D.F.; Antonelli, J.; McCulloch, C.; Barr, L.C.; McDonald, N.A.; Dhaliwal, K.; Jones, R.O.; et al. C5a-Mediated Neutrophil Dysfunction Is RhoA-Dependent and Predicts Infection in Critically Ill Patients. Blood 2011, 117, 5178–5188. [Google Scholar] [CrossRef]
- Wilkinson, T.S.; Conway Morris, A.; Kefala, K.; O’Kane, C.M.; Moore, N.R.; Booth, N.A.; McAuley, D.F.; Dhaliwal, K.; Walsh, T.S.; Haslett, C.; et al. Ventilator-Associated Pneumonia Is Characterized by Excessive Release of Neutrophil Proteases in the Lung. Chest 2012, 142, 1425–1432. [Google Scholar] [CrossRef]
- Kasten, K.R.; Muenzer, J.T.; Caldwell, C.C. Neutrophils Are Significant Producers of IL-10 during Sepsis. Biochem. Biophys. Res. Commun. 2010, 393, 28–31. [Google Scholar] [CrossRef]
- McDonald, B.; Urrutia, R.; Yipp, B.G.; Jenne, C.N.; Kubes, P. Intravascular Neutrophil Extracellular Traps Capture Bacteria from the Bloodstream during Sepsis. Cell Host Microbe 2012, 12, 324–333. [Google Scholar] [CrossRef] [PubMed]
- Meng, W.; Paunel-Görgülü, A.; Flohé, S.; Hoffmann, A.; Witte, I.; MacKenzie, C.; Baldus, S.E.; Windolf, J.; Lögters, T.T. Depletion of Neutrophil Extracellular Traps in Vivo Results in Hypersusceptibility to Polymicrobial Sepsis in Mice. Crit. Care 2012, 16, R137. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, O.E.; Borregaard, N. Neutrophil Extracellular Traps—The Dark Side of Neutrophils. J. Clin. Investig. 2016, 126, 1612–1620. [Google Scholar] [CrossRef]
- Warnatsch, A.; Ioannou, M.; Wang, Q.; Papayannopoulos, V. Neutrophil Extracellular Traps License Macrophages for Cytokine Production in Atherosclerosis. Science 2015, 349, 316–320. [Google Scholar] [CrossRef] [PubMed]
- Borella, R.; De Biasi, S.; Paolini, A.; Boraldi, F.; Lo Tartaro, D.; Mattioli, M.; Fidanza, L.; Neroni, A.; Caro-Maldonado, A.; Meschiari, M.; et al. Metabolic Reprograming Shapes Neutrophil Functions in Severe COVID-19. Eur. J. Immunol. 2022, 52, 484–502. [Google Scholar] [CrossRef] [PubMed]
- Delabranche, X.; Stiel, L.; Severac, F.; Galoisy, A.-C.; Mauvieux, L.; Zobairi, F.; Lavigne, T.; Toti, F.; Anglès-Cano, E.; Meziani, F.; et al. Evidence of Netosis in Septic Shock-Induced Disseminated Intravascular Coagulation. Shock 2017, 47, 313–317. [Google Scholar] [CrossRef] [PubMed]
- von Brühl, M.-L.; Stark, K.; Steinhart, A.; Chandraratne, S.; Konrad, I.; Lorenz, M.; Khandoga, A.; Tirniceriu, A.; Coletti, R.; Köllnberger, M.; et al. Monocytes, Neutrophils, and Platelets Cooperate to Initiate and Propagate Venous Thrombosis in Mice in Vivo. J. Exp. Med. 2012, 209, 819–835. [Google Scholar] [CrossRef]
- Fuchs, T.A.; Brill, A.; Duerschmied, D.; Schatzberg, D.; Monestier, M.; Myers, D.D.; Wrobleski, S.K.; Wakefield, T.W.; Hartwig, J.H.; Wagner, D.D. Extracellular DNA Traps Promote Thrombosis. Proc. Natl. Acad. Sci. USA 2010, 107, 15880–15885. [Google Scholar] [CrossRef]
- Massberg, S.; Grahl, L.; von Bruehl, M.-L.; Manukyan, D.; Pfeiler, S.; Goosmann, C.; Brinkmann, V.; Lorenz, M.; Bidzhekov, K.; Khandagale, A.B.; et al. Reciprocal Coupling of Coagulation and Innate Immunity via Neutrophil Serine Proteases. Nat. Med. 2010, 16, 887–896. [Google Scholar] [CrossRef]
- Shang, Y.X.; Zheng, Z.; Wang, M.; Guo, H.X.; Chen, Y.J.; Wu, Y.; Li, X.; Li, Q.; Cui, J.Y.; Ren, X.X.; et al. Diagnostic performance of Neutrophil CD64 index, procalcitonin, and C-reactive protein for early sepsis in hematological patients. World J. Clin. Cases 2022, 10, 2127–2137. [Google Scholar] [CrossRef]
- Davis, B.H. Improved Diagnostic Approaches to Infection/Sepsis Detection. Expert Rev. Mol. Diagn. 2005, 5, 193–207. [Google Scholar] [CrossRef]
- Song, S.H.; Kim, H.K.; Park, M.H.; Cho, H.-I. Neutrophil CD64 Expression Is Associated with Severity and Prognosis of Disseminated Intravascular Coagulation. Thromb. Res. 2008, 121, 499–507. [Google Scholar] [CrossRef]
- Patnaik, R.; Azim, A.; Agarwal, V. Neutrophil CD64 a Diagnostic and Prognostic Marker of Sepsis in Adult Critically Ill Patients: A Brief Review. Indian J. Crit. Care Med. 2020, 24, 1242–1250. [Google Scholar] [CrossRef] [PubMed]
- Hashem, H.E.; El Masry, S.A.; Mokhtar, A.M.; Ismail, E.A.; Abdelaal, N.M. Valuable Role of Neutrophil CD64 and Highly Sensitive CRP Biomarkers for Diagnostic, Monitoring, and Prognostic Evaluations of Sepsis Patients in Neonatal ICUs. BioMed Res. Int. 2020, 2020, 6214363. [Google Scholar] [CrossRef]
- Dal Ponte, S.T.; Alegretti, A.P.; Pilger, D.A.; Rezende, G.P.; Andrioli, G.; Ludwig, H.C.; Diogo, L.; Goldani, L.Z.; Loreto, M.; Machado, P.S.; et al. Diagnostic Accuracy of CD64 for Sepsis in Emergency Department. J. Glob. Infect. Dis. 2018, 10, 42–46. [Google Scholar] [CrossRef]
- van der Meer, W.; van Dun, L.; Gunnewiek, J.K.; Roemer, B.; Scott, C.S. Simultaneous Determination of Membrane CD64 and HLA-DR Expression by Blood Neutrophils and Monocytes Using the Monoclonal Antibody Fluorescence Capability of a Routine Haematology Analyser. J. Immunol. Methods 2006, 311, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Pillay, J.; Kamp, V.M.; van Hoffen, E.; Visser, T.; Tak, T.; Lammers, J.-W.; Ulfman, L.H.; Leenen, L.P.; Pickkers, P.; Koenderman, L. A Subset of Neutrophils in Human Systemic Inflammation Inhibits T Cell Responses through Mac-1. J. Clin. Investig. 2012, 122, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Leliefeld, P.H.C.; Pillay, J.; Vrisekoop, N.; Heeres, M.; Tak, T.; Kox, M.; Rooijakkers, S.H.M.; Kuijpers, T.W.; Pickkers, P.; Leenen, L.P.H.; et al. Differential Antibacterial Control by Neutrophil Subsets. Blood Adv. 2018, 2, 1344–1355. [Google Scholar] [CrossRef] [PubMed]
- Fingerle, G.; Pforte, A.; Passlick, B.; Blumenstein, M.; Ströbel, M.; Ziegler-Heitbrock, H.W. The Novel Subset of CD14+/CD16+ Blood Monocytes Is Expanded in Sepsis Patients. Blood 1993, 82, 3170–3176. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, R.; Kanti Barman, P.; Kumar Thatoi, P.; Tripathy, R.; Kumar Das, B.; Ravindran, B. Non-Classical Monocytes Display Inflammatory Features: Validation in Sepsis and Systemic Lupus Erythematous. Sci. Rep. 2015, 5, 13886. [Google Scholar] [CrossRef]
- Hortová-Kohoutková, M.; Lázničková, P.; Bendíčková, K.; De Zuani, M.; Andrejčinová, I.; Tomášková, V.; Suk, P.; Šrámek, V.; Helán, M.; Frič, J. Differences in Monocyte Subsets Are Associated with Short-Term Survival in Patients with Septic Shock. J. Cell. Mol. Med. 2020, 24, 12504–12512. [Google Scholar] [CrossRef]
- Bodinier, M.; Peronnet, E.; Brengel-Pesce, K.; Conti, F.; Rimmelé, T.; Textoris, J.; Vedrine, C.; Quemeneur, L.; Griffiths, A.D.; Tan, L.K.; et al. Monocyte Trajectories Endotypes Are Associated With Worsening in Septic Patients. Front. Immunol. 2021, 12, 795052. [Google Scholar] [CrossRef]
- Leijte, G.P.; Rimmelé, T.; Kox, M.; Bruse, N.; Monard, C.; Gossez, M.; Monneret, G.; Pickkers, P.; Venet, F. Monocytic HLA-DR Expression Kinetics in Septic Shock Patients with Different Pathogens, Sites of Infection and Adverse Outcomes. Crit. Care 2020, 24, 110. [Google Scholar] [CrossRef]
- Gouel-Chéron, A.; Venet, F.; Allaouchiche, B.; Monneret, G. CD4+ T-Lymphocyte Alterations in Trauma Patients. Crit. Care 2012, 16, 432. [Google Scholar] [CrossRef]
- van Dissel, J.T.; van Langevelde, P.; Westendorp, R.G.; Kwappenberg, K.; Frölich, M. Anti-Inflammatory Cytokine Profile and Mortality in Febrile Patients. Lancet 1998, 351, 950–953. [Google Scholar] [CrossRef]
- Monneret, G.; Finck, M.-E.; Venet, F.; Debard, A.-L.; Bohé, J.; Bienvenu, J.; Lepape, A. The Anti-Inflammatory Response Dominates after Septic Shock: Association of Low Monocyte HLA-DR Expression and High Interleukin-10 Concentration. Immunol. Lett. 2004, 95, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Osuchowski, M.F.; Winkler, M.S.; Skirecki, T.; Cajander, S.; Shankar-Hari, M.; Lachmann, G.; Monneret, G.; Venet, F.; Bauer, M.; Brunkhorst, F.M.; et al. The COVID-19 Puzzle: Deciphering Pathophysiology and Phenotypes of a New Disease Entity. Lancet Respir. Med. 2021, 9, 622–642. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.; Svabek, C.; Vazquez-Guillamet, C.; Sato, B.; Rasche, D.; Wilson, S.; Robbins, P.; Ulbrandt, N.; Suzich, J.; Green, J.; et al. Targeting the Programmed Cell Death 1: Programmed Cell Death Ligand 1 Pathway Reverses T Cell Exhaustion in Patients with Sepsis. Crit. Care 2014, 18, R3. [Google Scholar] [CrossRef] [PubMed]
- Guignant, C.; Lepape, A.; Huang, X.; Kherouf, H.; Denis, L.; Poitevin, F.; Malcus, C.; Chéron, A.; Allaouchiche, B.; Gueyffier, F.; et al. Programmed Death-1 Levels Correlate with Increased Mortality, Nosocomial Infection and Immune Dysfunctions in Septic Shock Patients. Crit. Care 2011, 15, R99. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, J.; Lou, J.; Zhou, Y.; Bo, L.; Zhu, J.; Zhu, K.; Wan, X.; Cai, Z.; Deng, X. Upregulation of Programmed Death-1 on T Cells and Programmed Death Ligand-1 on Monocytes in Septic Shock Patients. Crit. Care 2011, 15, R70. [Google Scholar] [CrossRef]
- Santos, S.S.; Carmo, A.M.; Brunialti, M.K.C.; Machado, F.R.; Azevedo, L.C.; Assunção, M.; Trevelin, S.C.; Cunha, F.Q.; Salomao, R. Modulation of Monocytes in Septic Patients: Preserved Phagocytic Activity, Increased ROS and NO Generation, and Decreased Production of Inflammatory Cytokines. Intensive Care Med. Exp. 2016, 4, 5. [Google Scholar] [CrossRef]
- Ferreira da Mota, N.V.; Brunialti, M.K.C.; Santos, S.S.; Machado, F.R.; Assuncao, M.; Azevedo, L.C.P.; Salomao, R. Immunophenotyping of Monocytes During Human Sepsis Shows Impairment in Antigen Presentation: A Shift Toward Nonclassical Differentiation and Upregulation of FCγRi-Receptor. Shock 2018, 50, 293–300. [Google Scholar] [CrossRef]
- Cavaillon, J.-M.; Adib-Conquy, M. Bench-to-Bedside Review: Endotoxin Tolerance as a Model of Leukocyte Reprogramming in Sepsis. Crit. Care 2006, 10, 233. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Biswas, S.K.; Lopez-Collazo, E. Endotoxin Tolerance: New Mechanisms, Molecules and Clinical Significance. Trends Immunol. 2009, 30, 475–487. [Google Scholar] [CrossRef] [PubMed]
- Brunialti, M.K.C.; Martins, P.S.; Barbosa de Carvalho, H.; Machado, F.R.; Barbosa, L.M.; Salomao, R. TLR2, TLR4, CD14, CD11B, and CD11C Expressions on Monocytes Surface and Cytokine Production in Patients with Sepsis, Severe Sepsis, and Septic Shock. Shock 2006, 25, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Salomao, R.; Brunialti, M.K.C.; Kallás, E.G.; Martins, P.S.; Rigato, O.; Freudenberg, M. Lipopolysaccharide-Cell Interaction and Induced Cellular Activation in Whole Blood of Septic Patients. J. Endotoxin Res. 2002, 8, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Martins, P.S.; Brunialti, M.K.C.; Martos, L.S.W.; Machado, F.R.; Assunçao, M.S.; Blecher, S.; Salomao, R. Expression of Cell Surface Receptors and Oxidative Metabolism Modulation in the Clinical Continuum of Sepsis. Crit. Care 2008, 12, R25. [Google Scholar] [CrossRef] [PubMed]
- Santos, S.S.; Brunialti, M.K.C.; Rigato, O.; Machado, F.R.; Silva, E.; Salomao, R. Generation of Nitric Oxide and Reactive Oxygen Species by Neutrophils and Monocytes from Septic Patients and Association with Outcomes. Shock 2012, 38, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Monneret, G.; Venet, F.; Pachot, A.; Lepape, A. Monitoring Immune Dysfunctions in the Septic Patient: A New Skin for the Old Ceremony. Mol. Med. 2008, 14, 64–78. [Google Scholar] [CrossRef]
- Venet, F.; Lukaszewicz, A.-C.; Payen, D.; Hotchkiss, R.; Monneret, G. Monitoring the Immune Response in Sepsis: A Rational Approach to Administration of Immunoadjuvant Therapies. Curr. Opin. Immunol. 2013, 25, 477–483. [Google Scholar] [CrossRef]
- Liu, S.; Luo, W.; Szatmary, P.; Zhang, X.; Lin, J.W.; Chen, L.; Liu, D.; Sutton, R.; Xia, Q.; Jin, T.; et al. Monocytic HLA-DR Expression in Immune Responses of Acute Pancreatitis and COVID-19. Int. J. Mol. Sci. 2023, 24, 3246. [Google Scholar] [CrossRef]
- Sinha, P.; Meyer, N.J.; Calfee, C.S. Biological Phenotyping in Sepsis and Acute Respiratory Distress Syndrome. Annu. Rev. Med. 2023, 74, 457–471. [Google Scholar] [CrossRef] [PubMed]
- Scicluna, B.P.; van Vught, L.A.; Zwinderman, A.H.; Wiewel, M.A.; Davenport, E.E.; Burnham, K.L.; Nürnberg, P.; Schultz, M.J.; Horn, J.; Cremer, O.L.; et al. Classification of Patients with Sepsis According to Blood Genomic Endotype: A Prospective Cohort Study. Lancet Respir. Med. 2017, 5, 816–826. [Google Scholar] [CrossRef]
- Sinistro, A.; Almerighi, C.; Ciaprini, C.; Natoli, S.; Sussarello, E.; Di Fino, S.; Calò-Carducci, F.; Rocchi, G.; Bergamini, A. Downregulation of CD40 Ligand Response in Monocytes from Sepsis Patients. Clin. Vaccine Immunol. 2008, 15, 1851–1858. [Google Scholar] [CrossRef] [PubMed]
- Reyes, M.; Filbin, M.R.; Bhattacharyya, R.P.; Billman, K.; Eisenhaure, T.; Hung, D.T.; Levy, B.D.; Baron, R.M.; Blainey, P.C.; Goldberg, M.B.; et al. An Immune-Cell Signature of Bacterial Sepsis. Nat. Med. 2020, 26, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Chiche, L.; Forel, J.-M.; Papazian, L. The Role of Viruses in Nosocomial Pneumonia. Curr. Opin. Infect. Dis. 2011, 24, 152–156. [Google Scholar] [CrossRef]
- Souza-Fonseca-Guimaraes, F.; Parlato, M.; Philippart, F.; Misset, B.; Cavaillon, J.-M.; Adib-Conquy, M.; Captain Study Group. Toll-like Receptors Expression and Interferon-γ Production by NK Cells in Human Sepsis. Crit. Care 2012, 16, R206. [Google Scholar] [CrossRef]
- Weissler, J.C.; Nicod, L.P.; Lipscomb, M.F.; Toews, G.B. Natural Killer Cell Function in Human Lung Is Compartmentalized. Am. Rev. Respir. Dis. 1987, 135, 941–949. [Google Scholar] [CrossRef]
- Grégoire, C.; Chasson, L.; Luci, C.; Tomasello, E.; Geissmann, F.; Vivier, E.; Walzer, T. The Trafficking of Natural Killer Cells. Immunol. Rev. 2007, 220, 169–182. [Google Scholar] [CrossRef]
- de Pablo, R.; Monserrat, J.; Torrijos, C.; Martín, M.; Prieto, A.; Alvarez-Mon, M. The Predictive Role of Early Activation of Natural Killer Cells in Septic Shock. Crit. Care 2012, 16, 413. [Google Scholar] [CrossRef][Green Version]
- Gogos, C.; Kotsaki, A.; Pelekanou, A.; Giannikopoulos, G.; Vaki, I.; Maravitsa, P.; Adamis, S.; Alexiou, Z.; Andrianopoulos, G.; Antonopoulou, A.; et al. Early Alterations of the Innate and Adaptive Immune Statuses in Sepsis According to the Type of Underlying Infection. Crit. Care 2010, 14, R96. [Google Scholar] [CrossRef]
- Giannikopoulos, G.; Antonopoulou, A.; Kalpakou, G.; Makaritsis, K.; Panou, C.; Papadomichelakis, E.; Sinapidis, D.; Theodotou, A.; Tzagkaraki, A.; Giamarellos-Bourboulis, E.J. The Functional Role of Natural Killer Cells Early in Clinical Sepsis. Acta Pathol. Microbiol. Immunol. Scand. 2013, 121, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Elemam, N.M.; Ramakrishnan, R.K.; Hundt, J.E.; Halwani, R.; Maghazachi, A.A.; Hamid, Q. Innate Lymphoid Cells and Natural Killer Cells in Bacterial Infections: Function, Dysregulation, and Therapeutic Targets. Front. Cell. Infect. Microbiol. 2021, 11, 733564. [Google Scholar] [CrossRef] [PubMed]
- Nomura, T.; Kawamura, I.; Tsuchiya, K.; Kohda, C.; Baba, H.; Ito, Y.; Kimoto, T.; Watanabe, I.; Mitsuyama, M. Essential Role of Interleukin-12 (IL-12) and IL-18 for Gamma Interferon Production Induced by Listeriolysin O in Mouse Spleen Cells. Infect. Immun. 2002, 70, 1049–1055. [Google Scholar] [CrossRef] [PubMed]
- Yao, C.; Bora, S.A.; Parimon, T.; Zaman, T.; Friedman, O.A.; Palatinus, J.A.; Surapaneni, N.S.; Matusov, Y.P.; Cerro Chiang, G.; Kassar, A.G.; et al. Cell-Type-Specific Immune Dysregulation in Severely Ill COVID-19 Patients. Cell Rep. 2021, 34, 108590. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Gao, Y.; Wang, G.; Song, G.; Liu, S.; Sun, D.; Xu, Y.; Tian, Z. Functional Exhaustion of Antiviral Lymphocytes in COVID-19 Patients. Cell. Mol. Immunol. 2020, 17, 533–535. [Google Scholar] [CrossRef] [PubMed]
- Yaqinuddin, A.; Kashir, J. Innate Immunity in COVID-19 Patients Mediated by NKG2A Receptors, and Potential Treatment Using Monalizumab, Cholroquine, and Antiviral Agents. Med. Hypotheses 2020, 140, 109777. [Google Scholar] [CrossRef] [PubMed]
- Venet, F.; Davin, F.; Guignant, C.; Larue, A.; Cazalis, M.-A.; Darbon, R.; Allombert, C.; Mougin, B.; Malcus, C.; Poitevin-Later, F.; et al. Early Assessment of Leukocyte Alterations at Diagnosis of Septic Shock. Shock 2010, 34, 358–363. [Google Scholar] [CrossRef] [PubMed]
- Forel, J.-M.; Chiche, L.; Thomas, G.; Mancini, J.; Farnarier, C.; Cognet, C.; Guervilly, C.; Daumas, A.; Vély, F.; Xéridat, F.; et al. Phenotype and Functions of Natural Killer Cells in Critically-Ill Septic Patients. PLoS ONE 2012, 7, e50446. [Google Scholar] [CrossRef]
- Holub, M.; Klučková, Z.; Helcl, M.; Příhodov, J.; Rokyta, R.; Beran, O. Lymphocyte Subset Numbers Depend on the Bacterial Origin of Sepsis. Clin. Microbiol. Infect. 2003, 9, 202–211. [Google Scholar] [CrossRef]
- Guo, Y.; Patil, N.K.; Luan, L.; Bohannon, J.K.; Sherwood, E.R. The Biology of Natural Killer Cells during Sepsis. Immunology 2018, 153, 190–202. [Google Scholar] [CrossRef]
- Souza-Fonseca-Guimaraes, F.; Adib-Conquy, M.; Cavaillon, J.-M. Natural Killer (NK) Cells in Antibacterial Innate Immunity: Angels or Devils? Mol. Med. 2012, 18, 270–285. [Google Scholar] [CrossRef]
- Souza-Fonseca-Guimaraes, F.; Parlato, M.; Fitting, C.; Cavaillon, J.-M.; Adib-Conquy, M. NK Cell Tolerance to TLR Agonists Mediated by Regulatory T Cells after Polymicrobial Sepsis. J. Immunol. 2012, 188, 5850–5858. [Google Scholar] [CrossRef] [PubMed]
- Chiche, L.; Forel, J.-M.; Thomas, G.; Farnarier, C.; Cognet, C.; Guervilly, C.; Zandotti, C.; Vély, F.; Roch, A.; Vivier, E.; et al. Interferon-γ Production by Natural Killer Cells and Cytomegalovirus in Critically Ill Patients. Crit. Care Med. 2012, 40, 3162–3169. [Google Scholar] [CrossRef] [PubMed]
- Luyt, C.-E.; Combes, A.; Deback, C.; Aubriot-Lorton, M.-H.; Nieszkowska, A.; Trouillet, J.-L.; Capron, F.; Agut, H.; Gibert, C.; Chastre, J. Herpes Simplex Virus Lung Infection in Patients Undergoing Prolonged Mechanical Ventilation. Am. J. Respir. Crit. Care Med. 2007, 175, 935–942. [Google Scholar] [CrossRef] [PubMed]
- Limaye, A.P.; Kirby, K.A.; Rubenfeld, G.D.; Leisenring, W.M.; Bulger, E.M.; Neff, M.J.; Gibran, N.S.; Huang, M.-L.; Santo Hayes, T.K.; Corey, L.; et al. Cytomegalovirus Reactivation in Critically Ill Immunocompetent Patients. JAMA 2008, 300, 413–422. [Google Scholar] [CrossRef] [PubMed]
- Cook, C.H.; Trgovcich, J. Cytomegalovirus Reactivation in Critically Ill Immunocompetent Hosts: A Decade of Progress and Remaining Challenges. Antivir. Res. 2011, 90, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Maucourant, C.; Filipovic, I.; Ponzetta, A.; Aleman, S.; Cornillet, M.; Hertwig, L.; Strunz, B.; Lentini, A.; Reinius, B.; Brownlie, D.; et al. Natural Killer Cell Immunotypes Related to COVID-19 Disease Severity. Sci. Immunol. 2020, 5, eabd6832. [Google Scholar] [CrossRef] [PubMed]
- Jämsä, J.; Syrjälä, H.; Huotari, V.; Savolainen, E.-R.; Ala-Kokko, T. Monocyte and Lymphocyte Surface Molecules in Severe Sepsis and Non-Septic Critically Ill Patients. Acta Pathol. Microbiol. Immunol. Scand. 2017, 125, 536–543. [Google Scholar] [CrossRef] [PubMed]
- Vantourout, P.; Hayday, A. Six-of-the-Best: Unique Contributions of Γδ T Cells to Immunology. Nat. Rev. Immunol. 2013, 13, 88–100. [Google Scholar] [CrossRef]
- Lee, H.W.; Chung, Y.S.; Kim, T.J. Heterogeneity of Human Γδ T Cells and Their Role in Cancer Immunity. Immune Netw. 2020, 20, e5. [Google Scholar] [CrossRef]
- Ogiku, M.; Kono, H.; Hara, M.; Tsuchiya, M.; Fujii, H. Interleukin-17A Plays a Pivotal Role in Polymicrobial Sepsis According to Studies Using IL-17A Knockout Mice. J. Surg. Res. 2012, 174, 142–149. [Google Scholar] [CrossRef] [PubMed]
- de Souza Costa, M.F.; Bastos Trigo de Negreiros, C.; Ugarte Bornstein, V.; Hemmi Valente, R.; Mengel, J.; Henriques, M.d.G.; Farias Benjamim, C.; Penido, C. Murine IL-17+ Vγ4 T Lymphocytes Accumulate in the Lungs and Play a Protective Role during Severe Sepsis. BMC Immunol. 2015, 16, 36. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.-L.; Feng, T.; Zhang, J.-Q.; Cao, X.; Wu, Q.-H.; Xie, Z.-C.; Kang, Y.; Li, H. Phenotypic Changes and Impaired Function of Peripheral Γδ T Cells in Patients With Sepsis. Shock 2017, 48, 321–328. [Google Scholar] [CrossRef]
- Andreu-Ballester, J.C.; Arribas, M.A.; Rico, M.; García-Ballesteros, C.; Galindo-Regal, L.; Sorando-Serra, R.; Albert, L.; Navarro, A.; López-Chuliá, F.; Peydró, F.; et al. Changes of CD3+CD56+ Γδ T Cell Number and Apoptosis during Hospital Admission Are Related to Mortality in Septic Patients. Clin. Immunol. 2022, 236, 108956. [Google Scholar] [CrossRef]
- Cheng, Z.; Abrams, S.T.; Toh, J.; Wang, S.S.; Wang, Z.; Yu, Q.; Yu, W.; Toh, C.H.; Wang, G. The Critical Roles and Mechanisms of Immune Cell Death in Sepsis. Front. Immunol. 2020, 11, 1918. [Google Scholar] [CrossRef]
- Andreu-Ballester, J.C.; Tormo-Calandín, C.; Garcia-Ballesteros, C.; Pérez-Griera, J.; Amigó, V.; Almela-Quilis, A.; Ruiz del Castillo, J.; Peñarroja-Otero, C.; Ballester, F. Association of Γδ T Cells with Disease Severity and Mortality in Septic Patients. Clin. Vaccine Immunol. 2013, 20, 738–746. [Google Scholar] [CrossRef] [PubMed]
- Treiner, E.; Duban, L.; Bahram, S.; Radosavljevic, M.; Wanner, V.; Tilloy, F.; Affaticati, P.; Gilfillan, S.; Lantz, O. Selection of Evolutionarily Conserved Mucosal-Associated Invariant T Cells by MR1. Nature 2003, 422, 164–169. [Google Scholar] [CrossRef]
- Kjer-Nielsen, L.; Patel, O.; Corbett, A.J.; Le Nours, J.; Meehan, B.; Liu, L.; Bhati, M.; Chen, Z.; Kostenko, L.; Reantragoon, R.; et al. MR1 Presents Microbial Vitamin B Metabolites to MAIT Cells. Nature 2012, 491, 717–723. [Google Scholar] [CrossRef]
- Corbett, A.J.; Eckle, S.B.G.; Birkinshaw, R.W.; Liu, L.; Patel, O.; Mahony, J.; Chen, Z.; Reantragoon, R.; Meehan, B.; Cao, H.; et al. T-Cell Activation by Transitory Neo-Antigens Derived from Distinct Microbial Pathways. Nature 2014, 509, 361–365. [Google Scholar] [CrossRef]
- De Biasi, S.; Gibellini, L.; Lo Tartaro, D.; Puccio, S.; Rabacchi, C.; Mazza, E.M.C.; Brummelman, J.; Williams, B.; Kaihara, K.; Forcato, M.; et al. Circulating Mucosal-Associated Invariant T Cells Identify Patients Responding to Anti-PD-1 Therapy. Nat. Commun. 2021, 12, 1669. [Google Scholar] [CrossRef] [PubMed]
- Godfrey, D.I.; Koay, H.-F.; McCluskey, J.; Gherardin, N.A. The Biology and Functional Importance of MAIT Cells. Nat. Immunol. 2019, 20, 1110–1128. [Google Scholar] [CrossRef] [PubMed]
- Martin, E.; Treiner, E.; Duban, L.; Guerri, L.; Laude, H.; Toly, C.; Premel, V.; Devys, A.; Moura, I.C.; Tilloy, F.; et al. Stepwise Development of MAIT Cells in Mouse and Human. PLoS Biol. 2009, 7, e1000054. [Google Scholar] [CrossRef] [PubMed]
- Ibidapo-Obe, O.; Stengel, S.; Köse-Vogel, N.; Quickert, S.; Reuken, P.A.; Busch, M.; Bauer, M.; Stallmach, A.; Bruns, T. Mucosal-Associated Invariant T Cells Redistribute to the Peritoneal Cavity During Spontaneous Bacterial Peritonitis and Contribute to Peritoneal Inflammation. Cell. Mol. Gastroenterol. Hepatol. 2020, 9, 661–677. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, S.; Labuz, D.; Anderson, C.P.; Araujo, C.V.; Blair, A.; Middleton, E.A.; Jensen, O.; Tran, A.; Mulvey, M.A.; Campbell, R.A.; et al. Mucosal-Associated Invariant T (MAIT) Cells Mediate Protective Host Responses in Sepsis. eLife 2020, 9, e55615. [Google Scholar] [CrossRef]
- Tian, L.; Xu, J.; Chen, C.; Lin, J.; Ju, L.; Chen, L.; Zhang, Y.; Han, X.; Liu, L. HLA-DR+ Mucosal-Associated Invariant T Cells Predict Poor Prognosis in Patients with Sepsis: A Prospective Observational Study. Scand. J. Immunol. 2023, 98, e13286. [Google Scholar] [CrossRef]
- Gutcher, I.; Becher, B. APC-Derived Cytokines and T Cell Polarization in Autoimmune Inflammation. J. Clin. Investig. 2007, 117, 1119–1127. [Google Scholar] [CrossRef]
- Cheadle, W.G.; Pemberton, R.M.; Robinson, D.; Livingston, D.H.; Rodriguez, J.L.; Polk, H.C. Lymphocyte Subset Responses to Trauma and Sepsis. J. Trauma 1993, 35, 844–849. [Google Scholar] [CrossRef]
- Heffernan, D.S.; Monaghan, S.F.; Thakkar, R.K.; Machan, J.T.; Cioffi, W.G.; Ayala, A. Failure to Normalize Lymphopenia Following Trauma Is Associated with Increased Mortality, Independent of the Leukocytosis Pattern. Crit. Care 2012, 16, R12. [Google Scholar] [CrossRef] [PubMed]
- Hotchkiss, R.S.; Sherwood, E.R. Getting Sepsis Therapy Right. Science 2015, 347, 1201–1202. [Google Scholar] [CrossRef] [PubMed]
- Inoue, S.; Suzuki-Utsunomiya, K.; Okada, Y.; Taira, T.; Iida, Y.; Miura, N.; Tsuji, T.; Yamagiwa, T.; Morita, S.; Chiba, T.; et al. Reduction of Immunocompetent T Cells Followed by Prolonged Lymphopenia in Severe Sepsis in the Elderly. Crit. Care Med. 2013, 41, 810–819. [Google Scholar] [CrossRef]
- Le Tulzo, Y.; Pangault, C.; Gacouin, A.; Guilloux, V.; Tribut, O.; Amiot, L.; Tattevin, P.; Thomas, R.; Fauchet, R.; Drénou, B. Early Circulating Lymphocyte Apoptosis in Human Septic Shock Is Associated with Poor Outcome. Shock 2002, 18, 487–494. [Google Scholar] [CrossRef]
- Brummelman, J.; Pilipow, K.; Lugli, E. The Single-Cell Phenotypic Identity of Human CD8+ and CD4+ T Cells. In International Review of Cell and Molecular Biology; Elsevier: Amsterdam, The Netherlands, 2018; Volume 341, pp. 63–124. [Google Scholar] [CrossRef]
- Boomer, J.S.; To, K.; Chang, K.C.; Takasu, O.; Osborne, D.F.; Walton, A.H.; Bricker, T.L.; Jarman, S.D.; Kreisel, D.; Krupnick, A.S.; et al. Immunosuppression in Patients Who Die of Sepsis and Multiple Organ Failure. JAMA 2011, 306, 2594–2605. [Google Scholar] [CrossRef]
- Hotchkiss, R.S.; Tinsley, K.W.; Swanson, P.E.; Schmieg, R.E.; Hui, J.J.; Chang, K.C.; Osborne, D.F.; Freeman, B.D.; Cobb, J.P.; Buchman, T.G.; et al. Sepsis-Induced Apoptosis Causes Progressive Profound Depletion of B and CD4+ T Lymphocytes in Humans. J. Immunol. 2001, 166, 6952–6963. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, S.T.; Lederer, J.A.; Horgan, A.F.; Chin, D.H.; Mannick, J.A.; Rodrick, M.L. Major Injury Leads to Predominance of the T Helper-2 Lymphocyte Phenotype and Diminished Interleukin-12 Production Associated with Decreased Resistance to Infection. Ann. Surg. 1995, 222, 482–490; discussion 490–492. [Google Scholar] [CrossRef] [PubMed]
- Pachot, A.; Monneret, G.; Voirin, N.; Leissner, P.; Venet, F.; Bohé, J.; Payen, D.; Bienvenu, J.; Mougin, B.; Lepape, A. Longitudinal Study of Cytokine and Immune Transcription Factor mRNA Expression in Septic Shock. Clin. Immunol. 2005, 114, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Smeekens, S.P.; Ng, A.; Kumar, V.; Johnson, M.D.; Plantinga, T.S.; van Diemen, C.; Arts, P.; Verwiel, E.T.P.; Gresnigt, M.S.; Fransen, K.; et al. Functional Genomics Identifies Type I Interferon Pathway as Central for Host Defense against Candida albicans. Nat. Commun. 2013, 4, 1342. [Google Scholar] [CrossRef] [PubMed]
- Unsinger, J.; McGlynn, M.; Kasten, K.R.; Hoekzema, A.S.; Watanabe, E.; Muenzer, J.T.; McDonough, J.S.; Tschoep, J.; Ferguson, T.A.; McDunn, J.E.; et al. IL-7 Promotes T Cell Viability, Trafficking, and Functionality and Improves Survival in Sepsis. J. Immunol. 2010, 184, 3768–3779. [Google Scholar] [CrossRef] [PubMed]
- Venet, F.; Monneret, G. Advances in the Understanding and Treatment of Sepsis-Induced Immunosuppression. Nat. Rev. Nephrol. 2018, 14, 121–137. [Google Scholar] [CrossRef] [PubMed]
- Boomer, J.S.; Shuherk-Shaffer, J.; Hotchkiss, R.S.; Green, J.M. A Prospective Analysis of Lymphocyte Phenotype and Function over the Course of Acute Sepsis. Crit. Care 2012, 16, R112. [Google Scholar] [CrossRef] [PubMed]
- Takahama, M.; Patil, A.; Richey, G.; Cipurko, D.; Johnson, K.; Carbonetto, P.; Plaster, M.; Pandey, S.; Cheronis, K.; Ueda, T.; et al. A pairwise cytokine code explains the organism-wide response to sepsis. Nat. Immunol. 2024, 25, 226–239. [Google Scholar] [CrossRef] [PubMed]
- Avendaño-Ortiz, J.; Lozano-Rodríguez, R.; Martín-Quirós, A.; Maroun-Eid, C.; Terrón, V.; Valentín, J.; Montalbán-Hernández, K.; Ruiz de la Bastida, F.; García-Garrido, M.A.; Cubillos-Zapata, C.; et al. Proteins from SARS-CoV-2 Reduce T Cell Proliferation: A Mirror Image of Sepsis. Heliyon 2020, 6, e05635. [Google Scholar] [CrossRef] [PubMed]
- Kaech, S.M.; Hemby, S.; Kersh, E.; Ahmed, R. Molecular and Functional Profiling of Memory CD8 T Cell Differentiation. Cell 2002, 111, 837–851. [Google Scholar] [CrossRef] [PubMed]
- Brady, J.; Horie, S.; Laffey, J.G. Role of the Adaptive Immune Response in Sepsis. Intensive Care Med. Exp. 2020, 8, 20. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Rodríguez, R.; Avendaño-Ortíz, J.; Montalbán-Hernández, K.; Ruiz-Rodríguez, J.C.; Ferrer, R.; Martín-Quirós, A.; Maroun-Eid, C.; González-López, J.J.; Fàbrega, A.; Terrón-Arcos, V.; et al. The Prognostic Impact of SIGLEC5-Induced Impairment of CD8+ T Cell Activation in Sepsis. eBioMedicine 2023, 97, 104841. [Google Scholar] [CrossRef] [PubMed]
- Berton, R.R.; McGonagil, P.W.; Jensen, I.J.; Ybarra, T.K.; Bishop, G.A.; Harty, J.T.; Griffith, T.S.; Badovinac, V.P. Sepsis Leads to Lasting Changes in Phenotype and Function of Naïve CD8 T Cells. PLoS Pathog. 2023, 19, e1011720. [Google Scholar] [CrossRef]
- Walton, A.H.; Muenzer, J.T.; Rasche, D.; Boomer, J.S.; Sato, B.; Brownstein, B.H.; Pachot, A.; Brooks, T.L.; Deych, E.; Shannon, W.D.; et al. Reactivation of Multiple Viruses in Patients with Sepsis. PLoS ONE 2014, 9, e98819. [Google Scholar] [CrossRef]
- Gatto, I.; Biagioni, E.; Coloretti, I.; Farinelli, C.; Avoni, C.; Caciagli, V.; Busani, S.; Sarti, M.; Pecorari, M.; Gennari, W.; et al. Cytomegalovirus Blood Reactivation in COVID-19 Critically Ill Patients: Risk Factors and Impact on Mortality. Intensive Care Med. 2022, 48, 706–713. [Google Scholar] [CrossRef]
- Choi, Y.J.; Kim, S.B.; Kim, J.H.; Park, S.-H.; Park, M.S.; Kim, J.M.; Han, S.H.; Shin, E.-C. Impaired Polyfunctionality of CD8+ T Cells in Severe Sepsis Patients with Human Cytomegalovirus Reactivation. Exp. Mol. Med. 2017, 49, e382. [Google Scholar] [CrossRef][Green Version]
- Cao, C.; Chai, Y.; Shou, S.; Wang, J.; Huang, Y.; Ma, T. Toll-like Receptor 4 Deficiency Increases Resistance in Sepsis-Induced Immune Dysfunction. Int. Immunopharmacol. 2018, 54, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Venet, F.; Chung, C.-S.; Kherouf, H.; Geeraert, A.; Malcus, C.; Poitevin, F.; Bohé, J.; Lepape, A.; Ayala, A.; Monneret, G. Increased Circulating Regulatory T Cells (CD4+CD25+CD127−) Contribute to Lymphocyte Anergy in Septic Shock Patients. Intensive Care Med. 2009, 35, 678–686. [Google Scholar] [CrossRef]
- Venet, F.; Pachot, A.; Debard, A.-L.; Bohé, J.; Bienvenu, J.; Lepape, A.; Monneret, G. Increased Percentage of CD4+CD25+ Regulatory T Cells during Septic Shock Is Due to the Decrease of CD4+CD25− Lymphocytes. Crit. Care Med. 2004, 32, 2329–2331. [Google Scholar] [CrossRef] [PubMed]
- Brinkhoff, A.; Sieberichs, A.; Engler, H.; Dolff, S.; Benson, S.; Korth, J.; Schedlowski, M.; Kribben, A.; Witzke, O.; Wilde, B. Pro-Inflammatory Th1 and Th17 Cells Are Suppressed During Human Experimental Endotoxemia Whereas Anti-Inflammatory IL-10 Producing T-Cells Are Unaffected. Front. Immunol. 2018, 9, 1133. [Google Scholar] [CrossRef] [PubMed]
- Conway Morris, A.; Datta, D.; Shankar-Hari, M.; Stephen, J.; Weir, C.J.; Rennie, J.; Antonelli, J.; Bateman, A.; Warner, N.; Judge, K.; et al. Cell-Surface Signatures of Immune Dysfunction Risk-Stratify Critically Ill Patients: INFECT Study. Intensive Care Med. 2018, 44, 627–635. [Google Scholar] [CrossRef] [PubMed]
- Gustave, C.-A.; Gossez, M.; Demaret, J.; Rimmelé, T.; Lepape, A.; Malcus, C.; Poitevin-Later, F.; Jallades, L.; Textoris, J.; Monneret, G.; et al. Septic Shock Shapes B Cell Response toward an Exhausted-like/Immunoregulatory Profile in Patients. J. Immunol. 2018, 200, 2418–2425. [Google Scholar] [CrossRef]
- Andaluz-Ojeda, D.; Iglesias, V.; Bobillo, F.; Almansa, R.; Rico, L.; Gandía, F.; Loma, A.M.; Nieto, C.; Diego, R.; Ramos, E.; et al. Early Natural Killer Cell Counts in Blood Predict Mortality in Severe Sepsis. Crit. Care 2011, 15, R243. [Google Scholar] [CrossRef]
- Monserrat, J.; de Pablo, R.; Diaz-Martín, D.; Rodríguez-Zapata, M.; de la Hera, A.; Prieto, A.; Alvarez-Mon, M. Early Alterations of B Cells in Patients with Septic Shock. Crit. Care 2013, 17, R105. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Inoue, S.; Kametani, Y.; Komori, Y.; Chiba, S.; Sato, T.; Inokuchi, S.; Ogura, S. Reduced Immunocompetent B Cells and Increased Secondary Infection in Elderly Patients With Severe Sepsis. Shock 2016, 46, 270–278. [Google Scholar] [CrossRef]
- Wilson, J.K.; Zhao, Y.; Singer, M.; Spencer, J.; Shankar-Hari, M. Lymphocyte Subset Expression and Serum Concentrations of PD-1/PD-L1 in Sepsis—Pilot Study. Crit. Care 2018, 22, 95. [Google Scholar] [CrossRef]
- Brinkhoff, A.; Zeng, Y.; Sieberichs, A.; Dolff, S.; Shilei, X.; Sun, M.; Engler, H.; Benson, S.; Korth, J.; Schedlowski, M.; et al. B-Cell Dynamics during Experimental Endotoxemia in Humans. Biosci. Rep. 2019, 39, BSR20182347. [Google Scholar] [CrossRef]
- Dong, X.; Liu, Q.; Zheng, Q.; Liu, X.; Wang, Y.; Xie, Z.; Liu, T.; Yang, F.; Gao, W.; Bai, X.; et al. Alterations of B Cells in Immunosuppressive Phase of Septic Shock Patients. Crit. Care Med. 2020, 48, 815–821. [Google Scholar] [CrossRef]
- Duan, S.; Jiao, Y.; Wang, J.; Tang, D.; Xu, S.; Wang, R.; Jiang, T.; Shao, J.; He, Z.; Yu, W. Impaired B-Cell Maturation Contributes to Reduced B Cell Numbers and Poor Prognosis in Sepsis. Shock 2020, 54, 70–77. [Google Scholar] [CrossRef]
- Tian, L.; Zhu, J.; Jin, J.; Tong, C.; Zeng, W.; Deng, S.; Zou, S. Prognostic Value of Circulating Lymphocyte B and Plasma Immunoglobulin M on Septic Shock and Sepsis: A Systematic Review and Meta-Analysis. Am. J. Transl. Res. 2019, 11, 7223–7232. [Google Scholar]
- Krautz, C.; Maier, S.L.; Brunner, M.; Langheinrich, M.; Giamarellos-Bourboulis, E.J.; Gogos, C.; Armaganidis, A.; Kunath, F.; Grützmann, R.; Weber, G.F. Reduced Circulating B Cells and Plasma IgM Levels Are Associated with Decreased Survival in Sepsis—A Meta-Analysis. J. Crit. Care 2018, 45, 71–75. [Google Scholar] [CrossRef]
- Hohlstein, P.; Gussen, H.; Bartneck, M.; Warzecha, K.T.; Roderburg, C.; Buendgens, L.; Trautwein, C.; Koch, A.; Tacke, F. Prognostic Relevance of Altered Lymphocyte Subpopulations in Critical Illness and Sepsis. J. Clin. Med. 2019, 8, 353. [Google Scholar] [CrossRef]
- Schenz, J.; Tamulyte, S.; Nusshag, C.; Brenner, T.; Poschet, G.; Weigand, M.A.; Uhle, F. Population-Specific Metabolic Alterations in Professional Antigen-Presenting Cells Contribute to Sepsis-Associated Immunosuppression. Shock 2020, 53, 5–15. [Google Scholar] [CrossRef]
- Hotchkiss, R.S.; Swanson, P.E.; Cobb, J.P.; Jacobson, A.; Buchman, T.G.; Karl, I.E. Apoptosis in Lymphoid and Parenchymal Cells during Sepsis: Findings in Normal and T- and B-Cell-Deficient Mice. Crit. Care Med. 1997, 25, 1298–1307. [Google Scholar] [CrossRef] [PubMed]
- Ayala, A.; Xin Xu, Y.; Ayala, C.A.; Sonefeld, D.E.; Karr, S.M.; Evans, T.A.; Chaudry, I.H. Increased Mucosal B-Lymphocyte Apoptosis during Polymicrobial Sepsis Is a Fas Ligand but Not an Endotoxin-Mediated Process. Blood 1998, 91, 1362–1372. [Google Scholar] [CrossRef]
- Hotchkiss, R.S.; Swanson, P.E.; Freeman, B.D.; Tinsley, K.W.; Cobb, J.P.; Matuschak, G.M.; Buchman, T.G.; Karl, I.E. Apoptotic Cell Death in Patients with Sepsis, Shock, and Multiple Organ Dysfunction. Crit. Care Med. 1999, 27, 1230–1251. [Google Scholar] [CrossRef] [PubMed]
- Hotchkiss, R.S.; Osmon, S.B.; Chang, K.C.; Wagner, T.H.; Coopersmith, C.M.; Karl, I.E. Accelerated Lymphocyte Death in Sepsis Occurs by Both the Death Receptor and Mitochondrial Pathways. J. Immunol. 2005, 174, 5110–5118. [Google Scholar] [CrossRef] [PubMed]
- Ordoñez, C.; Savage, H.P.; Tarajia, M.; Rivera, R.; Weeks-Galindo, C.; Sambrano, D.; Riley, L.; Fernandez, P.L.; Baumgarth, N.; Goodridge, A. Both B-1a and B-1b Cells Exposed to Mycobacterium tuberculosis Lipids Differentiate into IgM Antibody-Secreting Cells. Immunology 2018, 154, 613–623. [Google Scholar] [CrossRef]
- Smith, F.L.; Baumgarth, N. B-1 Cell Responses to Infections. Curr. Opin. Immunol. 2019, 57, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Aziz, M.; Holodick, N.E.; Rothstein, T.L.; Wang, P. The Role of B-1 Cells in Inflammation. Immunol. Res. 2015, 63, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Girardis, M.; Cossarizza, A. Early Alterations of B Cells in Patients with Septic Shock: Another Piece in the Complex Puzzle of the Immune Response in Sepsis. Crit. Care 2013, 17, 162. [Google Scholar] [CrossRef]
- Ma, C.; Liu, H.; Yang, S.; Li, H.; Liao, X.; Kang, Y. The Emerging Roles and Therapeutic Potential of B Cells in Sepsis. Front. Pharmacol. 2022, 13, 1034667. [Google Scholar] [CrossRef] [PubMed]
- Knippenberg, S.; Peelen, E.; Smolders, J.; Thewissen, M.; Menheere, P.; Cohen Tervaert, J.W.; Hupperts, R.; Damoiseaux, J. Reduction in IL-10 Producing B Cells (Breg) in Multiple Sclerosis Is Accompanied by a Reduced Naïve/Memory Breg Ratio during a Relapse but Not in Remission. J. Neuroimmunol. 2011, 239, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Xu, H.; Gao, R.; Leng, F.; Huo, F.; Li, Y.; Liu, S.; Xu, M.; Bai, J. CD19+CD24hiCD38hi Regulatory B Cells Deficiency Revealed Severity and Poor Prognosis in Patients with Sepsis. BMC Immunol. 2022, 23, 54. [Google Scholar] [CrossRef] [PubMed]
- Giamarellos-Bourboulis, E.J.; Apostolidou, E.; Lada, M.; Perdios, I.; Gatselis, N.K.; Tsangaris, I.; Georgitsi, M.; Bristianou, M.; Kanni, T.; Sereti, K.; et al. Kinetics of Circulating Immunoglobulin M in Sepsis: Relationship with Final Outcome. Crit. Care 2013, 17, R247. [Google Scholar] [CrossRef]
- Akatsuka, M.; Tatsumi, H.; Sonoda, T.; Masuda, Y. Low Immunoglobulin G Level Is Associated with Poor Outcomes in Patients with Sepsis and Septic Shock. J. Microbiol. Immunol. Infect. 2021, 54, 728–732. [Google Scholar] [CrossRef]
- Alagna, L.; Meessen, J.M.T.A.; Bellani, G.; Albiero, D.; Caironi, P.; Principale, I.; Vivona, L.; Grasselli, G.; Motta, F.; Agnelli, N.M.; et al. Higher Levels of IgA and IgG at Sepsis Onset Are Associated with Higher Mortality: Results from the Albumin Italian Outcome Sepsis (ALBIOS) Trial. Ann. Intensive Care 2021, 11, 161. [Google Scholar] [CrossRef]
- Chang, L.-Y.; Li, Y.; Kaplan, D.E. Endotoxemia Contributes to CD27+ Memory B-Cell Apoptosis via Enhanced Sensitivity to Fas Ligation in Patients with Cirrhosis. Sci. Rep. 2016, 6, 36862. [Google Scholar] [CrossRef]
- Chung, C.-S.; Xu, Y.X.; Wang, W.; Chaudry, I.H.; Ayala, A. Is Fas Ligand or Endotoxin Responsible for Mucosal Lymphocyte Apoptosis in Sepsis? Arch. Surg. 1998, 133, 1213–1220. [Google Scholar] [CrossRef]
- Chung, C.S.; Wang, W.; Chaudry, I.H.; Ayala, A. Increased Apoptosis in Lamina Propria B Cells during Polymicrobial Sepsis Is FasL but Not Endotoxin Mediated. Am. J. Physiol. Gastrointest. Liver Physiol. 2001, 280, G812–G818. [Google Scholar] [CrossRef] [PubMed]
- Rauch, P.J.; Chudnovskiy, A.; Robbins, C.S.; Weber, G.F.; Etzrodt, M.; Hilgendorf, I.; Tiglao, E.; Figueiredo, J.-L.; Iwamoto, Y.; Theurl, I.; et al. Innate Response Activator B Cells Protect against Microbial Sepsis. Science 2012, 335, 597–601. [Google Scholar] [CrossRef] [PubMed]
- Weber, G.F.; Chousterman, B.G.; He, S.; Fenn, A.M.; Nairz, M.; Anzai, A.; Brenner, T.; Uhle, F.; Iwamoto, Y.; Robbins, C.S.; et al. Interleukin-3 Amplifies Acute Inflammation and Is a Potential Therapeutic Target in Sepsis. Science 2015, 347, 1260–1265. [Google Scholar] [CrossRef]
- Honda, S.-I.; Sato, K.; Totsuka, N.; Fujiyama, S.; Fujimoto, M.; Miyake, K.; Nakahashi-Oda, C.; Tahara-Hanaoka, S.; Shibuya, K.; Shibuya, A. Marginal Zone B Cells Exacerbate Endotoxic Shock via Interleukin-6 Secretion Induced by Fcα/μR-Coupled TLR4 Signalling. Nat. Commun. 2016, 7, 11498. [Google Scholar] [CrossRef]
- Tsay, G.J.; Zouali, M. The Interplay Between Innate-Like B Cells and Other Cell Types in Autoimmunity. Front. Immunol. 2018, 9, 1064. [Google Scholar] [CrossRef]
- Lo, L.-W.; Chang, C.-W.; Chiang, M.-F.; Lin, I.-Y.; Lin, K.-I. Marginal Zone B Cells Assist With Neutrophil Accumulation to Fight Against Systemic Staphylococcus aureus Infection. Front. Immunol. 2021, 12, 636818. [Google Scholar] [CrossRef] [PubMed]
- MacIver, N.J.; Michalek, R.D.; Rathmell, J.C. Metabolic Regulation of T Lymphocytes. Annu. Rev. Immunol. 2013, 31, 259–283. [Google Scholar] [CrossRef]
- Stienstra, R.; Netea-Maier, R.T.; Riksen, N.P.; Joosten, L.A.B.; Netea, M.G. Specific and Complex Reprogramming of Cellular Metabolism in Myeloid Cells during Innate Immune Responses. Cell Metab. 2017, 26, 142–156. [Google Scholar] [CrossRef]
- Singer, M.; Brealey, D. Mitochondrial Dysfunction in Sepsis. Biochem. Soc. Symp. 1999, 66, 149–166. [Google Scholar] [CrossRef]
- Van den Berghe, G.; de Zegher, F.; Bouillon, R. Acute and Prolonged Critical Illness as Different Neuroendocrine Paradigms. J. Clin. Endocrinol. Metab. 1998, 83, 1827–1834. [Google Scholar] [CrossRef]
- Singer, M. Mitochondrial Function in Sepsis: Acute Phase versus Multiple Organ Failure. Crit. Care Med. 2007, 35 (Suppl. S9), S441–S448. [Google Scholar] [CrossRef]
- Kushimoto, S.; Akaishi, S.; Sato, T.; Nomura, R.; Fujita, M.; Kudo, D.; Kawazoe, Y.; Yoshida, Y.; Miyagawa, N. Lactate, a Useful Marker for Disease Mortality and Severity but an Unreliable Marker of Tissue Hypoxia/Hypoperfusion in Critically Ill Patients. Acute Med. Surg. 2016, 3, 293–297. [Google Scholar] [CrossRef]
- Moran, J.L.; Santamaria, J. Reconsidering Lactate as a Sepsis Risk Biomarker. PLoS ONE 2017, 12, e0185320. [Google Scholar] [CrossRef]
- Sauer, C.M.; Gómez, J.; Botella, M.R.; Ziehr, D.R.; Oldham, W.M.; Gavidia, G.; Rodríguez, A.; Elbers, P.; Girbes, A.; Bodi, M.; et al. Understanding Critically Ill Sepsis Patients with Normal Serum Lactate Levels: Results from U.S. and European ICU Cohorts. Sci. Rep. 2021, 11, 20076. [Google Scholar] [CrossRef]
- Zhang, H.; Feng, Y.-W.; Yao, Y.-M. Potential Therapy Strategy: Targeting Mitochondrial Dysfunction in Sepsis. Mil. Med. Res. 2018, 5, 41. [Google Scholar] [CrossRef] [PubMed]
- Singer, M. The Role of Mitochondrial Dysfunction in Sepsis-Induced Multi-Organ Failure. Virulence 2014, 5, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Correa, T.; Jakob, S.; Takala, J. Mitochondrial Function in Sepsis. Crit. Care Horiz. 2015, 1, 31. [Google Scholar]
- Fink, M.P. Bench-to-Bedside Review: Cytopathic Hypoxia. Crit. Care 2002, 6, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Larsen, F.J.; Schiffer, T.A.; Weitzberg, E.; Lundberg, J.O. Regulation of Mitochondrial Function and Energetics by Reactive Nitrogen Oxides. Free Radic. Biol. Med. 2012, 53, 1919–1928. [Google Scholar] [CrossRef] [PubMed]
- Carré, J.E.; Orban, J.-C.; Re, L.; Felsmann, K.; Iffert, W.; Bauer, M.; Suliman, H.B.; Piantadosi, C.A.; Mayhew, T.M.; Breen, P.; et al. Survival in Critical Illness Is Associated with Early Activation of Mitochondrial Biogenesis. Am. J. Respir. Crit. Care Med. 2010, 182, 745–751. [Google Scholar] [CrossRef]
- Haden, D.W.; Suliman, H.B.; Carraway, M.S.; Welty-Wolf, K.E.; Ali, A.S.; Shitara, H.; Yonekawa, H.; Piantadosi, C.A. Mitochondrial Biogenesis Restores Oxidative Metabolism during Staphylococcus aureus Sepsis. Am. J. Respir. Crit. Care Med. 2007, 176, 768–777. [Google Scholar] [CrossRef]
- Singer, M.; De Santis, V.; Vitale, D.; Jeffcoate, W. Multiorgan Failure Is an Adaptive, Endocrine-Mediated, Metabolic Response to Overwhelming Systemic Inflammation. Lancet 2004, 364, 545–548. [Google Scholar] [CrossRef]
- Arulkumaran, N.; Deutschman, C.S.; Pinsky, M.R.; Zuckerbraun, B.; Schumacker, P.T.; Gomez, H.; Gomez, A.; Murray, P.; Kellum, J.A. Mitochondrial Function in Sepsis. Shock 2016, 45, 271–281. [Google Scholar] [CrossRef]
- Vary, T.C.; Drnevich, D.; Jurasinski, C.; Brennan, W.A. Mechanisms Regulating Skeletal Muscle Glucose Metabolism in Sepsis. Shock 1995, 3, 403–410. [Google Scholar] [PubMed]
- Zhang, Q.; Hu, Y.; Zhang, J.; Deng, C. iTRAQ-Based Proteomic Analysis of Endotoxin Tolerance Induced by Lipopolysaccharide. Mol. Med. Rep. 2019, 20, 584–592. [Google Scholar] [CrossRef] [PubMed]
- Morris, M.; Li, L. Molecular Mechanisms and Pathological Consequences of Endotoxin Tolerance and Priming. Arch. Immunol. Ther. Exp. 2012, 60, 13–18. [Google Scholar] [CrossRef]
- Liu, T.F.; Vachharajani, V.T.; Yoza, B.K.; McCall, C.E. NAD+-Dependent Sirtuin 1 and 6 Proteins Coordinate a Switch from Glucose to Fatty Acid Oxidation during the Acute Inflammatory Response. J. Biol. Chem. 2012, 287, 25758–25769. [Google Scholar] [CrossRef]
- Papathanakos, G.; Andrianopoulos, I.; Xenikakis, M.; Papathanasiou, A.; Koulenti, D.; Blot, S.; Koulouras, V. Clinical Sepsis Phenotypes in Critically Ill Patients. Microorganisms 2023, 11, 2165. [Google Scholar] [CrossRef]
- Wong, H.R. Personalized Medicine, Endotypes, and Intensive Care Medicine. Intensive Care Med. 2015, 41, 1138–1140. [Google Scholar] [CrossRef] [PubMed]
- Rhee, C.; Kadri, S.S.; Danner, R.L.; Suffredini, A.F.; Massaro, A.F.; Kitch, B.T.; Lee, G.; Klompas, M. Diagnosing Sepsis Is Subjective and Highly Variable: A Survey of Intensivists Using Case Vignettes. Crit. Care 2016, 20, 89. [Google Scholar] [CrossRef] [PubMed]
- Kellum, J.A.; Formeck, C.L.; Kernan, K.F.; Gómez, H.; Carcillo, J.A. Subtypes and Mimics of Sepsis. Crit. Care Clin. 2022, 38, 195–211. [Google Scholar] [CrossRef] [PubMed]
- Shah, F.A.; Meyer, N.J.; Angus, D.C.; Awdish, R.; Azoulay, É.; Calfee, C.S.; Clermont, G.; Gordon, A.C.; Kwizera, A.; Leligdowicz, A.; et al. A Research Agenda for Precision Medicine in Sepsis and Acute Respiratory Distress Syndrome: An Official American Thoracic Society Research Statement. Am. J. Respir. Crit. Care Med. 2021, 204, 891–901. [Google Scholar] [CrossRef] [PubMed]
- Evans, L.; Rhodes, A.; Alhazzani, W.; Antonelli, M.; Coopersmith, C.M.; French, C.; Machado, F.R.; Mcintyre, L.; Ostermann, M.; Prescott, H.C.; et al. Surviving Sepsis Campaign: International Guidelines for Management of Sepsis and Septic Shock 2021. Intensive Care Med. 2021, 47, 1181–1247. [Google Scholar] [CrossRef] [PubMed]
- Castela Forte, J.; Perner, A.; van der Horst, I.C.C. The Use of Clustering Algorithms in Critical Care Research to Unravel Patient Heterogeneity. Intensive Care Med. 2019, 45, 1025–1028. [Google Scholar] [CrossRef]
- Vincent, J.-L.; Singer, M.; Einav, S.; Moreno, R.; Wendon, J.; Teboul, J.-L.; Bakker, J.; Hernandez, G.; Annane, D.; de Man, A.M.E.; et al. Equilibrating SSC Guidelines with Individualized Care. Crit. Care 2021, 25, 397. [Google Scholar] [CrossRef]
- Evans, L.; Rhodes, A.; Alhazzani, W.; Antonelli, M.; Coopersmith, C.M.; French, C.; Machado, F.R.; Mcintyre, L.; Ostermann, M.; Prescott, H.C.; et al. Executive Summary: Surviving Sepsis Campaign: International Guidelines for the Management of Sepsis and Septic Shock 2021. Crit. Care Med. 2021, 49, 1974–1982. [Google Scholar] [CrossRef]
- Seymour, C.W.; Gomez, H.; Chang, C.-C.H.; Clermont, G.; Kellum, J.A.; Kennedy, J.; Yende, S.; Angus, D.C. Precision Medicine for All? Challenges and Opportunities for a Precision Medicine Approach to Critical Illness. Crit. Care 2017, 21, 257. [Google Scholar] [CrossRef]
- Beutler, B.; Du, X.; Hoebe, K. From Phenomenon to Phenotype and from Phenotype to Gene: Forward Genetics and the Problem of Sepsis. J. Infect. Dis. 2003, 187 (Suppl. S2), S321–S326. [Google Scholar] [CrossRef]
- Ward, N.; Levy, M. Sepsis: Definitions, Pathophysiology and the Challenge of Bedside Management; Humana: Cham, Switzerland, 2017. [Google Scholar] [CrossRef]
- Ke, X.; Zhang, F.; Huang, G.; Wang, A. Interpretable Machine Learning to Optimize Early In-Hospital Mortality Prediction for Elderly Patients with Sepsis: A Discovery Study. Comput. Math. Methods Med. 2022, 2022, 4820464. [Google Scholar] [CrossRef] [PubMed]
- Vranas, K.C.; Jopling, J.K.; Sweeney, T.E.; Ramsey, M.C.; Milstein, A.S.; Slatore, C.G.; Escobar, G.J.; Liu, V.X. Identifying Distinct Subgroups of ICU Patients: A Machine Learning Approach. Crit. Care Med. 2017, 45, 1607–1615. [Google Scholar] [CrossRef]
- Qin, Y.; Caldino Bohn, R.I.; Sriram, A.; Kernan, K.F.; Carcillo, J.A.; Kim, S.; Park, H.J. Refining Empiric Subgroups of Pediatric Sepsis Using Machine-Learning Techniques on Observational Data. Front. Pediatr. 2023, 11, 1035576. [Google Scholar] [CrossRef] [PubMed]
- Komorowski, M.; Green, A.; Tatham, K.C.; Seymour, C.; Antcliffe, D. Sepsis Biomarkers and Diagnostic Tools with a Focus on Machine Learning. EBioMedicine 2022, 86, 104394. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Markal, A.; Balch, J.A.; Loftus, T.J.; Efron, P.A.; Ozrazgat-Baslanti, T.; Bihorac, A. Methods for Phenotyping Adult Patients in Sepsis and Septic Shock: A Scoping Review. Crit. Care Explor. 2022, 4, e0672. [Google Scholar] [CrossRef]
- Sharma, M.; Taweesedt, P.T.; Surani, S. Utilizing Artificial Intelligence in Critical Care: Adding A Handy Tool to Our Armamentarium. Cureus 2021, 13, e15531. [Google Scholar] [CrossRef]
- Loftus, T.J.; Shickel, B.; Balch, J.A.; Tighe, P.J.; Abbott, K.L.; Fazzone, B.; Anderson, E.M.; Rozowsky, J.; Ozrazgat-Baslanti, T.; Ren, Y.; et al. Phenotype Clustering in Health Care: A Narrative Review for Clinicians. Front. Artif. Intell. 2022, 5, 842306. [Google Scholar] [CrossRef]
- Baek, M.S.; Kim, J.H.; Kwon, Y.S. Cluster Analysis Integrating Age and Body Temperature for Mortality in Patients with Sepsis: A Multicenter Retrospective Study. Sci. Rep. 2022, 12, 1090. [Google Scholar] [CrossRef]
- Zhu, J.-L.; Yuan, S.-Q.; Huang, T.; Zhang, L.-M.; Xu, X.-M.; Yin, H.-Y.; Wei, J.-R.; Lyu, J. Influence of Systolic Blood Pressure Trajectory on In-Hospital Mortality in Patients with Sepsis. BMC Infect. Dis. 2023, 23, 90. [Google Scholar] [CrossRef]
- Daulasim, A.; Vieillard-Baron, A.; Geri, G. Hemodynamic Clinical Phenotyping in Septic Shock. Curr. Opin. Crit. Care 2021, 27, 290–297. [Google Scholar] [CrossRef]
- Geri, G.; Vignon, P.; Aubry, A.; Fedou, A.-L.; Charron, C.; Silva, S.; Repessé, X.; Vieillard-Baron, A. Cardiovascular Clusters in Septic Shock Combining Clinical and Echocardiographic Parameters: A Post Hoc Analysis. Intensive Care Med. 2019, 45, 657–667. [Google Scholar] [CrossRef]
- Knox, D.B.; Lanspa, M.J.; Kuttler, K.G.; Brewer, S.C.; Brown, S.M. Phenotypic Clusters within Sepsis-Associated Multiple Organ Dysfunction Syndrome. Intensive Care Med. 2015, 41, 814–822. [Google Scholar] [CrossRef]
- Ibrahim, Z.M.; Wu, H.; Hamoud, A.; Stappen, L.; Dobson, R.J.B.; Agarossi, A. On Classifying Sepsis Heterogeneity in the ICU: Insight Using Machine Learning. J. Am. Med. Inform. Assoc. 2020, 27, 437–443. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, G.; Goyal, H.; Mo, L.; Hong, Y. Identification of Subclasses of Sepsis That Showed Different Clinical Outcomes and Responses to Amount of Fluid Resuscitation: A Latent Profile Analysis. Crit. Care 2018, 22, 347. [Google Scholar] [CrossRef]
- Seymour, C.W.; Kennedy, J.N.; Wang, S.; Chang, C.-C.H.; Elliott, C.F.; Xu, Z.; Berry, S.; Clermont, G.; Cooper, G.; Gomez, H.; et al. Derivation, Validation, and Potential Treatment Implications of Novel Clinical Phenotypes for Sepsis. JAMA 2019, 321, 2003–2017. [Google Scholar] [CrossRef]
- Shald, E.A.; Erdman, M.J.; Ferreira, J.A. Impact of Clinical Sepsis Phenotypes on Mortality and Fluid Status in Critically Ill Patients. Shock 2022, 57, 57–62. [Google Scholar] [CrossRef]
- Wang, M.; Zhu, B.; Jiang, L.; Luo, X.; Wang, N.; Zhu, Y.; Xi, X. Association between Latent Trajectories of Fluid Balance and Clinical Outcomes in Critically Ill Patients with Acute Kidney Injury: A Prospective Multicenter Observational Study. Kidney Dis. 2022, 8, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Liu, J.; Shen, F.; Liao, X.; Xiu, M.; Zhao, H.; Zhao, M.; Xie, J.; Wang, P.; Huang, M.; et al. Individualized Resuscitation Strategy for Septic Shock Formalized by Finite Mixture Modeling and Dynamic Treatment Regimen. Crit. Care 2021, 25, 243. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.P.; Bray, B.C.; Chou, S.-H.; Burns, R.; Kowalkowski, M.A. Clinical Subtypes of Sepsis Survivors Predict Readmission and Mortality after Hospital Discharge. Ann. Am. Thorac. Soc. 2022, 19, 1355–1363. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Han, D.; Zhang, L.; Huang, T.; Xu, F.; Zheng, S.; Yin, H.; Lyu, J. Analysis of the Correlation between the Longitudinal Trajectory of SOFA Scores and Prognosis in Patients with Sepsis at 72 Hour after Admission Based on Group Trajectory Modeling. J. Intensive Med. 2022, 2, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Soussi, S.; Sharma, D.; Jüni, P.; Lebovic, G.; Brochard, L.; Marshall, J.C.; Lawler, P.R.; Herridge, M.; Ferguson, N.; Del Sorbo, L.; et al. Identifying Clinical Subtypes in Sepsis-Survivors with Different One-Year Outcomes: A Secondary Latent Class Analysis of the FROG-ICU Cohort. Crit. Care 2022, 26, 114. [Google Scholar] [CrossRef]
- Cajander, S.; Kox, M.; Scicluna, B.P.; Weigand, M.A.; Mora, R.A.; Flohé, S.B.; Martin-Loeches, I.; Lachmann, G.; Girardis, M.; Garcia-Salido, A.; et al. Profiling the Dysregulated Immune Response in Sepsis: Overcoming Challenges to Achieve the Goal of Precision Medicine. Lancet Respir. Med. 2023. [Google Scholar] [CrossRef] [PubMed]
- Wong, H.R.; Salisbury, S.; Xiao, Q.; Cvijanovich, N.Z.; Hall, M.; Allen, G.L.; Thomas, N.J.; Freishtat, R.J.; Anas, N.; Meyer, K.; et al. The Pediatric Sepsis Biomarker Risk Model. Crit. Care 2012, 16, R174. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, T.E.; Azad, T.D.; Donato, M.; Haynes, W.A.; Perumal, T.M.; Henao, R.; Bermejo-Martin, J.F.; Almansa, R.; Tamayo, E.; Howrylak, J.A.; et al. Unsupervised Analysis of Transcriptomics in Bacterial Sepsis Across Multiple Datasets Reveals Three Robust Clusters. Crit. Care Med. 2018, 46, 915–925. [Google Scholar] [CrossRef]
- Marutescu, L.G. Current and Future Flow Cytometry Applications Contributing to Antimicrobial Resistance Control. Microorganisms 2023, 11, 1300. [Google Scholar] [CrossRef] [PubMed]
- Fisher, C.J.; Agosti, J.M.; Opal, S.M.; Lowry, S.F.; Balk, R.A.; Sadoff, J.C.; Abraham, E.; Schein, R.M.; Benjamin, E. Treatment of Septic Shock with the Tumor Necrosis Factor Receptor:Fc Fusion Protein. N. Engl. J. Med. 1996, 334, 1697–1702. [Google Scholar] [CrossRef] [PubMed]
- Patel, G.P.; Balk, R.A. Systemic Steroids in Severe Sepsis and Septic Shock. Am. J. Respir. Crit. Care Med. 2012, 185, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Schumer, W. Steroids in the Treatment of Clinical Septic Shock. Ann. Surg. 1976, 184, 333–341. [Google Scholar] [CrossRef]
- Vincent, J.-L.; Martinez, E.O.; Silva, E. Evolving Concepts in Sepsis Definitions. Crit. Care Nurs. Clin. N. Am. 2011, 23, 29–39. [Google Scholar] [CrossRef]
- Munford, R.S. Severe Sepsis and Septic Shock: The Role of Gram-Negative Bacteremia. Annu. Rev. Pathol. 2006, 1, 467–496. [Google Scholar] [CrossRef]
- Drazen, J.M. Transparency for Clinical Trials--the TEST Act. N. Engl. J. Med. 2012, 367, 863–864. [Google Scholar] [CrossRef]
- Calvano, S.E.; Xiao, W.; Richards, D.R.; Felciano, R.M.; Baker, H.V.; Cho, R.J.; Chen, R.O.; Brownstein, B.H.; Cobb, J.P.; Tschoeke, S.K.; et al. A Network-Based Analysis of Systemic Inflammation in Humans. Nature 2005, 437, 1032–1037. [Google Scholar] [CrossRef]
- Kamisoglu, K.; Sleight, K.E.; Calvano, S.E.; Coyle, S.M.; Corbett, S.A.; Androulakis, I.P. Temporal Metabolic Profiling of Plasma during Endotoxemia in Humans. Shock 2013, 40, 519–526. [Google Scholar] [CrossRef]
- Seok, J.; Warren, H.S.; Cuenca, A.G.; Mindrinos, M.N.; Baker, H.V.; Xu, W.; Richards, D.R.; McDonald-Smith, G.P.; Gao, H.; Hennessy, L.; et al. Genomic Responses in Mouse Models Poorly Mimic Human Inflammatory Diseases. Proc. Natl. Acad. Sci. USA 2013, 110, 3507–3512. [Google Scholar] [CrossRef] [PubMed]
- Osuchowski, M.F.; Remick, D.G.; Lederer, J.A.; Lang, C.H.; Aasen, A.O.; Aibiki, M.; Azevedo, L.C.; Bahrami, S.; Boros, M.; Cooney, R.; et al. Abandon the Mouse Research Ship? Not Just Yet! Shock 2014, 41, 463–475. [Google Scholar] [CrossRef] [PubMed]
- Bode, C.; Weis, S.; Sauer, A.; Wendel-Garcia, P.; David, S. Targeting the host response in sepsis: Current approaches and future evidence. Crit. Care 2023, 27, 478. [Google Scholar] [CrossRef] [PubMed]
- Lombardo, E.; van der Poll, T.; DelaRosa, O.; Dalemans, W. Mesenchymal Stem Cells as a Therapeutic Tool to Treat Sepsis. World J. Stem Cells 2015, 7, 368–379. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.D.; Wilson, J.G.; Zhuo, H.; Caballero, L.; McMillan, M.L.; Fang, X.; Cosgrove, K.; Calfee, C.S.; Lee, J.-W.; Kangelaris, K.N.; et al. Design and Implementation of the START (STem Cells for ARDS Treatment) Trial, a Phase 1/2 Trial of Human Mesenchymal Stem/Stromal Cells for the Treatment of Moderate-Severe Acute Respiratory Distress Syndrome. Ann. Intensive Care 2014, 4, 22. [Google Scholar] [CrossRef] [PubMed]
- Abraham, E. New Definitions for Sepsis and Septic Shock: Continuing Evolution but With Much Still to Be Done. JAMA 2016, 315, 757–759. [Google Scholar] [CrossRef] [PubMed]
- Loupy, A.; Lefaucheur, C.; Vernerey, D.; Chang, J.; Hidalgo, L.G.; Beuscart, T.; Verine, J.; Aubert, O.; Dubleumortier, S.; Duong van Huyen, J.-P.; et al. Molecular Microscope Strategy to Improve Risk Stratification in Early Antibody-Mediated Kidney Allograft Rejection. J. Am. Soc. Nephrol. 2014, 25, 2267–2277. [Google Scholar] [CrossRef]
- Davenport, E.E.; Burnham, K.L.; Radhakrishnan, J.; Humburg, P.; Hutton, P.; Mills, T.C.; Rautanen, A.; Gordon, A.C.; Garrard, C.; Hill, A.V.S.; et al. Genomic Landscape of the Individual Host Response and Outcomes in Sepsis: A Prospective Cohort Study. Lancet Respir. Med. 2016, 4, 259–271. [Google Scholar] [CrossRef]
- Semmler, A.; Prost, J.-C.; Smulders, Y.; Smith, D.; Blom, H.; Bigler, L.; Linnebank, M. Methylation Metabolism in Sepsis and Systemic Inflammatory Response Syndrome. Scand. J. Clin. Lab. Investig. 2013, 73, 368–372. [Google Scholar] [CrossRef] [PubMed]
- Nicodeme, E.; Jeffrey, K.L.; Schaefer, U.; Beinke, S.; Dewell, S.; Chung, C.-W.; Chandwani, R.; Marazzi, I.; Wilson, P.; Coste, H.; et al. Suppression of Inflammation by a Synthetic Histone Mimic. Nature 2010, 468, 1119–1123. [Google Scholar] [CrossRef] [PubMed]
- Tacke, F.; Roderburg, C.; Benz, F.; Cardenas, D.V.; Luedde, M.; Hippe, H.-J.; Frey, N.; Vucur, M.; Gautheron, J.; Koch, A.; et al. Levels of Circulating miR-133a Are Elevated in Sepsis and Predict Mortality in Critically Ill Patients. Crit. Care Med. 2014, 42, 1096–1104. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Vilanova, D.; Atalar, K.; Delfour, O.; Edgeworth, J.; Ostermann, M.; Hernandez-Fuentes, M.; Razafimahatratra, S.; Michot, B.; Persing, D.H.; et al. Genome-Wide Sequencing of Cellular microRNAs Identifies a Combinatorial Expression Signature Diagnostic of Sepsis. PLoS ONE 2013, 8, e75918. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, P.; Chen, W.; Feng, D.; Jia, Y.; Xie, L. Serum microRNA Signatures Identified by Solexa Sequencing Predict Sepsis Patients’ Mortality: A Prospective Observational Study. PLoS ONE 2012, 7, e38885. [Google Scholar] [CrossRef]
- Camussi, G.; Deregibus, M.C.; Bruno, S.; Cantaluppi, V.; Biancone, L. Exosomes/Microvesicles as a Mechanism of Cell-to-Cell Communication. Kidney Int. 2010, 78, 838–848. [Google Scholar] [CrossRef]
- Gambim, M.H.; do Carmo, A.d.O.; Marti, L.; Veríssimo-Filho, S.; Lopes, L.R.; Janiszewski, M. Platelet-Derived Exosomes Induce Endothelial Cell Apoptosis through Peroxynitrite Generation: Experimental Evidence for a Novel Mechanism of Septic Vascular Dysfunction. Crit. Care 2007, 11, R107. [Google Scholar] [CrossRef]
- Azevedo, L.C.P.; Janiszewski, M.; Pontieri, V.; Pedro, M.d.A.; Bassi, E.; Tucci, P.J.F.; Laurindo, F.R.M. Platelet-Derived Exosomes from Septic Shock Patients Induce Myocardial Dysfunction. Crit. Care 2007, 11, R120. [Google Scholar] [CrossRef]
- Dakhlallah, D.A.; Wisler, J.; Gencheva, M.; Brown, C.M.; Leatherman, E.R.; Singh, K.; Brundage, K.; Karsies, T.; Dakhlallah, A.; Witwer, K.W.; et al. Circulating Extracellular Vesicle Content Reveals de Novo DNA Methyltransferase Expression as a Molecular Method to Predict Septic Shock. J. Extracell. Vesicles 2019, 8, 1669881. [Google Scholar] [CrossRef]
- Timár, C.I.; Lorincz, A.M.; Csépányi-Kömi, R.; Vályi-Nagy, A.; Nagy, G.; Buzás, E.I.; Iványi, Z.; Kittel, A.; Powell, D.W.; McLeish, K.R.; et al. Antibacterial Effect of Microvesicles Released from Human Neutrophilic Granulocytes. Blood 2013, 121, 510–518. [Google Scholar] [CrossRef]
- Morris, D.C.; Jaehne, A.K.; Chopp, M.; Zhang, Z.; Poisson, L.; Chen, Y.; Datta, I.; Rivers, E.P. Proteomic Profiles of Exosomes of Septic Patients Presenting to the Emergency Department Compared to Healthy Controls. J. Clin. Med. 2020, 9, 2930. [Google Scholar] [CrossRef]
- Real, J.M.; Ferreira, L.R.P.; Esteves, G.H.; Koyama, F.C.; Dias, M.V.S.; Bezerra-Neto, J.E.; Cunha-Neto, E.; Machado, F.R.; Salomão, R.; Azevedo, L.C.P. Exosomes from Patients with Septic Shock Convey miRNAs Related to Inflammation and Cell Cycle Regulation: New Signaling Pathways in Sepsis? Crit. Care 2018, 22, 68. [Google Scholar] [CrossRef]
- Eken, C.; Gasser, O.; Zenhaeusern, G.; Oehri, I.; Hess, C.; Schifferli, A. Polymorphonuclear Neutrophil-Derived Ectosomes Interfere with the Maturation of Monocyte-Derived Dendritic Cells. J. Immunol. 2008, 180, 817–824. [Google Scholar] [CrossRef]
- Gasser, O.; Schifferli, J.A. Activated Polymorphonuclear Neutrophils Disseminate Anti-Inflammatory Microparticles by Ectocytosis. Blood 2004, 104, 2543–2548. [Google Scholar] [CrossRef]
- Dalli, J.; Norling, L.V.; Renshaw, D.; Cooper, D.; Leung, K.-Y.; Perretti, M. Annexin 1 Mediates the Rapid Anti-Inflammatory Effects of Neutrophil-Derived Microparticles. Blood 2008, 112, 2512–2519. [Google Scholar] [CrossRef]
- Eken, C.; Martin, P.J.; Sadallah, S.; Treves, S.; Schaller, M.; Schifferli, J.A. Ectosomes Released by Polymorphonuclear Neutrophils Induce a MerTK-Dependent Anti-Inflammatory Pathway in Macrophages. J. Biol. Chem. 2010, 285, 39914–39921. [Google Scholar] [CrossRef]
- Dalli, J.; Montero-Melendez, T.; Norling, L.V.; Yin, X.; Hinds, C.; Haskard, D.; Mayr, M.; Perretti, M. Heterogeneity in Neutrophil Microparticles Reveals Distinct Proteome and Functional Properties. Mol. Cell. Proteom. 2013, 12, 2205–2219. [Google Scholar] [CrossRef] [PubMed]
- Dalli, J.; Norling, L.V.; Montero-Melendez, T.; Canova, D.F.; Lashin, H.; Pavlov, A.M.; Sukhorukov, G.B.; Hinds, C.J.; Perretti, M. Microparticle Alpha-2-Macroglobulin Enhances pro-Resolving Responses and Promotes Survival in Sepsis. EMBO Mol. Med. 2014, 6, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Wisler, J.R.; Singh, K.; Mccarty, A.R.; Abouhashem, A.S.E.; Christman, J.W.; Sen, C.K. Proteomic Pathway Analysis of Monocyte-Derived Exosomes during Surgical Sepsis Identifies Immunoregulatory Functions. Surg. Infect. 2020, 21, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.; Liu, Y.; Qiu, X.; Tang, Y.; Wang, H.; Xiao, X.; Chen, F.; Lu, B. Bacteria-Released Outer Membrane Vesicles Promote Disseminated Intravascular Coagulation. Thromb. Res. 2019, 178, 26–33. [Google Scholar] [CrossRef]
- Sui, X.; Liu, W.; Liu, Z. Exosomes Derived from LPS-Induced MHs Cells Prompted an Inflammatory Response in Sepsis-Induced Acute Lung Injury. Respir. Physiol. Neurobiol. 2021, 292, 103711. [Google Scholar] [CrossRef] [PubMed]
- Nair, R.R.; Mazza, D.; Brambilla, F.; Gorzanelli, A.; Agresti, A.; Bianchi, M.E. LPS-Challenged Macrophages Release Microvesicles Coated With Histones. Front. Immunol. 2018, 9, 1463. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Huang, W.; Wang, S.; Wang, J.; Cui, W.; Zhang, W.; Lou, A.; Geng, S.; Li, X. Macrophagic Extracellular Vesicle CXCL2 Recruits and Activates the Neutrophil CXCR2/PKC/NOX4 Axis in Sepsis. J. Immunol. 2021, 207, 2118–2128. [Google Scholar] [CrossRef] [PubMed]
- Murao, A.; Tan, C.; Jha, A.; Wang, P.; Aziz, M. Exosome-Mediated eCIRP Release From Macrophages to Induce Inflammation in Sepsis. Front. Pharmacol. 2021, 12, 791648. [Google Scholar] [CrossRef] [PubMed]
- Barbora, K.; Jinbong, P.; Woon-Yong, K.; Barbora, V.; Quanzhi, Z.; Wei, H.; Hyo In, K.; Michael, B.Y.; Leo, E.O.; Kiyoshi, I.; et al. Monocyte Exocytosis of Mitochondrial Danger-Associated Molecular Patterns in Sepsis Suppresses Neutrophil Chemotaxis. J. Trauma Acute Care Surg. 2021, 90, 46–53. [Google Scholar] [CrossRef]
- Yang, K.; Fan, M.; Wang, X.; Xu, J.; Wang, Y.; Tu, F.; Gill, P.S.; Ha, T.; Liu, L.; Williams, D.L.; et al. Lactate Promotes Macrophage HMGB1 Lactylation, Acetylation, and Exosomal Release in Polymicrobial Sepsis. Cell Death Differ. 2022, 29, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Kumari, P.; Vasudevan, S.O.; Russo, A.J.; Wright, S.S.; Fraile-Ágreda, V.; Krajewski, D.; Jellison, E.R.; Rubio, I.; Bauer, M.; Shimoyama, A.; et al. Host Extracellular Vesicles Confer Cytosolic Access to Systemic LPS Licensing Non-Canonical Inflammasome Sensing and Pyroptosis. Nat. Cell Biol. 2023, 25, 1860–1872. [Google Scholar] [CrossRef] [PubMed]
- Monneret, G.; Gossez, M.; Aghaeepour, N.; Gaudilliere, B.; Venet, F. How Clinical Flow Cytometry Rebooted Sepsis Immunology. Cytom. Part A J. Int. Soc. Anal. Cytol. 2019, 95, 431–441. [Google Scholar] [CrossRef]
- Ruiz-Rodriguez, J.C.; Plata-Menchaca, E.P.; Chiscano-Camón, L.; Ruiz-Sanmartin, A.; Pérez-Carrasco, M.; Palmada, C.; Ribas, V.; Martínez-Gallo, M.; Hernández-González, M.; Gonzalez-Lopez, J.J.; et al. Precision Medicine in Sepsis and Septic Shock: From Omics to Clinical Tools. World J. Crit. Care Med. 2022, 11, 1–21. [Google Scholar] [CrossRef]
- Yoon, J.H.; Pinsky, M.R.; Clermont, G. Artificial Intelligence in Critical Care Medicine. Crit. Care 2022, 26, 75. [Google Scholar] [CrossRef] [PubMed]
- Ren, T.; Chen, C.; Danilov, A.V.; Liu, S.; Guan, X.; Du, S.; Wu, X.; Sherman, M.H.; Spellman, P.T.; Coussens, L.M.; et al. Supervised learning of high-confidence phenotypic subpopulations from single-cell data. Nat. Mach. Intell. 2023, 5, 528–541. [Google Scholar] [CrossRef]
- Hédou, J.; Marić, I.; Bellan, G.; Einhaus, J.; Gaudillière, D.K.; Ladant, F.-X.; Verdonk, F.; Stelzer, I.A.; Feyaerts, D.; Tsai, A.S.; et al. Discovery of sparse, reliable omic biomarkers with Stabl. Nat. Biotechnol. 2024. [Google Scholar] [CrossRef] [PubMed]
- Persson, I.; Macura, A.; Becedas, D.; Sjövall, F. Early prediction of sepsis in intensive care patients using the machine learning algorithm NAVOY® Sepsis, a prospective randomized clinical validation study. J. Crit. Care 2024, 80, 154400. [Google Scholar] [CrossRef]

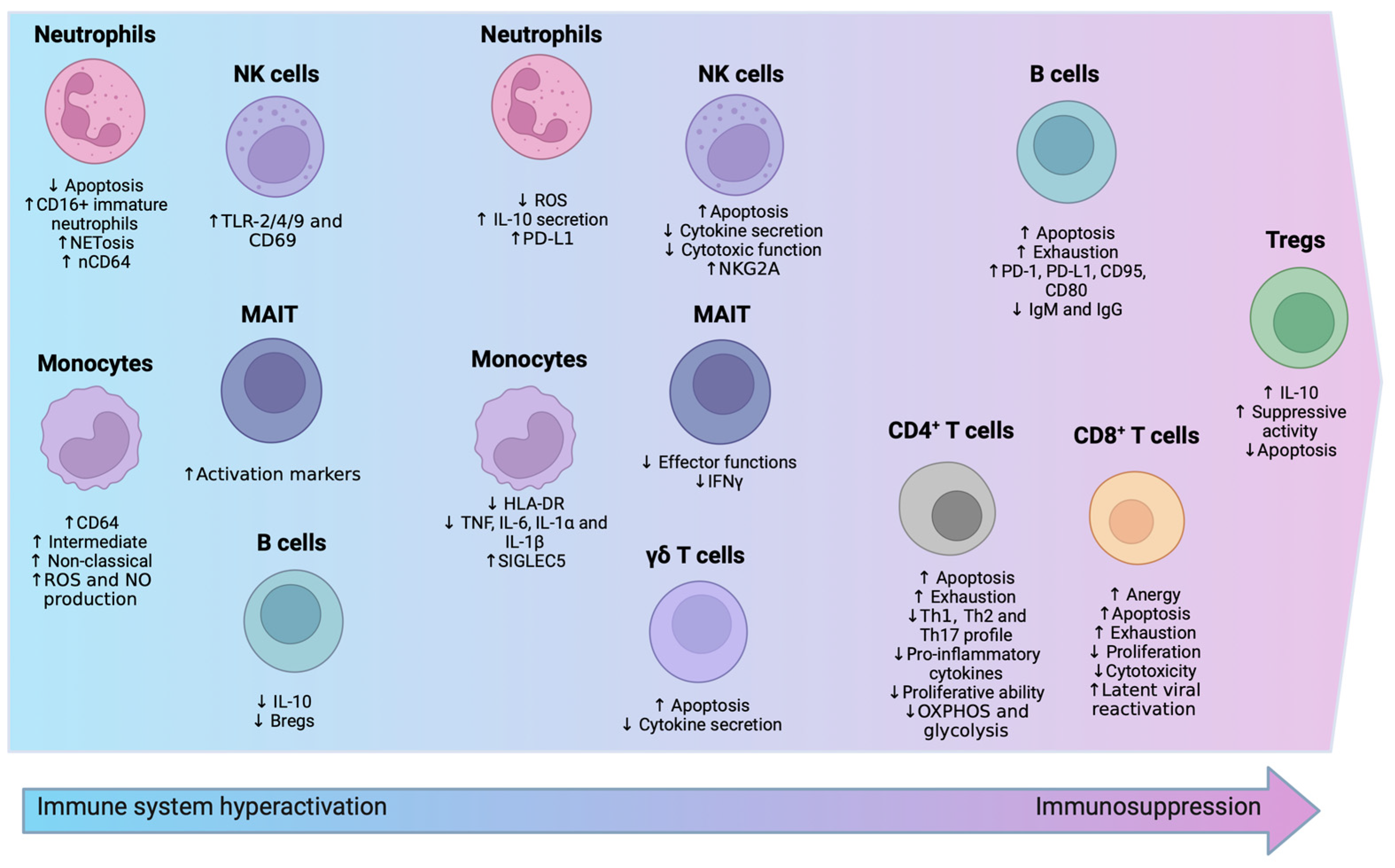



| Alterations in Sepsis | Main Findings in Septic Patients | Refs. |
|---|---|---|
| Immune Cells Alterations | ||
| Neutrophils | reduced apoptosis and ROS production | [59,61,62,63,69,70] |
| upregulation of CD64 and PD-L1 expression | [58,83,84,85,87,88,90] | |
| upregulation of CD16+ immature neutrophils | [63,64,65] | |
| enhanced IL-10 production and NETosis | [32,73,76,77] | |
| Monocytes | enhanced CD64 expression | [104,105] |
| reduced expression of HLA-DR | [94,95,96,97,98,99,100] | |
| upregulation of intermediate and non-classical phenotypes | [92] | |
| enhanced ROS and NO production | [104,108,109,110,111] | |
| upregulation of SIGLEC5 expression | [180] | |
| downregulation of TNF, IL-6, IL-1β, and IL-1α production | [106,107,112,113,114] | |
| NK cells | enhanced apoptosis | [134] |
| upregulation of TLR-2/4/9 and CD69 in the initial phase of sepsis | [123] | |
| upregulation of NKG2A expression | [129,130] | |
| reduced cytokine secretion | [120,135,136,137] | |
| impaired cytotoxic function | [128] | |
| γδT cells | enhanced apoptosis | [148] |
| reduced IFNγ secretion and CD69 expression | [147] | |
| MAIT cells | reduced effector functions | [152] |
| upregulation of HLA-DR and PD-L1 expression | [159] | |
| reduced IFNγ production | [158] | |
| upregulation of CD69, CD38 and CD137 expression | [158] | |
| CD4+ T cells | enhanced apoptosis | [167,168] |
| enhanced exhaustion | [10,174] | |
| reduced Th1, Th2, and Th17 populations | [169,170,171] | |
| reduced production of pro-inflammatory cytokines | [112,113,174,175] | |
| reduced proliferative ability | [176] | |
| reduced OXPHOS and glycolysis | [172] | |
| CD8+ T cells | augmented anergy | [179] |
| enhanced apoptosis | [178] | |
| enhanced exhaustion | [178] | |
| decreased naive CD8+ T cells and increased effector/effector memory CD8+ T cells in the first phase of sepsis | [180] | |
| reduced cell proliferation | [178] | |
| reduced cytotoxicity | [178] | |
| increased reactivation of latent viruses | [181,182,183] | |
| Tregs | augmented number of Tregs | [58,186] |
| constant IL-10 production | [187] | |
| reduced apoptosis | [184] | |
| B cells | enhanced apoptosis | [201,202,203,204] |
| enhanced exhaustion | [189,192] | |
| upregulation of PD-1, PD-L1, CD95, and CD80 expression | [191,193,200] (p. 2) | |
| reduced IL-10 production | [210] | |
| reduced IgM and IgG production | [35,192,195,212] | |
| reduced number of Bregs | [211] | |
| Metabolic Alterations | ||
| Mitochondrial dysfunction | reduced expression of genes encoding for proteins involved in electron transport chain (ECT) | [232,235,236] |
| lower copied of mtDNA | [237] | |
| decreased metabolic rate | [237] | |
| upregulated ROS production | [232,233,239] | |
| Glicolysis enhancement | augmented lactate production | [228,229,230] |
| increased GLUT1 expression | [240] (p. 199) | |
| Shift towards fatty acid oxidation (FAO) | increased levels of CD36 and CPT-1 | [243] |
| SIRT1 and SIRT6 cordinate the metabolic swith toward FAO | [243] | |
| Communication/Cell-Cell Crosstalk | ||
| miRNAs | upregulated levels of miR-15a, miR-16, miR-122, miR-133, miR-193, miR-223, and miR-483-5p | [299,300,301] |
| Extracellular vescicles (Evs) | increased number of EVs | [305,306] |
| EVs carrrying mRNA involved in antioxidand defence and oxidative stress | [308] | |
| upregulation of EVs carryng DNA methyltransferase (DNMT)1, DNMT3A, and DNMT3B mRNA | [305] | |
| neutrophil-derived EVs have anti-inflammatory effects | [309,310,311,312] | |
| pro-inflammatory response is enhanced by EVs carryng PAMPs, DAMPs like HMGB1, eCIRP, mtDAMPS | [315,316,317,318,319,320,321,322,323] (p. 20) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santacroce, E.; D’Angerio, M.; Ciobanu, A.L.; Masini, L.; Lo Tartaro, D.; Coloretti, I.; Busani, S.; Rubio, I.; Meschiari, M.; Franceschini, E.; et al. Advances and Challenges in Sepsis Management: Modern Tools and Future Directions. Cells 2024, 13, 439. https://doi.org/10.3390/cells13050439
Santacroce E, D’Angerio M, Ciobanu AL, Masini L, Lo Tartaro D, Coloretti I, Busani S, Rubio I, Meschiari M, Franceschini E, et al. Advances and Challenges in Sepsis Management: Modern Tools and Future Directions. Cells. 2024; 13(5):439. https://doi.org/10.3390/cells13050439
Chicago/Turabian StyleSantacroce, Elena, Miriam D’Angerio, Alin Liviu Ciobanu, Linda Masini, Domenico Lo Tartaro, Irene Coloretti, Stefano Busani, Ignacio Rubio, Marianna Meschiari, Erica Franceschini, and et al. 2024. "Advances and Challenges in Sepsis Management: Modern Tools and Future Directions" Cells 13, no. 5: 439. https://doi.org/10.3390/cells13050439
APA StyleSantacroce, E., D’Angerio, M., Ciobanu, A. L., Masini, L., Lo Tartaro, D., Coloretti, I., Busani, S., Rubio, I., Meschiari, M., Franceschini, E., Mussini, C., Girardis, M., Gibellini, L., Cossarizza, A., & De Biasi, S. (2024). Advances and Challenges in Sepsis Management: Modern Tools and Future Directions. Cells, 13(5), 439. https://doi.org/10.3390/cells13050439