Exploring the Interplay of RUNX2 and CXCR4 in Melanoma Progression
Abstract
1. Introduction
2. Material and Methods
2.1. Cell Cultures
2.2. RUNX2 Cloning and Transfection
2.3. Immunofluorescence
2.4. Three-Dimensional (3D) Cell Cultures
2.5. Total RNA Extraction and Reverse Transcription (RT)
2.6. Real-Time Quantitative PCR
2.7. Western Blotting
2.8. Statistical Analysis
3. Results
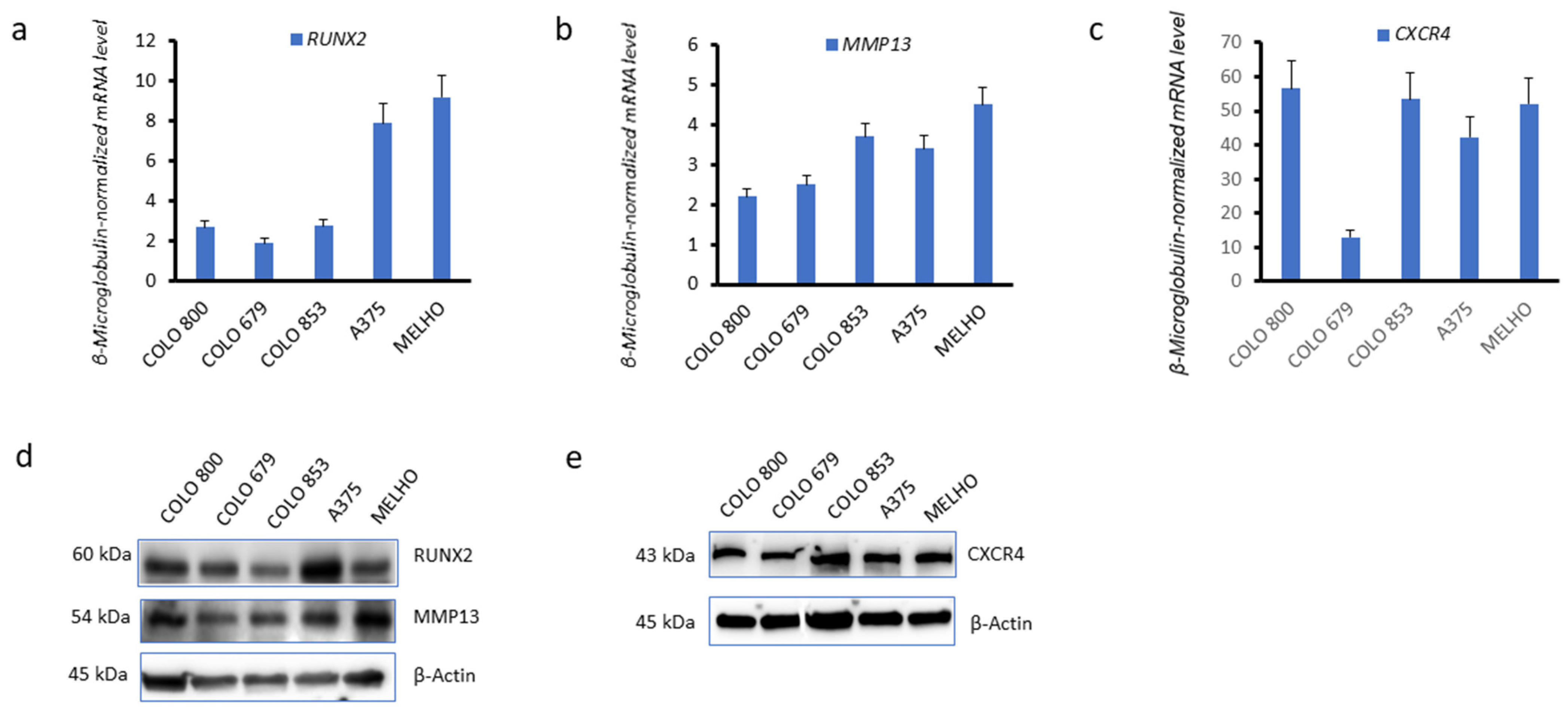
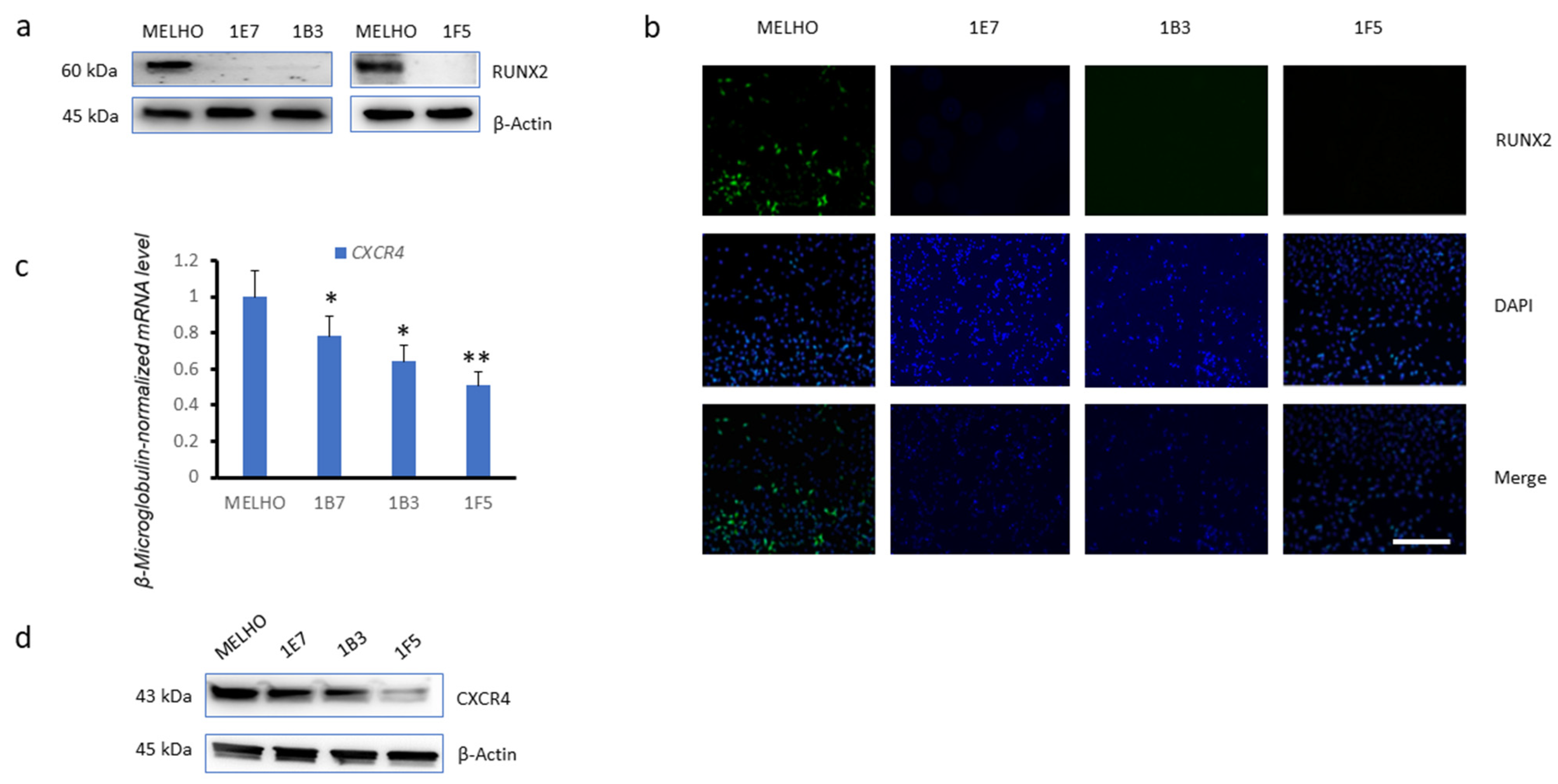
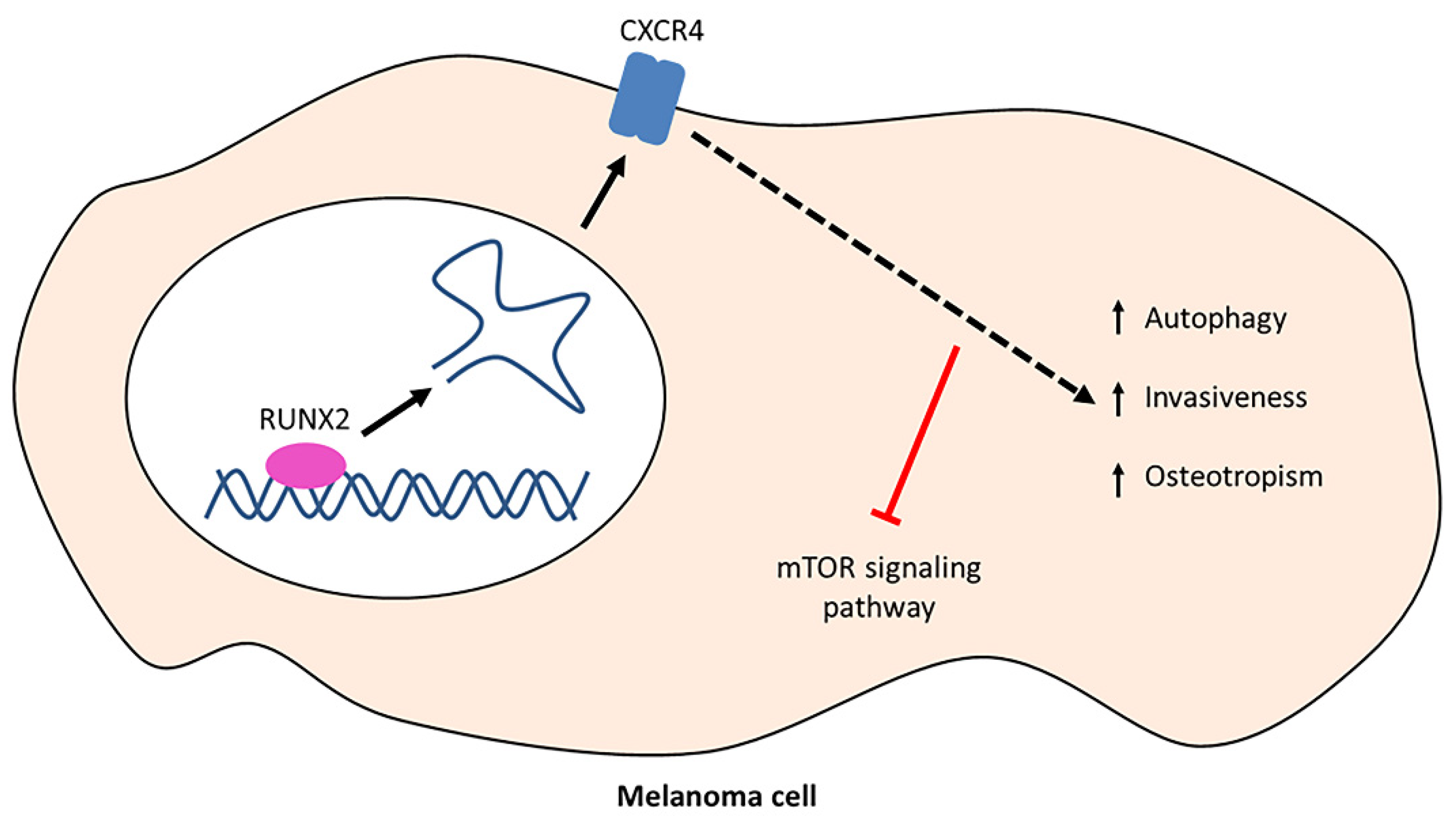
3.1. CXCR4 Is Associated with RUNX2 Expression, Melanoma Invasiveness, and Osteotropism
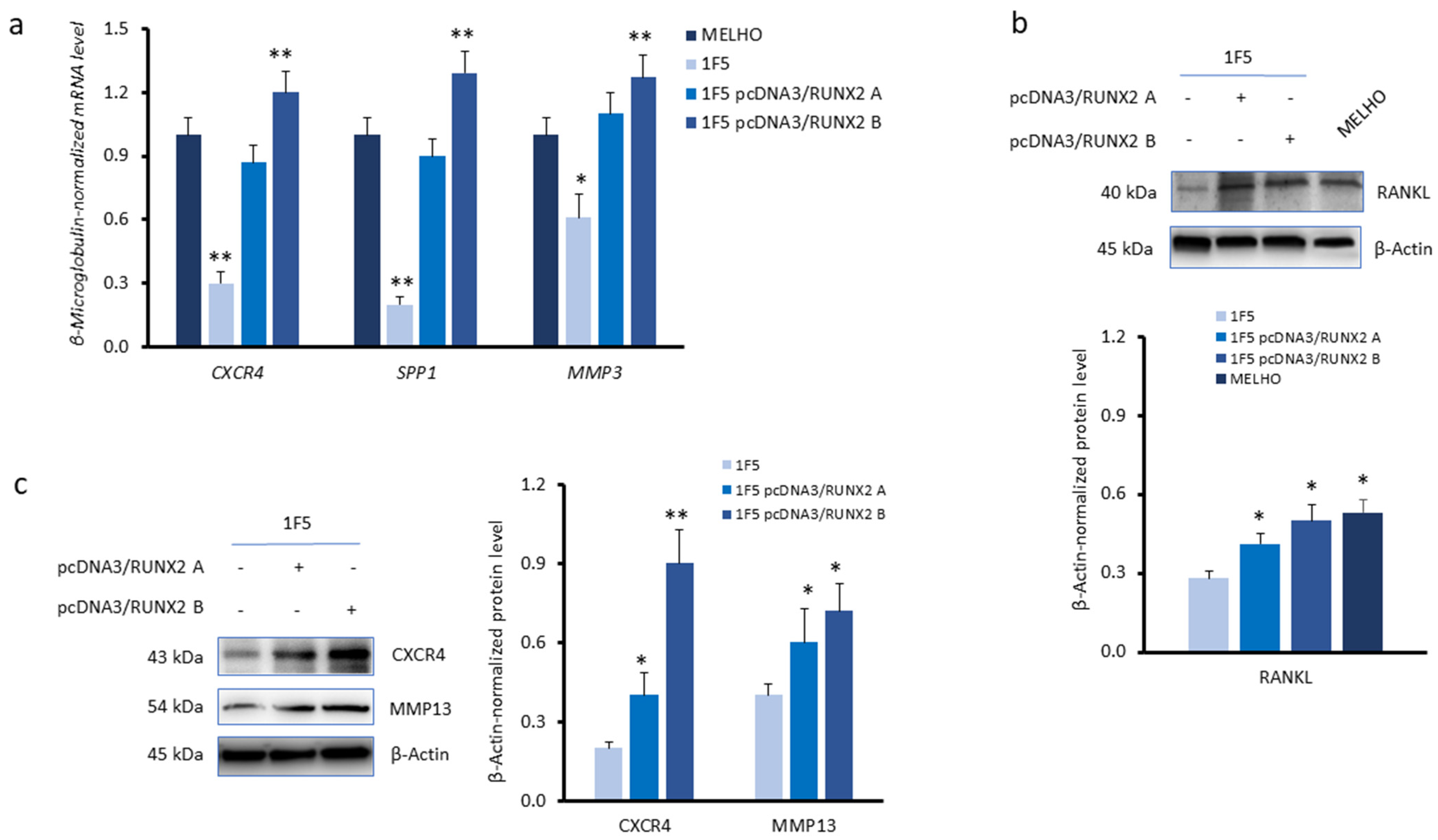
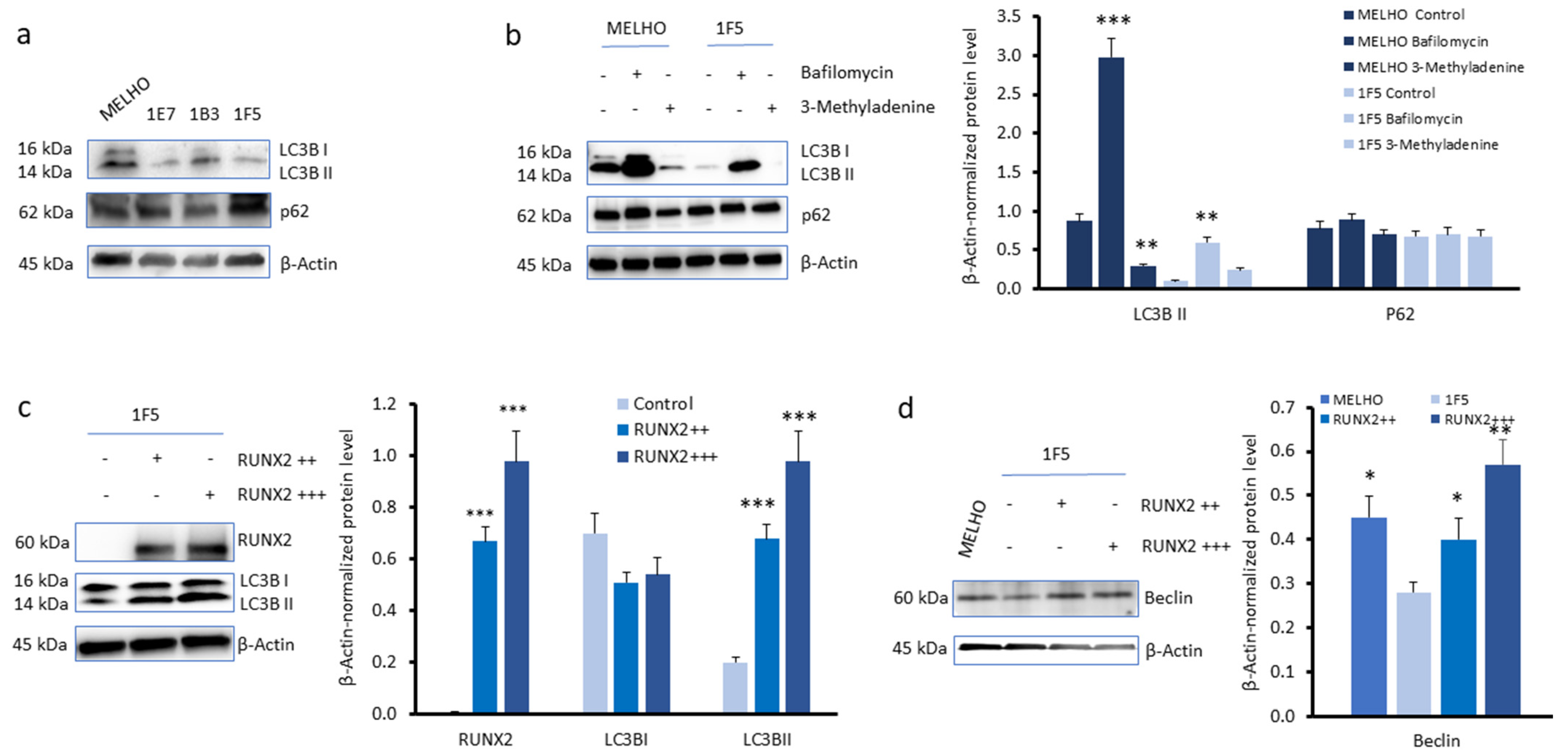
3.2. RUNX2 Expression Upregulates Autophagy Markers in Melanoma Cells
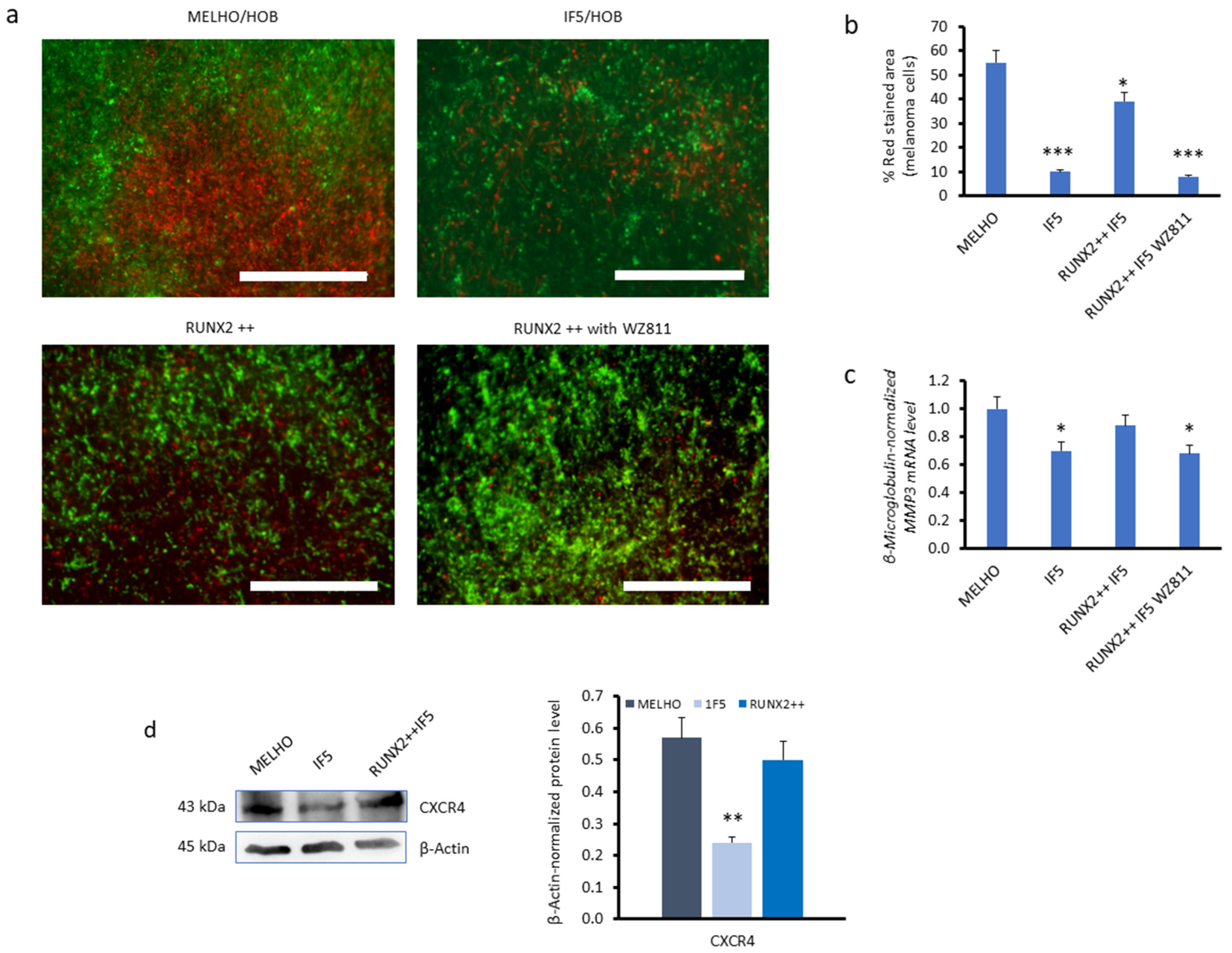
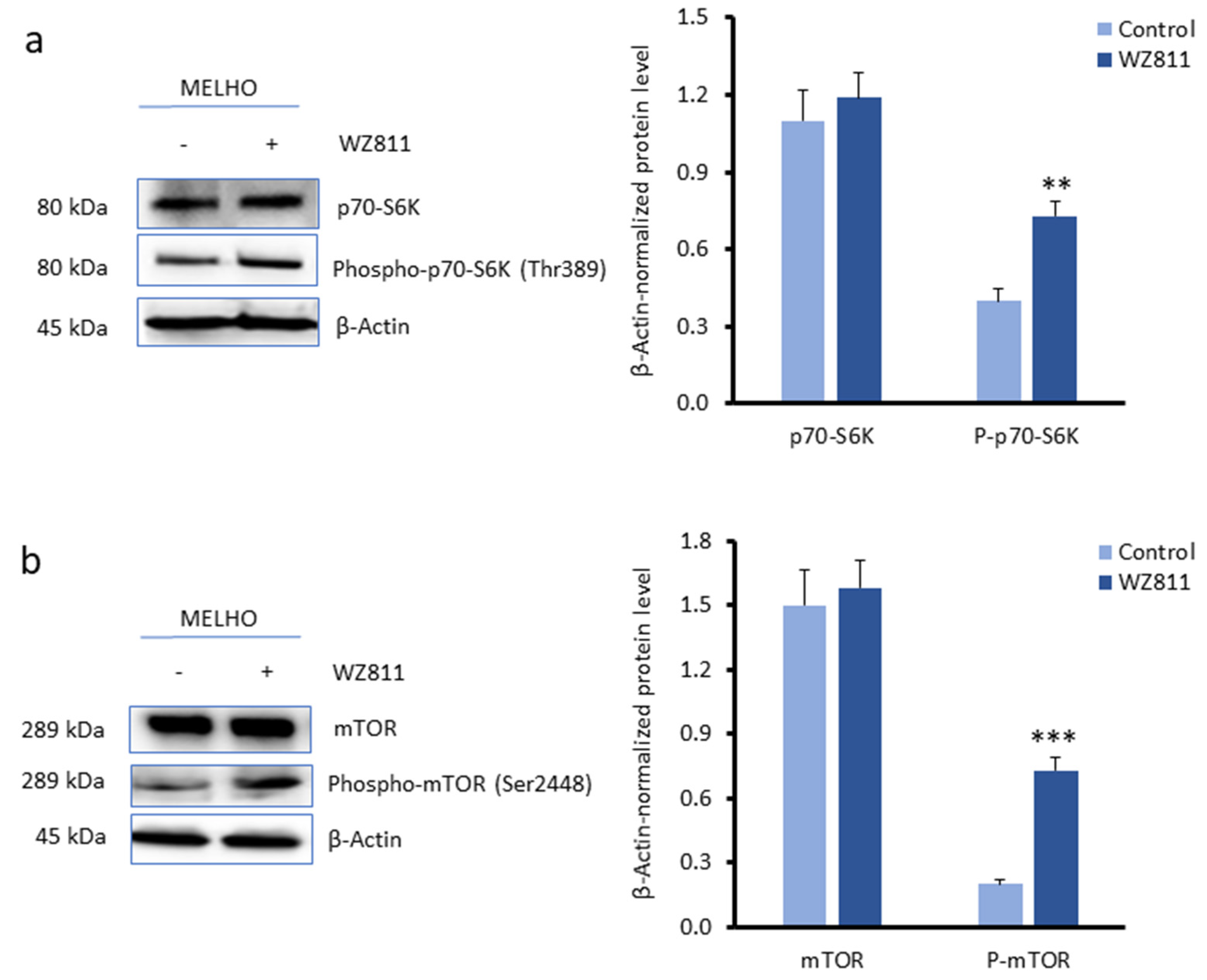
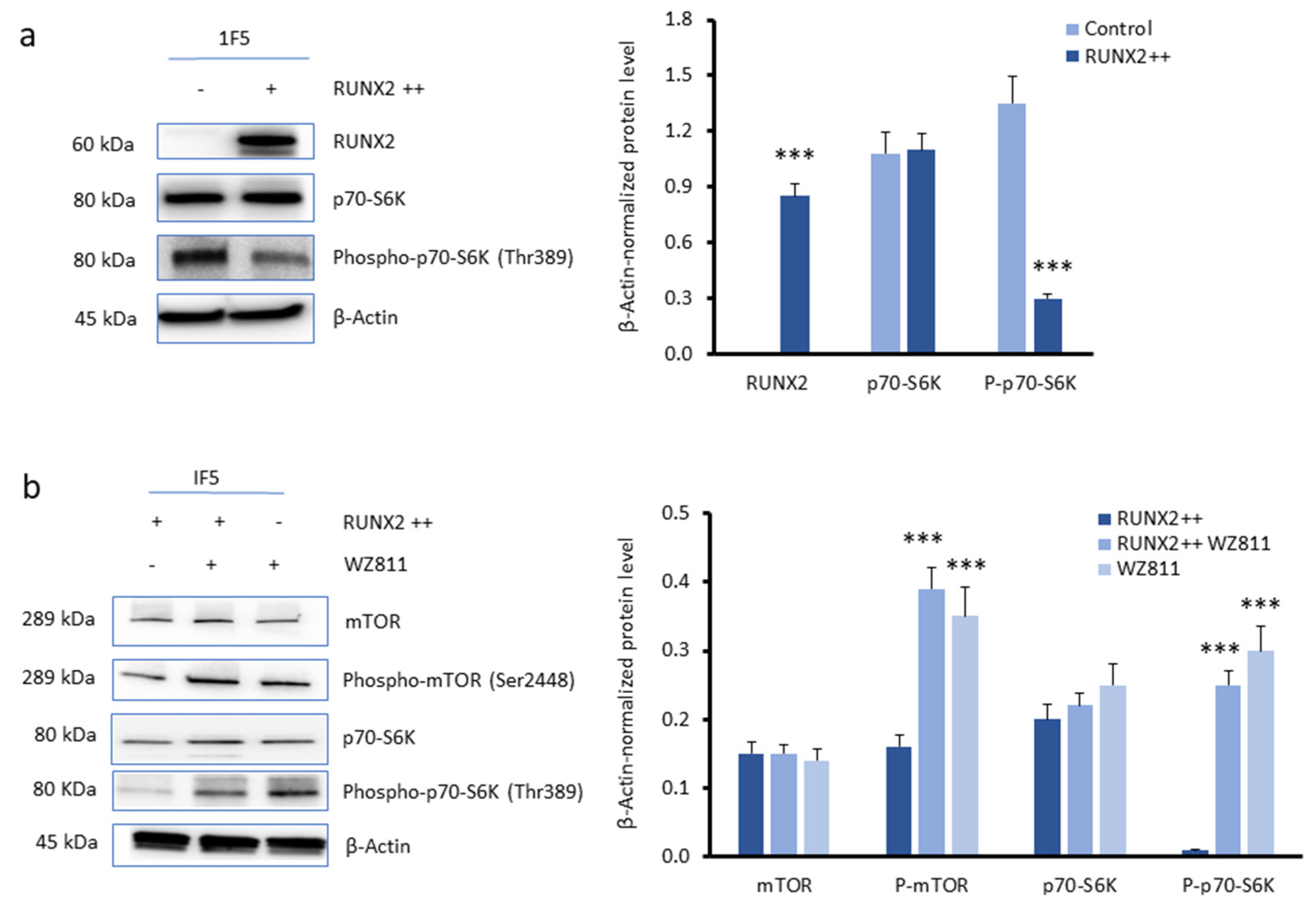
3.3. CXCR4 Is a Key Player in the RUNX2-Mediated Inhibition of the mTOR Signalling Pathway
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pratap, J.; Javed, A.; Languino, L.R.; Van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; Lian, J.B. The Runx2 osteogenic transcription factor regulates matrix metalloproteinase 9 in bone metastatic cancer cells and controls cell invasion. Mol. Cell. Biol. 2005, 25, 8581–8591. [Google Scholar] [CrossRef]
- Deiana, M.; Dalle Carbonare, L.; Serena, M.; Cheri, S.; Parolini, F.; Gandini, A.; Marchetto, G.; Innamorati, G.; Manfredi, M.; Marengo, E.; et al. New insights into the runt domain of RUNX2 in melanoma cell proliferation and migration. Cells 2018, 7, 220. [Google Scholar] [CrossRef]
- Cecconi, D.; Brandi, J.; Manfredi, M.; Serena, M.; Dalle Carbonare, L.; Deiana, M.; Cheri, S.; Parolini, F.; Gandini, A.; Marchetto, G.; et al. RUNX2 stimulates neoangiogenesis through the Runt domain in melanoma. Sci. Rep. 2019, 9, 8052. [Google Scholar] [CrossRef]
- Deiana, M.; Dalle Carbonare, L.; Serena, M.; Cheri, S.; Mutascio, S.; Gandini, A.; Innamorati, G.; Lorenzi, P.; Cumerlato, M.; Bertacco, J.; et al. A potential role of RUNX2-RUNT domain in modulating the expression of genes involved in bone metastases: An in vitro study with melanoma cells. Cells 2020, 9, 751. [Google Scholar] [CrossRef]
- Pulica, R.; Solal, K.C. Lasfar A: Role of RUNX2 in Melanoma: A New Player in Tumor Progression and Resistance to Therapy. In Melanoma; IntechOpen: London, UK, 2021. [Google Scholar]
- Cohen-Solal, K.A.; Kaufman, H.L.; Lasfar, A. Transcription factors as critical players in melanoma invasiveness, drug resistance, and opportunities for therapeutic drug development. Pigment Cell Melanoma Res. 2018, 31, 241–252. [Google Scholar] [CrossRef]
- Shakib, H.; Rajabi, S.; Dehghan, M.H.; Mashayekhi, F.J.; Safari-Alighiarloo, N.; Hedayati, M. Epithelial-to-mesenchymal transition in thyroid cancer: A comprehensive review. Endocrine 2019, 66, 435–455. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, S.; Saito, A.; Nagase, T. YAP/TAZ signaling as a molecular link between fibrosis and cancer. Int. J. Mol. Sci. 2018, 19, 3674. [Google Scholar] [CrossRef] [PubMed]
- Murakami, T.; Cardones, A.R.; Hwang, S.T. Chemokine receptors and melanoma metastasis. J. Dermatol. Sci. 2004, 36, 71–78. [Google Scholar] [CrossRef]
- Mendt, M.; Cardier, J.E. Activation of the CXCR4 chemokine receptor enhances biological functions associated with B16 melanoma liver metastasis. Melanoma Res. 2017, 27, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Balkwill, F. The significance of cancer cell expression of the chemokine receptor CXCR4. Semin. Cancer Biol. 2004, 14, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Domanska, U.M.; Kruizinga, R.C.; Nagengast, W.B.; Timmer-Bosscha, H.; Huls, G.; de Vries, E.G.; Walenkamp, A.M. A review on CXCR4/CXCL12 axis in oncology: No place to hide. Eur. J. Cancer 2013, 49, 219–230. [Google Scholar] [CrossRef]
- Guo, Z.-J.; Yang, L.; Qian, F.; Wang, Y.-X.; Yu, X.; Ji, C.-D.; Cui, W.; Xiang, D.-F.; Zhang, X.; Zhang, P.; et al. Transcription factor RUNX2 up-regulates chemokine receptor CXCR4 to promote invasive and metastatic potentials of human gastric cancer. Oncotarget 2016, 7, 20999. [Google Scholar] [CrossRef]
- Dalle Carbonare, L.; Bertacco, J.; Gaglio, S.C.; Minoia, A.; Cominacini, M.; Cheri, S.; Deiana, M.; Marchetto, G.; Bisognin, A.; Gandini, A.; et al. Fisetin: An Integrated Approach to Identify a Strategy Promoting Osteogenesis. Front Pharmacol 2022, 13, 890693. [Google Scholar] [CrossRef]
- Quan, Y.; Lei, H.; Wahafu, W.; Liu, Y.; Ping, H.; Zhang, X. Inhibition of autophagy enhances the anticancer effect of enzalutamide on bladder cancer. Biomed. Pharmacother. 2019, 120, 109490. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Gu, S.; Huang, J.; Chen, S.; Zhang, Z.; Xu, M. Inhibition of autophagy potentiates the efficacy of Gli inhibitor GANT-61 in MYCN-amplified neuroblastoma cells. BMC Cancer 2014, 14, 768. [Google Scholar] [CrossRef] [PubMed]
- Dalle Carbonare, L.; Gomez Lira, M.; Minoia, A.; Bertacco, J.; Orsi, S.; Lauriola, A.; Li Vigni, V.; Gandini, A.; Antoniazzi, F.; Zipeto, D.; et al. Expression of FBXW11 in normal and disease-associated osteogenic cells. J. Cell. Mol. Med. 2023, 27, 1580–1591. [Google Scholar] [CrossRef]
- Dalle Carbonare, L.; Bertacco, J.; Marchetto, G.; Cheri, S.; Deiana, M.; Minoia, A.; Tiso, N.; Mottes, M.; Valenti, M.T. Methylsulfonylmethane enhances MSC chondrogenic commitment and promotes pre-osteoblasts formation. Stem Cell Res. Ther. 2021, 12, 326. [Google Scholar] [CrossRef]
- Wai, P.Y.; Mi, Z.; Gao, C.; Guo, H.; Marroquin, C.; Kuo, P.C. Ets-1 and runx2 regulate transcription of a metastatic gene, osteopontin, in murine colorectal cancer cells. J. Biol. Chem. 2006, 281, 18973–18982. [Google Scholar] [CrossRef] [PubMed]
- Ganguly, S.S.; Hostetter, G.; Tang, L.; Frank, S.B.; Saboda, K.; Mehra, R.; Wang, L.; Li, X.; Keller, E.T.; Miranti, C.K. Notch3 promotes prostate cancer-induced bone lesion development via MMP-3. Oncogene 2020, 39, 204–218. [Google Scholar] [CrossRef]
- Othman, A.; Winogradzki, M.; Lee, L.; Tandon, M.; Blank, A.; Pratap, J. Bone metastatic breast cancer: Advances in cell signaling and autophagy related mechanisms. Cancers 2021, 13, 4310. [Google Scholar] [CrossRef]
- Mizushima, N.; Klionsky, D.J. Protein turnover via autophagy: Implications for metabolism. Annu. Rev. Nutr. 2007, 27, 19–40. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, J.; Lu, L.; Qiu, Z.-Y.; Zhang, X.; Yu, S.-B.; Wu, Y.-P.; Wang, M.-Q. Enhancement of chondrocyte autophagy is an early response in the degenerative cartilage of the temporomandibular joint to biomechanical dental stimulation. Apoptosis 2013, 18, 423–434. [Google Scholar] [CrossRef] [PubMed]
- Tandon, M.; Othman, A.H.; Ashok, V.; Stein, G.S.; Pratap, J. The role of Runx2 in facilitating autophagy in metastatic breast cancer cells. J. Cell. Physiol. 2018, 233, 559–571. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.S.; Vats, S.; Chia, A.Y.-Q.; Tan, T.Z.; Deng, S.; Ong, M.S.; Arfuso, F.; Yap, C.T.; Goh, B.C.; Sethi, G.; et al. Dual role of autophagy in hallmarks of cancer. Oncogene 2018, 37, 1142–1158. [Google Scholar] [CrossRef] [PubMed]
- Wiman, K.; Zhivotovsky, B. Understanding cell cycle and cell death regulation provides novel weapons against human diseases. J. Intern. Med. 2017, 281, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Jung, J.U. Autophagy genes as tumor suppressors. Curr. Opin. Cell Biol. 2010, 22, 226–233. [Google Scholar] [CrossRef] [PubMed]
- Degenhardt, K.; Mathew, R.; Beaudoin, B.; Bray, K.; Anderson, D.; Chen, G.; Mukherjee, C.; Shi, Y.; Gélinas, C.; Fan, Y.; et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 2006, 10, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Outschoorn, U.E.; Trimmer, C.; Lin, Z.; Whitaker-Menezes, D.; Chiavarina, B.; Zhou, J.; Wang, C.; Pavlides, S.; Martinez-Cantarin, M.P.; Capozza, F.; et al. Autophagy in cancer associated fibroblasts promotes tumor cell survival: Role of hypoxia, HIF1 induction and NFκB activation in the tumor stromal microenvironment. Cell Cycle 2010, 9, 3515–3533. [Google Scholar] [CrossRef] [PubMed]
- Kimmelman, A.C.; White, E. Autophagy and tumor metabolism. Cell Metab. 2017, 25, 1037–1043. [Google Scholar] [CrossRef]
- Műzes, G.; Sipos, F. Autoimmunity and Carcinogenesis: Their Relationship under the Umbrella of Autophagy. Biomedicines 2023, 11, 1130. [Google Scholar] [CrossRef]
- Di Leo, L.; Bodemeyer, V.; De Zio, D. The complex role of autophagy in melanoma evolution: New perspectives from mouse models. Front. Oncol. 2020, 9, 1506. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Koh, J.Y.; Price, S.; White, E.; Mehnert, J.M. Atg7 overcomes senescence and promotes growth of Braf V600E-driven melanoma. Cancer Discov. 2015, 5, 410–423. [Google Scholar] [CrossRef] [PubMed]
- Ravikumar, B.; Sarkar, S.; Davies, J.E.; Futter, M.; Garcia-Arencibia, M.; Green-Thompson, Z.W.; Jimenez-Sanchez, M.; Korolchuk, V.I.; Lichtenberg, M.; Luo, S.; et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol. Rev. 2010, 90, 1383–1435. [Google Scholar] [CrossRef] [PubMed]
- Corazzari, M.; Fimia, G.M.; Lovat, P.; Piacentini, M. Why is autophagy important for melanoma? Molecular mechanisms and therapeutic implications. Semin. Cancer Biol. 2013, 23, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Arya, M.; Patel, H.R.; McGurk, C.; Tatoud, R.; Klocker, H.; Masters, J.; Williamson, M. The importance of the CXCL12-CXCR4 chemokine ligand-receptor interaction in prostate cancer metastasis. J. Exp. Ther. Oncol. 2004, 4, 291. [Google Scholar]
- Shi, Y.; Riese, D.J.; Shen, J. The role of the CXCL12/CXCR4/CXCR7 chemokine axis in cancer. Front. Pharmacol. 2020, 11, 574667. [Google Scholar] [CrossRef]
- Mitchell, B.; Mahalingam, M. The CXCR4/CXCL12 Axis in Cutaneous Malignancies with an Emphasis on Melanoma. Histol Histopathol. 2014, 29, 1539–1546. [Google Scholar] [PubMed]
- Toyozawa, S.; Kaminaka, C.; Furukawa, F.; Nakamura, Y.; Matsunaka, H.; Yamamoto, Y. Chemokine receptor CXCR4 is a novel marker for the progression of cutaneous malignant melanomas. Acta Histochem. Cytochem. 2012, 45, 293–299. [Google Scholar] [CrossRef]
- Gatti, M.; Pattarozzi, A.; Bajetto, A.; Würth, R.; Daga, A.; Fiaschi, P.; Zona, G.; Florio, T.; Barbieri, F. Inhibition of CXCL12/CXCR4 autocrine/paracrine loop reduces viability of human glioblastoma stem-like cells affecting self-renewal activity. Toxicology 2013, 314, 209–220. [Google Scholar] [CrossRef]
- Ganju, R.K.; Brubaker, S.A.; Meyer, J.; Dutt, P.; Yang, Y.; Qin, S.; Newman, W.; Groopman, J.E. The α-chemokine, stromal cell-derived factor-1α, binds to the transmembrane G-protein-coupled CXCR-4 receptor and activates multiple signal transduction pathways. J. Biol. Chem. 1998, 273, 23169–23175. [Google Scholar] [CrossRef]
- Yu, X.; Shi, W.; Zhang, Y.; Wang, X.; Sun, S.; Song, Z.; Liu, M.; Zeng, Q.; Cui, S.; Qu, X. CXCL12/CXCR4 axis induced miR-125b promotes invasion and confers 5-fluorouracil resistance through enhancing autophagy in colorectal cancer. Sci. Rep. 2017, 7, 42226. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, H.; Cai, L.; Guo, D.; Zhang, D.; Zhou, X.; Xie, J. SDF-1α Promotes Chondrocyte Autophagy through CXCR4/mTOR Signaling Axis. Int. J. Mol. Sci. 2023, 24, 1710. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dalle Carbonare, L.; Minoia, A.; Vareschi, A.; Piritore, F.C.; Zouari, S.; Gandini, A.; Meneghel, M.; Elia, R.; Lorenzi, P.; Antoniazzi, F.; et al. Exploring the Interplay of RUNX2 and CXCR4 in Melanoma Progression. Cells 2024, 13, 408. https://doi.org/10.3390/cells13050408
Dalle Carbonare L, Minoia A, Vareschi A, Piritore FC, Zouari S, Gandini A, Meneghel M, Elia R, Lorenzi P, Antoniazzi F, et al. Exploring the Interplay of RUNX2 and CXCR4 in Melanoma Progression. Cells. 2024; 13(5):408. https://doi.org/10.3390/cells13050408
Chicago/Turabian StyleDalle Carbonare, Luca, Arianna Minoia, Anna Vareschi, Francesca Cristiana Piritore, Sharazed Zouari, Alberto Gandini, Mirko Meneghel, Rossella Elia, Pamela Lorenzi, Franco Antoniazzi, and et al. 2024. "Exploring the Interplay of RUNX2 and CXCR4 in Melanoma Progression" Cells 13, no. 5: 408. https://doi.org/10.3390/cells13050408
APA StyleDalle Carbonare, L., Minoia, A., Vareschi, A., Piritore, F. C., Zouari, S., Gandini, A., Meneghel, M., Elia, R., Lorenzi, P., Antoniazzi, F., Pessoa, J., Zipeto, D., Romanelli, M. G., Guardavaccaro, D., & Valenti, M. T. (2024). Exploring the Interplay of RUNX2 and CXCR4 in Melanoma Progression. Cells, 13(5), 408. https://doi.org/10.3390/cells13050408











