Spatio-Temporal Regulation of Notch Activation in Asymmetrically Dividing Sensory Organ Precursor Cells in Drosophila melanogaster Epithelium
Abstract
1. Introduction
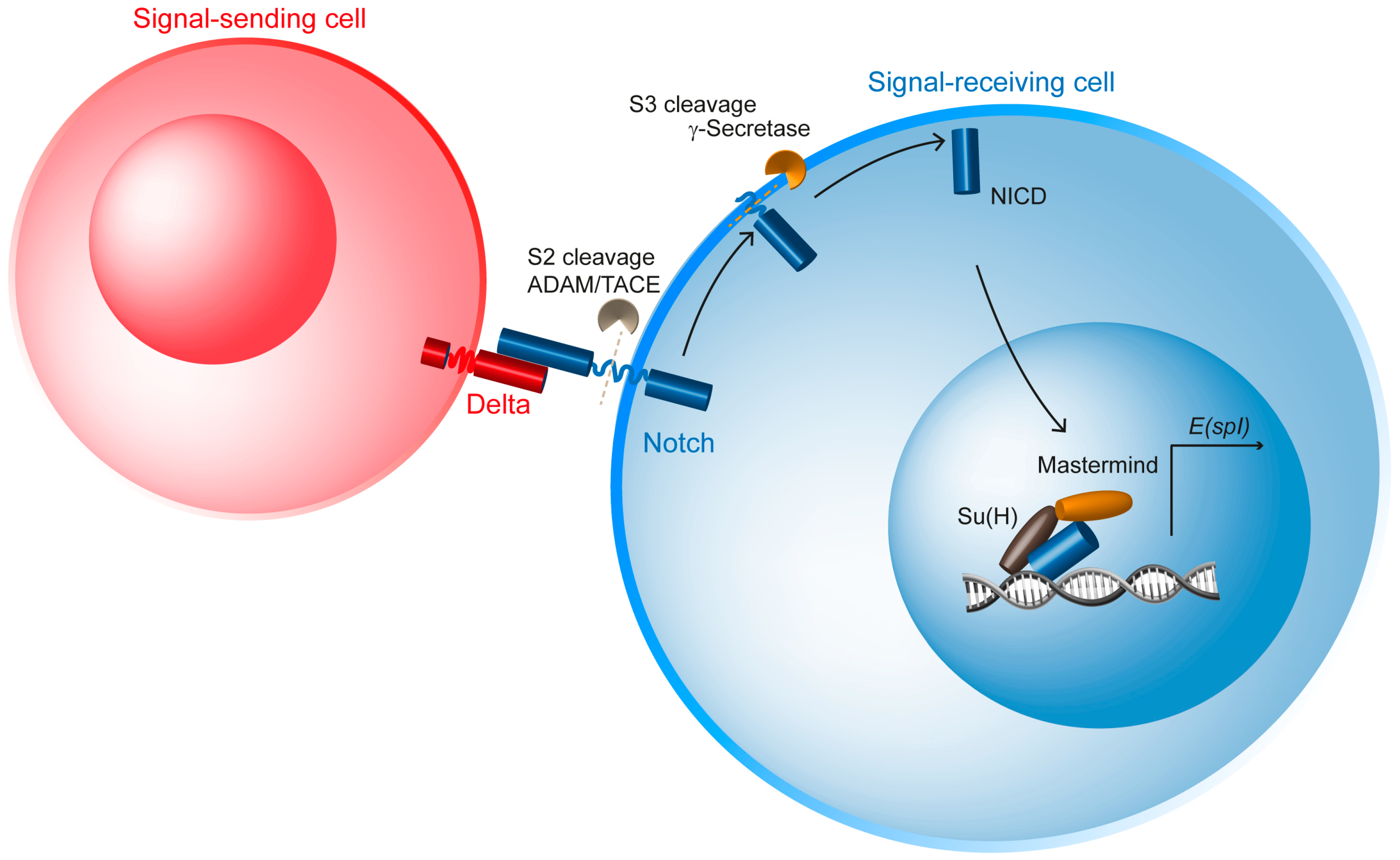
2. Discovery and Overview of the Notch Signaling Pathway
3. Establishment of the Asymmetry of Cell Fate Determinants
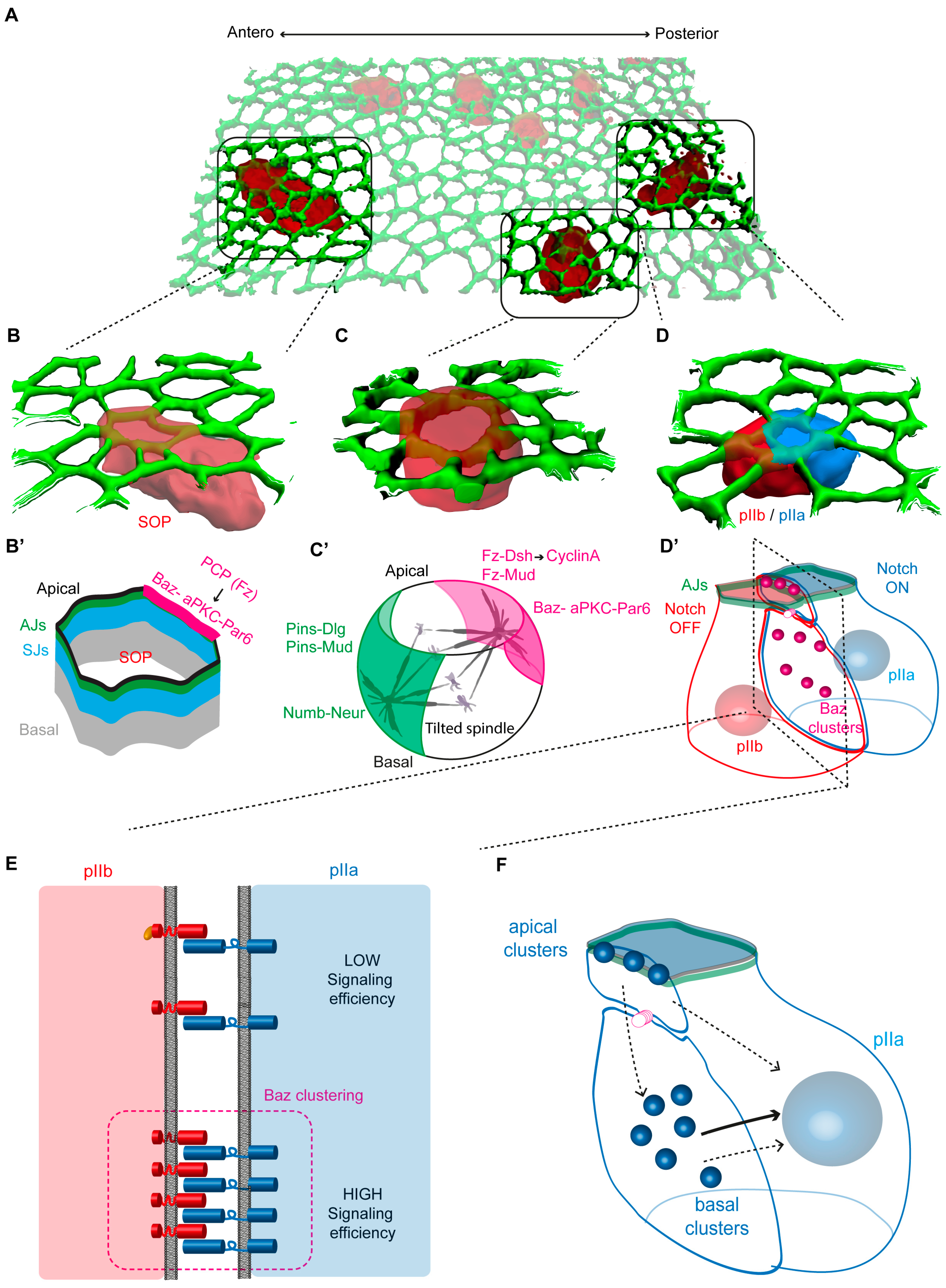
3.1. From Polarization to Asymmetric Division of Sensory Organ Precursor Cells
3.2. Polarization Drives Asymmetric Localization of Cell Fate Determinants
4. Remodeling of Cell–Cell Contacts during SOP Cytokinesis
4.1. Alignment of the Mitotic Spindle with Cortical Polarity
4.2. Activation of Notch Takes Place during SOP Cytokinesis
4.3. Assembly of the Notch Signalling Interface at SOP Cytokinesis
5. Role of Polarized Trafficking and Endocytosis in Notch Activation
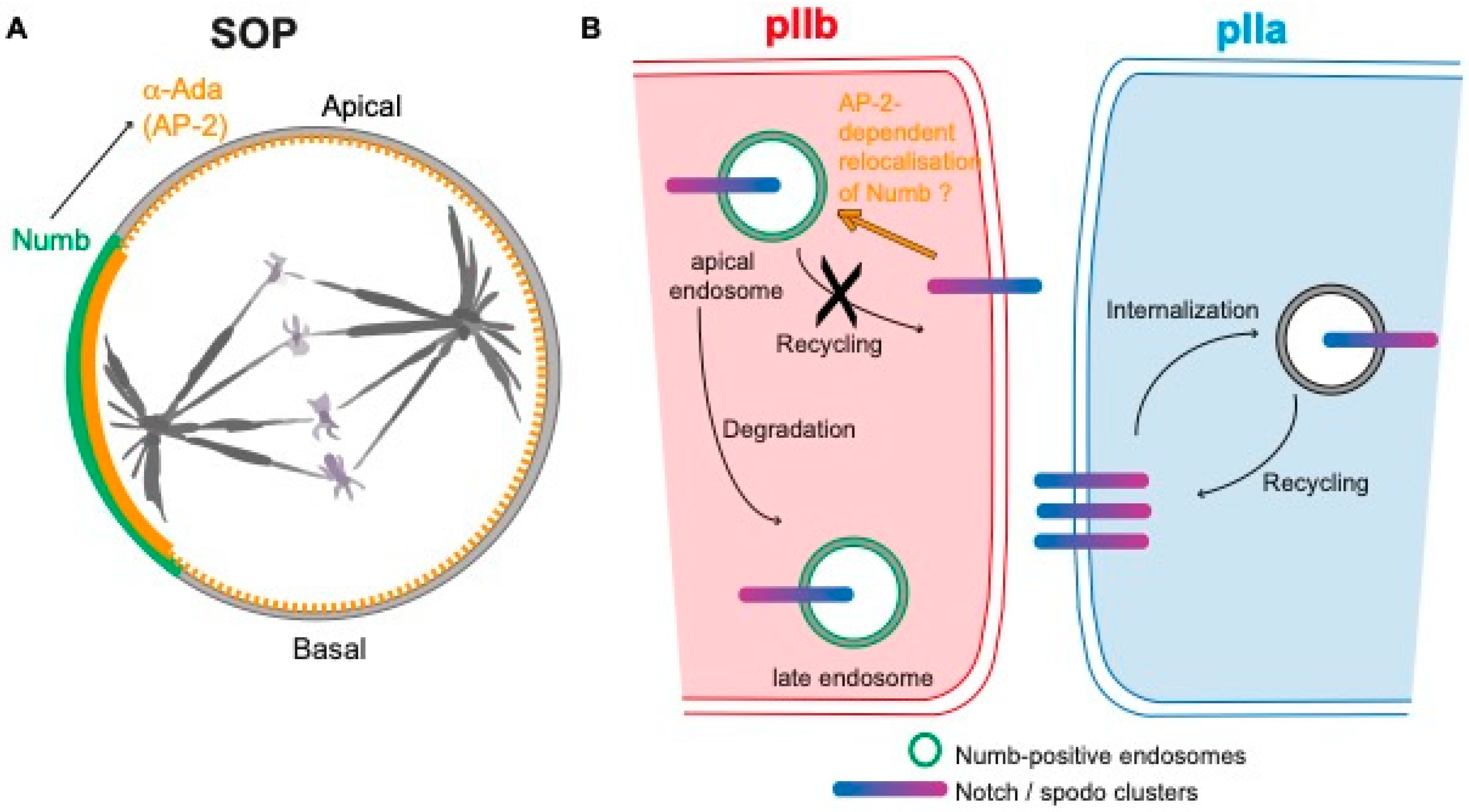
5.1. Site of Production of the Notch Intracellular Domain (NICD)
5.2. Mechanism of Notch Inhibition by Numb in the pIIb Cell
5.3. Mechanisms of Action of Neuralized in the pIIb Cell
5.4. Additional Contribution of Membrane Traffic in Binary Cell Fate Acquisition
6. Actual Limitations and Concluding Remarks
Funding
Acknowledgments
Conflicts of Interest
References
- Artavanis-Tsakonas, S.; Muskavitch, M.A. Notch: The past, the present, and the future. Curr. Top. Dev. Biol. 2010, 92, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Sato, C.; Cerletti, M.; Wagers, A. Notch signaling in the regulation of stem cell self-renewal and differentiation. Curr. Top. Dev. Biol. 2010, 92, 367–409. [Google Scholar] [CrossRef] [PubMed]
- Koch, U.; Radtke, F. Notch signaling in solid tumors. Curr. Top. Dev. Biol. 2010, 92, 411–455. [Google Scholar] [CrossRef] [PubMed]
- Masek, J.; Andersson, E.R. The developmental biology of genetic Notch disorders. Development 2017, 144, 1743–1763. [Google Scholar] [CrossRef]
- Zhou, B.; Lin, W.; Long, Y.; Yang, Y.; Zhang, H.; Wu, K.; Chu, Q. Notch signaling pathway: Architecture, disease, and therapeutics. Signal Transduct. Target. Ther. 2022, 7, 95. [Google Scholar] [CrossRef]
- Kopan, R. Notch signaling. Cold Spring Harb. Perspect. Biol. 2012, 4, a011213. [Google Scholar] [CrossRef]
- Gho, M.; Bellaiche, Y.; Schweisguth, F. Revisiting the Drosophila microchaete lineage: A novel intrinsically asymmetric cell division generates a glial cell. Development 1999, 126, 3573–3584. [Google Scholar] [CrossRef]
- Hartenstein, V.; Posakony, J.W. Development of adult sensilla on the wing and notum of Drosophila melanogaster. Development 1989, 107, 389–405. [Google Scholar] [CrossRef]
- Schweisguth, F. Asymmetric cell division in the Drosophila bristle lineage: From the polarization of sensory organ precursor cells to Notch-mediated binary fate decision. Wiley Interdiscip. Rev. Dev. Biol. 2015, 4, 299–309. [Google Scholar] [CrossRef]
- Simpson, P. Lateral inhibition and the development of the sensory bristles of the adult peripheral nervous system of Drosophila. Development 1990, 109, 509–519. [Google Scholar] [CrossRef]
- Gho, M.; Schweisguth, F. Frizzled signalling controls orientation of asymmetric sense organ precursor cell divisions in Drosophila. Nature 1998, 393, 178–181. [Google Scholar] [CrossRef] [PubMed]
- Rhyu, M.S.; Jan, L.Y.; Jan, Y.N. Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell 1994, 76, 477–491. [Google Scholar] [CrossRef]
- Le Borgne, R.; Schweisguth, F. Notch signaling: Endocytosis makes delta signal better. Curr. Biol. CB 2003, 13, R273–R275. [Google Scholar] [CrossRef]
- Deblandre, G.A.; Lai, E.C.; Kintner, C. Xenopus neuralized is a ubiquitin ligase that interacts with XDelta1 and regulates Notch signaling. Dev. Cell 2001, 1, 795–806. [Google Scholar] [CrossRef] [PubMed]
- Lai, E.C.; Deblandre, G.A.; Kintner, C.; Rubin, G.M. Drosophila neuralized is a ubiquitin ligase that promotes the internalization and degradation of delta. Dev. Cell 2001, 1, 783–794. [Google Scholar] [CrossRef]
- Pavlopoulos, E.; Pitsouli, C.; Klueg, K.M.; Muskavitch, M.A.; Moschonas, N.K.; Delidakis, C. neuralized Encodes a peripheral membrane protein involved in delta signaling and endocytosis. Dev. Cell 2001, 1, 807–816. [Google Scholar] [CrossRef] [PubMed]
- Frise, E.; Knoblich, J.A.; Younger-Shepherd, S.; Jan, L.Y.; Jan, Y.N. The Drosophila Numb protein inhibits signaling of the Notch receptor during cell-cell interaction in sensory organ lineage. Proc. Natl. Acad. Sci. USA 1996, 93, 11925–11932. [Google Scholar] [CrossRef] [PubMed]
- Cotton, M.; Benhra, N.; Le Borgne, R. Numb inhibits the recycling of Sanpodo in Drosophila sensory organ precursor. Curr. Biol. CB 2013, 23, 581–587. [Google Scholar] [CrossRef]
- Couturier, L.; Mazouni, K.; Schweisguth, F. Numb localizes at endosomes and controls the endosomal sorting of notch after asymmetric division in Drosophila. Curr. Biol. CB 2013, 23, 588–593. [Google Scholar] [CrossRef]
- Inaba, M.; Yamashita, Y.M. Asymmetric stem cell division: Precision for robustness. Cell Stem Cell 2012, 11, 461–469. [Google Scholar] [CrossRef]
- Tajbakhsh, S.; Rocheteau, P.; Le Roux, I. Asymmetric cell divisions and asymmetric cell fates. Annu. Rev. Cell Dev. Biol. 2009, 25, 671–699. [Google Scholar] [CrossRef]
- Gozlan, O.; Sprinzak, D. Notch signaling in development and homeostasis. Development 2023, 150, 201138. [Google Scholar] [CrossRef] [PubMed]
- Poulson, D.F. Chromosomal Deficiencies and the Embryonic Development of Drosophila Melanogaster. Proc. Natl. Acad. Sci. USA 1937, 23, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Mohr, O.L. Character Changes Caused by Mutation of an Entire Region of a Chromosome in Drosophila. Genetics 1919, 4, 275–282. [Google Scholar] [CrossRef]
- Lehmann, R.; Jimenez, F.; Dietrich, U.; Campos-Ortega, J.A. On the phenotype and development of mutants of early neurogenesis inDrosophila melanogaster. Wilehm Roux Arch. Dev. Biol. 1983, 192, 62–74. [Google Scholar] [CrossRef] [PubMed]
- Nusslein-Volhard, C.; Wieschaus, E. Mutations affecting segment number and polarity in Drosophila. Nature 1980, 287, 795–801. [Google Scholar] [CrossRef] [PubMed]
- Nusslein-Volhard, C.; Wieschaus, E.; Kluding, H. Mutations affecting the pattern of the larval cuticle inDrosophila melanogaster: I. Zygotic loci on the second chromosome. Wilehm Roux’s Arch. Dev. Biol. 1984, 193, 267–282. [Google Scholar] [CrossRef] [PubMed]
- Artavanis-Tsakonas, S.; Muskavitch, M.A.; Yedvobnick, B. Molecular cloning of Notch, a locus affecting neurogenesis in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 1983, 80, 1977–1981. [Google Scholar] [CrossRef]
- Kidd, S.; Lockett, T.J.; Young, M.W. The Notch locus of Drosophila melanogaster. Cell 1983, 34, 421–433. [Google Scholar] [CrossRef]
- Kidd, S.; Kelley, M.R.; Young, M.W. Sequence of the notch locus of Drosophila melanogaster: Relationship of the encoded protein to mammalian clotting and growth factors. Mol. Cell. Biol. 1986, 6, 3094–3108. [Google Scholar] [CrossRef]
- Greenwald, I.; Coulson, A.; Sulston, J.; Priess, J. Correlation of the physical and genetic maps in the lin-12 region of Caenorhabditis elegans. Nucleic Acids Res. 1987, 15, 2295–2307. [Google Scholar] [CrossRef] [PubMed]
- Yochem, J.; Weston, K.; Greenwald, I. The Caenorhabditis elegans lin-12 gene encodes a transmembrane protein with overall similarity to Drosophila Notch. Nature 1988, 335, 547–550. [Google Scholar] [CrossRef] [PubMed]
- Austin, J.; Kimble, J. glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell 1987, 51, 589–599. [Google Scholar] [CrossRef] [PubMed]
- Austin, J.; Kimble, J. Transcript analysis of glp-1 and lin-12, homologous genes required for cell interactions during development of C. elegans. Cell 1989, 58, 565–571. [Google Scholar] [CrossRef] [PubMed]
- Greenwald, I.S.; Sternberg, P.W.; Horvitz, H.R. The lin-12 locus specifies cell fates in Caenorhabditis elegans. Cell 1983, 34, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Coffman, C.; Harris, W.; Kintner, C. Xotch, the Xenopus homolog of Drosophila notch. Science 1990, 249, 1438–1441. [Google Scholar] [CrossRef] [PubMed]
- Stubbs, J.D.; Lekutis, C.; Singer, K.L.; Bui, A.; Yuzuki, D.; Srinivasan, U.; Parry, G. cDNA cloning of a mouse mammary epithelial cell surface protein reveals the existence of epidermal growth factor-like domains linked to factor VIII-like sequences. Proc. Natl. Acad. Sci. USA 1990, 87, 8417–8421. [Google Scholar] [CrossRef] [PubMed]
- Fortini, M.E. Notch and presenilin: A proteolytic mechanism emerges. Curr. Opin. Cell Biol. 2001, 13, 627–634. [Google Scholar] [CrossRef] [PubMed]
- Kopan, R.; Ilagan, M.X. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell 2009, 137, 216–233. [Google Scholar] [CrossRef]
- Kopan, R.; Schroeter, E.H.; Weintraub, H.; Nye, J.S. Signal transduction by activated mNotch: Importance of proteolytic processing and its regulation by the extracellular domain. Proc. Natl. Acad. Sci. USA 1996, 93, 1683–1688. [Google Scholar] [CrossRef]
- Kidd, S.; Lieber, T. Furin cleavage is not a requirement for Drosophila Notch function. Mech. Dev. 2002, 115, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Lieber, T.; Kidd, S.; Young, M.W. kuzbanian-mediated cleavage of Drosophila Notch. Genes Dev. 2002, 16, 209–221. [Google Scholar] [CrossRef] [PubMed]
- Brou, C.; Logeat, F.; Gupta, N.; Bessia, C.; LeBail, O.; Doedens, J.R.; Cumano, A.; Roux, P.; Black, R.A.; Israel, A. A novel proteolytic cleavage involved in Notch signaling: The role of the disintegrin-metalloprotease TACE. Mol. Cell 2000, 5, 207–216. [Google Scholar] [CrossRef] [PubMed]
- De Strooper, B.; Annaert, W.; Cupers, P.; Saftig, P.; Craessaerts, K.; Mumm, J.S.; Schroeter, E.H.; Schrijvers, V.; Wolfe, M.S.; Ray, W.J.; et al. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature 1999, 398, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Struhl, G.; Greenwald, I. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature 1999, 398, 522–525. [Google Scholar] [CrossRef] [PubMed]
- Ray, W.J.; Yao, M.; Mumm, J.; Schroeter, E.H.; Saftig, P.; Wolfe, M.; Selkoe, D.J.; Kopan, R.; Goate, A.M. Cell surface presenilin-1 participates in the gamma-secretase-like proteolysis of Notch. J. Biol. Chem. 1999, 274, 36801–36807. [Google Scholar] [CrossRef] [PubMed]
- Jarriault, S.; Brou, C.; Logeat, F.; Schroeter, E.H.; Kopan, R.; Israel, A. Signalling downstream of activated mammalian Notch. Nature 1995, 377, 355–358. [Google Scholar] [CrossRef] [PubMed]
- Bray, S.; Bernard, F. Notch targets and their regulation. Curr. Top. Dev. Biol. 2010, 92, 253–275. [Google Scholar] [CrossRef]
- Cowan, C.R.; Hyman, A.A. Asymmetric cell division in C. elegans: Cortical polarity and spindle positioning. Annu. Rev. Cell Dev. Biol. 2004, 20, 427–453. [Google Scholar] [CrossRef]
- Knoblich, J.A. Mechanisms of asymmetric stem cell division. Cell 2008, 132, 583–597. [Google Scholar] [CrossRef]
- Motegi, F.; Seydoux, G. The PAR network: Redundancy and robustness in a symmetry-breaking system. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368, 20130010. [Google Scholar] [CrossRef] [PubMed]
- Gonczy, P. Mechanisms of asymmetric cell division: Flies and worms pave the way. Nat. Rev. Mol. Cell Biol. 2008, 9, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Founounou, N.; Loyer, N.; Le Borgne, R. Septins regulate the contractility of the actomyosin ring to enable adherens junction remodeling during cytokinesis of epithelial cells. Dev. Cell 2013, 24, 242–255. [Google Scholar] [CrossRef] [PubMed]
- Bellaiche, Y.; Radovic, A.; Woods, D.F.; Hough, C.D.; Parmentier, M.L.; O’Kane, C.J.; Bryant, P.J.; Schweisguth, F. The Partner of Inscuteable/Discs-large complex is required to establish planar polarity during asymmetric cell division in Drosophila. Cell 2001, 106, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Houssin, E.; Pinot, M.; Bellec, K.; Le Borgne, R. Par3 cooperates with Sanpodo for the assembly of Notch clusters following asymmetric division of Drosophila sensory organ precursor cells. eLife 2021, 10, 66659. [Google Scholar] [CrossRef] [PubMed]
- Bellaiche, Y.; Gho, M.; Kaltschmidt, J.A.; Brand, A.H.; Schweisguth, F. Frizzled regulates localization of cell-fate determinants and mitotic spindle rotation during asymmetric cell division. Nat. Cell Biol. 2001, 3, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Bilder, D.; Li, M.; Perrimon, N. Cooperative regulation of cell polarity and growth by Drosophila tumor suppressors. Science 2000, 289, 113–116. [Google Scholar] [CrossRef] [PubMed]
- Bilder, D.; Perrimon, N. Localization of apical epithelial determinants by the basolateral PDZ protein Scribble. Nature 2000, 403, 676–680. [Google Scholar] [CrossRef] [PubMed]
- Wirtz-Peitz, F.; Nishimura, T.; Knoblich, J.A. Linking cell cycle to asymmetric division: Aurora-A phosphorylates the Par complex to regulate Numb localization. Cell 2008, 135, 161–173. [Google Scholar] [CrossRef]
- Besson, C.; Bernard, F.; Corson, F.; Rouault, H.; Reynaud, E.; Keder, A.; Mazouni, K.; Schweisguth, F. Planar Cell Polarity Breaks the Symmetry of PAR Protein Distribution prior to Mitosis in Drosophila Sensory Organ Precursor Cells. Curr. Biol. CB 2015, 25, 1104–1110. [Google Scholar] [CrossRef]
- Smith, C.A.; Lau, K.M.; Rahmani, Z.; Dho, S.E.; Brothers, G.; She, Y.M.; Berry, D.M.; Bonneil, E.; Thibault, P.; Schweisguth, F.; et al. aPKC-mediated phosphorylation regulates asymmetric membrane localization of the cell fate determinant Numb. EMBO J. 2007, 26, 468–480. [Google Scholar] [CrossRef]
- Langevin, J.; Le Borgne, R.; Rosenfeld, F.; Gho, M.; Schweisguth, F.; Bellaiche, Y. Lethal giant larvae controls the localization of notch-signaling regulators numb, neuralized, and Sanpodo in Drosophila sensory-organ precursor cells. Curr. Biol. CB 2005, 15, 955–962. [Google Scholar] [CrossRef]
- Gomes, J.E.; Corado, M.; Schweisguth, F. Van Gogh and Frizzled act redundantly in the Drosophila sensory organ precursor cell to orient its asymmetric division. PLoS ONE 2009, 4, e4485. [Google Scholar] [CrossRef]
- Bowman, S.K.; Neumuller, R.A.; Novatchkova, M.; Du, Q.; Knoblich, J.A. The Drosophila NuMA Homolog Mud regulates spindle orientation in asymmetric cell division. Dev. Cell 2006, 10, 731–742. [Google Scholar] [CrossRef] [PubMed]
- Segalen, M.; Johnston, C.A.; Martin, C.A.; Dumortier, J.G.; Prehoda, K.E.; David, N.B.; Doe, C.Q.; Bellaiche, Y. The Fz-Dsh planar cell polarity pathway induces oriented cell division via Mud/NuMA in Drosophila and zebrafish. Dev. Cell 2010, 19, 740–752. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Du, Q.; Chen, X.; Zheng, Z.; Balsbaugh, J.L.; Maitra, S.; Shabanowitz, J.; Hunt, D.F.; Macara, I.G. Par3 controls epithelial spindle orientation by aPKC-mediated phosphorylation of apical Pins. Curr. Biol. CB 2010, 20, 1809–1818. [Google Scholar] [CrossRef] [PubMed]
- Guilgur, L.G.; Prudencio, P.; Ferreira, T.; Pimenta-Marques, A.R.; Martinho, R.G. Drosophila aPKC is required for mitotic spindle orientation during symmetric division of epithelial cells. Development 2012, 139, 503–513. [Google Scholar] [CrossRef]
- Johnston, C.A.; Hirono, K.; Prehoda, K.E.; Doe, C.Q. Identification of an Aurora-A/PinsLINKER/Dlg spindle orientation pathway using induced cell polarity in S2 cells. Cell 2009, 138, 1150–1163. [Google Scholar] [CrossRef] [PubMed]
- Johnston, C.A.; Manning, L.; Lu, M.S.; Golub, O.; Doe, C.Q.; Prehoda, K.E. Formin-mediated actin polymerization cooperates with Mushroom body defect (Mud)-Dynein during Frizzled-Dishevelled spindle orientation. J. Cell Sci. 2013, 126, 4436–4444. [Google Scholar] [CrossRef]
- Darnat, P.; Burg, A.; Salle, J.; Lacoste, J.; Louvet-Vallee, S.; Gho, M.; Audibert, A. Cortical Cyclin A controls spindle orientation during asymmetric cell divisions in Drosophila. Nat. Commun. 2022, 13, 2723. [Google Scholar] [CrossRef]
- Daeden, A.; Mietke, A.; Derivery, E.; Seum, C.; Julicher, F.; Gonzalez-Gaitan, M. Polarized branched Actin modulates cortical mechanics to produce unequal-size daughters during asymmetric division. Nat. Cell Biol. 2023, 25, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Couturier, L.; Vodovar, N.; Schweisguth, F. Endocytosis by Numb breaks Notch symmetry at cytokinesis. Nat. Cell Biol. 2012, 14, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Trylinski, M.; Mazouni, K.; Schweisguth, F. Intra-lineage Fate Decisions Involve Activation of Notch Receptors Basal to the Midbody in Drosophila Sensory Organ Precursor Cells. Curr. Biol. CB 2017, 27, 2239–2247.e3. [Google Scholar] [CrossRef] [PubMed]
- Trylinski, M.; Schweisguth, F. Activation of Arp2/3 by WASp Is Essential for the Endocytosis of Delta Only during Cytokinesis in Drosophila. Cell Rep. 2019, 28, 1–10.e3. [Google Scholar] [CrossRef] [PubMed]
- Benhra, N.; Lallet, L.; Cotton, M.; Le Bras, S.; Dussert, A.; Le Borgne, R. AP-1 controls the trafficking of Notch and Sanpodo toward E-Cadherin junctions in sensory organ precursors. Curr. Biol. CB 2011, 21, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Bellec, K.; Gicquel, I.; Le Borgne, R. Stratum recruits Rab8 at Golgi exit sites to regulate the basolateral sorting of Notch and Sanpodo. Development 2018, 145, dev163469. [Google Scholar] [CrossRef]
- Bellec, K.; Pinot, M.; Gicquel, I.; Le Borgne, R. The Clathrin adaptor AP-1 and Stratum act in parallel pathways to control Notch activation in Drosophila sensory organ precursors cells. Development 2021, 148, dev191437. [Google Scholar] [CrossRef]
- Daniel, E.; Daude, M.; Kolotuev, I.; Charish, K.; Auld, V.; Le Borgne, R. Coordination of Septate Junctions Assembly and Completion of Cytokinesis in Proliferative Epithelial Tissues. Curr. Biol. CB 2018, 28, 1380–1391.e4. [Google Scholar] [CrossRef]
- Herszterg, S.; Leibfried, A.; Bosveld, F.; Martin, C.; Bellaiche, Y. Interplay between the dividing cell and its neighbors regulates adherens junction formation during cytokinesis in epithelial tissue. Dev. Cell 2013, 24, 256–270. [Google Scholar] [CrossRef]
- Johnson, S.A.; Zitserman, D.; Roegiers, F. Numb regulates the balance between Notch recycling and late-endosome targeting in Drosophila neural progenitor cells. Mol. Biol. Cell 2016, 27, 2857–2866. [Google Scholar] [CrossRef]
- Smith, C.A.; Dho, S.E.; Donaldson, J.; Tepass, U.; McGlade, C.J. The cell fate determinant numb interacts with EHD/Rme-1 family proteins and has a role in endocytic recycling. Mol. Biol. Cell 2004, 15, 3698–3708. [Google Scholar] [CrossRef] [PubMed]
- Santolini, E.; Puri, C.; Salcini, A.E.; Gagliani, M.C.; Pelicci, P.G.; Tacchetti, C.; Di Fiore, P.P. Numb is an endocytic protein. J. Cell Biol. 2000, 151, 1345–1352. [Google Scholar] [CrossRef] [PubMed]
- Berdnik, D.; Torok, T.; Gonzalez-Gaitan, M.; Knoblich, J.A. The endocytic protein alpha-Adaptin is required for numb-mediated asymmetric cell division in Drosophila. Dev. Cell 2002, 3, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Jan, L.Y.; Jan, Y.N. Control of daughter cell fates during asymmetric division: Interaction of Numb and Notch. Neuron 1996, 17, 27–41. [Google Scholar] [CrossRef] [PubMed]
- O’Connor-Giles, K.M.; Skeath, J.B. Numb inhibits membrane localization of Sanpodo, a four-pass transmembrane protein, to promote asymmetric divisions in Drosophila. Dev. Cell 2003, 5, 231–243. [Google Scholar] [CrossRef] [PubMed]
- Babaoglan, A.B.; O’Connor-Giles, K.M.; Mistry, H.; Schickedanz, A.; Wilson, B.A.; Skeath, J.B. Sanpodo: A context-dependent activator and inhibitor of Notch signaling during asymmetric divisions. Development 2009, 136, 4089–4098. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, A.; Kandachar, V.; Zitserman, D.; Tong, X.; Roegiers, F. Sanpodo controls sensory organ precursor fate by directing Notch trafficking and binding gamma-secretase. J. Cell Biol. 2013, 201, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Tong, X.; Zitserman, D.; Serebriiskii, I.; Andrake, M.; Dunbrack, R.; Roegiers, F. Numb independently antagonizes Sanpodo membrane targeting and Notch signaling in Drosophila sensory organ precursor cells. Mol. Biol. Cell 2010, 21, 802–810. [Google Scholar] [CrossRef] [PubMed]
- Hutterer, A.; Knoblich, J.A. Numb and alpha-Adaptin regulate Sanpodo endocytosis to specify cell fate in Drosophila external sensory organs. EMBO Rep. 2005, 6, 836–842. [Google Scholar] [CrossRef]
- Krieger, J.R.; Taylor, P.; Gajadhar, A.S.; Guha, A.; Moran, M.F.; McGlade, C.J. Identification and selected reaction monitoring (SRM) quantification of endocytosis factors associated with Numb. Mol. Cell. Proteom. MCP 2013, 12, 499–514. [Google Scholar] [CrossRef]
- Nilsson, L.; Conradt, B.; Ruaud, A.F.; Chen, C.C.; Hatzold, J.; Bessereau, J.L.; Grant, B.D.; Tuck, S. Caenorhabditis elegans num-1 negatively regulates endocytic recycling. Genetics 2008, 179, 375–387. [Google Scholar] [CrossRef] [PubMed]
- Le Borgne, R.; Schweisguth, F. Unequal segregation of Neuralized biases Notch activation during asymmetric cell division. Dev. Cell 2003, 5, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Daskalaki, A.; Shalaby, N.A.; Kux, K.; Tsoumpekos, G.; Tsibidis, G.D.; Muskavitch, M.A.; Delidakis, C. Distinct intracellular motifs of Delta mediate its ubiquitylation and activation by Mindbomb1 and Neuralized. J. Cell Biol. 2011, 195, 1017–1031. [Google Scholar] [CrossRef] [PubMed]
- Suarez Rodriguez, F.; Sanlidag, S.; Sahlgren, C. Mechanical regulation of the Notch signaling pathway. Curr. Opin. Cell Biol. 2023, 85, 102244. [Google Scholar] [CrossRef] [PubMed]
- Gordon, W.R.; Vardar-Ulu, D.; Histen, G.; Sanchez-Irizarry, C.; Aster, J.C.; Blacklow, S.C. Structural basis for autoinhibition of Notch. Nat. Struct. Mol. Biol. 2007, 14, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Tiyanont, K.; Wales, T.E.; Aste-Amezaga, M.; Aster, J.C.; Engen, J.R.; Blacklow, S.C. Evidence for increased exposure of the Notch1 metalloprotease cleavage site upon conversion to an activated conformation. Structure 2011, 19, 546–554. [Google Scholar] [CrossRef] [PubMed]
- Shergill, B.; Meloty-Kapella, L.; Musse, A.A.; Weinmaster, G.; Botvinick, E. Optical tweezers studies on Notch: Single-molecule interaction strength is independent of ligand endocytosis. Dev. Cell 2012, 22, 1313–1320. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, F.; Li, I.T.; Ngo, T.T.; Leslie, B.J.; Kim, B.C.; Sokoloski, J.E.; Weiland, E.; Wang, X.; Chemla, Y.R.; Lohman, T.M.; et al. Defining Single Molecular Forces Required for Notch Activation Using Nano Yoyo. Nano Lett. 2016, 16, 3892–3897. [Google Scholar] [CrossRef]
- Gordon, W.R.; Zimmerman, B.; He, L.; Miles, L.J.; Huang, J.; Tiyanont, K.; McArthur, D.G.; Aster, J.C.; Perrimon, N.; Loparo, J.J.; et al. Mechanical Allostery: Evidence for a Force Requirement in the Proteolytic Activation of Notch. Dev. Cell 2015, 33, 729–736. [Google Scholar] [CrossRef]
- Seo, D.; Southard, K.M.; Kim, J.W.; Lee, H.J.; Farlow, J.; Lee, J.U.; Litt, D.B.; Haas, T.; Alivisatos, A.P.; Cheon, J.; et al. A Mechanogenetic Toolkit for Interrogating Cell Signaling in Space and Time. Cell 2016, 165, 1507–1518. [Google Scholar] [CrossRef]
- Meloty-Kapella, L.; Shergill, B.; Kuon, J.; Botvinick, E.; Weinmaster, G. Notch ligand endocytosis generates mechanical pulling force dependent on dynamin, epsins, and actin. Dev. Cell 2012, 22, 1299–1312. [Google Scholar] [CrossRef] [PubMed]
- Langridge, P.D.; Struhl, G. Epsin-Dependent Ligand Endocytosis Activates Notch by Force. Cell 2017, 171, 1383–1396.e12. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Struhl, G. Drosophila Epsin mediates a select endocytic pathway that DSL ligands must enter to activate Notch. Development 2004, 131, 5367–5380. [Google Scholar] [CrossRef]
- Kaksonen, M.; Roux, A. Mechanisms of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 2018, 19, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Mund, M.; van der Beek, J.A.; Deschamps, J.; Dmitrieff, S.; Hoess, P.; Monster, J.L.; Picco, A.; Nedelec, F.; Kaksonen, M.; Ries, J. Systematic Nanoscale Analysis of Endocytosis Links Efficient Vesicle Formation to Patterned Actin Nucleation. Cell 2018, 174, 884–896. [Google Scholar] [CrossRef] [PubMed]
- Ben-Yaacov, S.; Le Borgne, R.; Abramson, I.; Schweisguth, F.; Schejter, E.D. Wasp, the Drosophila Wiskott-Aldrich syndrome gene homologue, is required for cell fate decisions mediated by Notch signaling. J. Cell Biol. 2001, 152, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Rajan, A.; Tien, A.C.; Haueter, C.M.; Schulze, K.L.; Bellen, H.J. The Arp2/3 complex and WASp are required for apical trafficking of Delta into microvilli during cell fate specification of sensory organ precursors. Nat. Cell Biol. 2009, 11, 815–824. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Slepnev, V.I.; Di Fiore, P.P.; De Camilli, P. The interaction of epsin and Eps15 with the clathrin adaptor AP-2 is inhibited by mitotic phosphorylation and enhanced by stimulation-dependent dephosphorylation in nerve terminals. J. Biol. Chem. 1999, 274, 3257–3260. [Google Scholar] [CrossRef] [PubMed]
- Kovall, R.A.; Gebelein, B.; Sprinzak, D.; Kopan, R. The Canonical Notch Signaling Pathway: Structural and Biochemical Insights into Shape, Sugar, and Force. Dev. Cell 2017, 41, 228–241. [Google Scholar] [CrossRef]
- Lai, E.C.; Roegiers, F.; Qin, X.; Jan, Y.N.; Rubin, G.M. The ubiquitin ligase Drosophila Mind bomb promotes Notch signaling by regulating the localization and activity of Serrate and Delta. Development 2005, 132, 2319–2332. [Google Scholar] [CrossRef]
- Kalodimou, K.; Stapountzi, M.; Vullings, N.; Seib, E.; Klein, T.; Delidakis, C. Separable Roles for Neur and Ubiquitin in Delta Signalling in the Drosophila CNS Lineages. Cells 2023, 12, 2833. [Google Scholar] [CrossRef] [PubMed]
- Chanet, S.; Schweisguth, F. Regulation of epithelial polarity by the E3 ubiquitin ligase Neuralized and the Bearded inhibitors in Drosophila. Nat. Cell Biol. 2012, 14, 467–476. [Google Scholar] [CrossRef] [PubMed]
- Perez-Mockus, G.; Roca, V.; Mazouni, K.; Schweisguth, F. Neuralized regulates Crumbs endocytosis and epithelium morphogenesis via specific Stardust isoforms. J. Cell Biol. 2017, 216, 1405–1420. [Google Scholar] [CrossRef] [PubMed]
- Perez-Mockus, G.; Mazouni, K.; Roca, V.; Corradi, G.; Conte, V.; Schweisguth, F. Spatial regulation of contractility by Neuralized and Bearded during furrow invagination in Drosophila. Nat. Commun. 2017, 8, 1594. [Google Scholar] [CrossRef] [PubMed]
- Coumailleau, F.; Furthauer, M.; Knoblich, J.A.; Gonzalez-Gaitan, M. Directional Delta and Notch trafficking in Sara endosomes during asymmetric cell division. Nature 2009, 458, 1051–1055. [Google Scholar] [CrossRef]
- Derivery, E.; Seum, C.; Daeden, A.; Loubery, S.; Holtzer, L.; Julicher, F.; Gonzalez-Gaitan, M. Polarized endosome dynamics by spindle asymmetry during asymmetric cell division. Nature 2015, 528, 280–285. [Google Scholar] [CrossRef] [PubMed]
- Loubery, S.; Seum, C.; Moraleda, A.; Daeden, A.; Furthauer, M.; Gonzalez-Gaitan, M. Uninflatable and Notch control the targeting of Sara endosomes during asymmetric division. Curr. Biol. CB 2014, 24, 2142–2148. [Google Scholar] [CrossRef]
- Couturier, L.; Trylinski, M.; Mazouni, K.; Darnet, L.; Schweisguth, F. A fluorescent tagging approach in Drosophila reveals late endosomal trafficking of Notch and Sanpodo. J. Cell Biol. 2014, 207, 351–363. [Google Scholar] [CrossRef]
- Emery, G.; Hutterer, A.; Berdnik, D.; Mayer, B.; Wirtz-Peitz, F.; Gaitan, M.G.; Knoblich, J.A. Asymmetric Rab 11 endosomes regulate delta recycling and specify cell fate in the Drosophila nervous system. Cell 2005, 122, 763–773. [Google Scholar] [CrossRef]
- Jafar-Nejad, H.; Andrews, H.K.; Acar, M.; Bayat, V.; Wirtz-Peitz, F.; Mehta, S.Q.; Knoblich, J.A.; Bellen, H.J. Sec15, a component of the exocyst, promotes notch signaling during the asymmetric division of Drosophila sensory organ precursors. Dev. Cell 2005, 9, 351–363. [Google Scholar] [CrossRef]
- Giagtzoglou, N.; Yamamoto, S.; Zitserman, D.; Graves, H.K.; Schulze, K.L.; Wang, H.; Klein, H.; Roegiers, F.; Bellen, H.J. dEHBP1 controls exocytosis and recycling of Delta during asymmetric divisions. J. Cell Biol. 2012, 196, 65–83. [Google Scholar] [CrossRef] [PubMed]
- Langevin, J.; Morgan, M.J.; Sibarita, J.B.; Aresta, S.; Murthy, M.; Schwarz, T.; Camonis, J.; Bellaiche, Y. Drosophila exocyst components Sec5, Sec6, and Sec15 regulate DE-Cadherin trafficking from recycling endosomes to the plasma membrane. Dev. Cell 2005, 9, 365–376. [Google Scholar] [CrossRef] [PubMed]
- Leibfried, A.; Fricke, R.; Morgan, M.J.; Bogdan, S.; Bellaiche, Y. Drosophila Cip4 and WASp define a branch of the Cdc42-Par6-aPKC pathway regulating E-cadherin endocytosis. Curr. Biol. CB 2008, 18, 1639–1648. [Google Scholar] [CrossRef] [PubMed]
- Georgiou, M.; Marinari, E.; Burden, J.; Baum, B. Cdc42, Par6, and aPKC regulate Arp2/3-mediated endocytosis to control local adherens junction stability. Curr. Biol. CB 2008, 18, 1631–1638. [Google Scholar] [CrossRef] [PubMed]
- Glashauser, J.; Camelo, C.; Hollmann, M.; Backer, W.; Jacobs, T.; Sanchez, J.I.; Schleutker, R.; Forster, D.; Berns, N.; Riechmann, V.; et al. Acute manipulation and real-time visualization of membrane trafficking and exocytosis in Drosophila. Dev. Cell 2023, 58, 709–723.e7. [Google Scholar] [CrossRef] [PubMed]
- Bruelle, C.; Pinot, M.; Daniel, E.; Daude, M.; Mathieu, J.; Le Borgne, R. Cell-intrinsic and -extrinsic roles of the ESCRT-III subunit Shrub in abscission of Drosophila sensory organ precursors. Development 2023, 150, dev201409. [Google Scholar] [CrossRef] [PubMed]
- Huenniger, K.; Kramer, A.; Soom, M.; Chang, I.; Kohler, M.; Depping, R.; Kehlenbach, R.H.; Kaether, C. Notch1 signaling is mediated by importins alpha 3, 4, and 7. Cell. Mol. Life Sci. CMLS 2010, 67, 3187–3196. [Google Scholar] [CrossRef]
- Sachan, N.; Mishra, A.K.; Mutsuddi, M.; Mukherjee, A. The Drosophila importin-alpha3 is required for nuclear import of notch in vivo and it displays synergistic effects with notch receptor on cell proliferation. PLoS ONE 2013, 8, e68247. [Google Scholar] [CrossRef]

| Function | Type | Invertebrates (Drosophila) | Vertebrates (Mammals) |
|---|---|---|---|
| Receptor | Notch | Notch 1, 2, 3 and 4 | |
| Ligand | Delta Serrate | Dll1, 3 and 4 Jagged1 and 2 | |
| Nuclear effectors | CSL DNA-binding transcription factor Transcriptional coactivator | Su(H) Mastermind | RBPjk/CBF-1 MAML1-3 |
| Receptor proteolysis | Site 1 cleavage Site 2 cleavage Site 3 cleavage | Kuzbanian Kuzbanian-like; TACE Presenlin, Nicastrin APH-1, PEN-2 | PC5/6, Furin ADAM10/Kuzbanian ADAM17/TACE Presenlin 1 and 2, Nicastrin APH-1a-c, PEN-2 |
| Membrane trafficking | E3 Ubiquitin ligase Negative regulators Neuralized inhibitors others | Mindbomb 1 and 2 Neuralized Numb Bearbed, Tom, M4 Sanpodo | Mindbomb, Skeletrophin Neuralized 1 and 2 Numb, Numb-like, ACBD3 |
| Canonical target bHLH repressor genes | E(spl) | HES/ESR/HEY |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pinot, M.; Le Borgne, R. Spatio-Temporal Regulation of Notch Activation in Asymmetrically Dividing Sensory Organ Precursor Cells in Drosophila melanogaster Epithelium. Cells 2024, 13, 1133. https://doi.org/10.3390/cells13131133
Pinot M, Le Borgne R. Spatio-Temporal Regulation of Notch Activation in Asymmetrically Dividing Sensory Organ Precursor Cells in Drosophila melanogaster Epithelium. Cells. 2024; 13(13):1133. https://doi.org/10.3390/cells13131133
Chicago/Turabian StylePinot, Mathieu, and Roland Le Borgne. 2024. "Spatio-Temporal Regulation of Notch Activation in Asymmetrically Dividing Sensory Organ Precursor Cells in Drosophila melanogaster Epithelium" Cells 13, no. 13: 1133. https://doi.org/10.3390/cells13131133
APA StylePinot, M., & Le Borgne, R. (2024). Spatio-Temporal Regulation of Notch Activation in Asymmetrically Dividing Sensory Organ Precursor Cells in Drosophila melanogaster Epithelium. Cells, 13(13), 1133. https://doi.org/10.3390/cells13131133





