Abstract
Peroxisome proliferator-activated receptor-γ (PPARγ) belongs to the superfamily of nuclear receptors that control the transcription of multiple genes. Although it is found in many cells and tissues, PPARγ is mostly expressed in the liver and adipose tissue. Preclinical and clinical studies show that PPARγ targets several genes implicated in various forms of chronic liver disease, including nonalcoholic fatty liver disease (NAFLD). Clinical trials are currently underway to investigate the beneficial effects of PPARγ agonists on NAFLD/nonalcoholic steatohepatitis. Understanding PPARγ regulators may therefore aid in unraveling the mechanisms governing the development and progression of NAFLD. Recent advances in high-throughput biology and genome sequencing have greatly facilitated the identification of epigenetic modifiers, including DNA methylation, histone modifiers, and non-coding RNAs as key factors that regulate PPARγ in NAFLD. In contrast, little is still known about the particular molecular mechanisms underlying the intricate relationships between these events. The paper that follows outlines our current understanding of the crosstalk between PPARγ and epigenetic regulators in NAFLD. Advances in this field are likely to aid in the development of early noninvasive diagnostics and future NAFLD treatment strategies based on PPARγ epigenetic circuit modification.
1. Introduction
Nonalcoholic fatty liver disease (NAFLD), which affects 25–30% of the global population, is currently one of the main public health and economic burdens [1]. Depending on the degree of liver abnormalities, the pathologic spectrum of NAFLD ranges from simple steatosis to its more aggressive form, nonalcoholic steatohepatitis (NASH), which can progress to advanced stages such as liver fibrosis, cirrhosis, and hepatocellular carcinoma (HCC) [2]. According to current knowledge, NAFLD is closely associated with several prevalent risk factors, including atherogenic dyslipidemia, type 2 diabetes mellitus (T2DM), metabolic syndrome, cardiovascular disease (CVD), and insulin resistance (IR) [3,4,5]. Epidemiology research shows that NAFLD is a metabolic disorder with a complex multifactorial pathogenesis and heterogeneous clinical manifestations [6], which varies between individuals with comparable lifestyles and metabolic abnormalities
Based on the heterogeneous aspect of this disease, an international panel of experts proposed a name change for NAFLD to Metabolic Associated Fatty Liver Disease (MAFLD) [7]. Although interesting, the newly proposed terminology has not been widely accepted and a debate on the subject is ongoing [8,9]. The pathogenesis of NAFLD is complicated and not well understood. Two hypotheses have been postulated to explain the onset and progression of this condition. The initial hypothesis representing the first “hit” is referred to as a “two-hit model”, in which liver fat accumulation and IR lead to liver damage in NASH [10,11]. Because NAFLD is well established as a multifactorial disorder in which multiple insults act together to induce pathology, the first hit may not be an accurate explanation of the pathogenesis of the disease. Therefore, scientists have turned to the second and increasingly accepted hypothesis, “multiple parallel hit”, which provides a more reliable explanation for the complex characteristics of NAFLD. This hypothesis suggests that, in addition to adipose tissue fat accumulation and lipotoxicity, other factors such as IR, mitochondrial dysfunction, increased endoplasmic reticulum stress, epigenetic alterations, and changes in the gut microbiota may act in concert in genetically predisposed individuals to promote liver inflammation and fibrosis [12,13,14].
There are limited therapeutic options and no approved drug that specifically targets NAFLD, despite intensive research on novel treatment for this condition and continuing clinical trials [15]. Lifestyle intervention, including diet and exercise, is still a preferred method. As IR is the most specific metabolic risk and pathologic hallmark of NAFLD, the use of PPARγ agonists, insulin-sensitizing thiazolidinedione (TZD) molecules, to treat NASH patients has also been investigated [16], as IR is the most specific metabolic risk and pathologic hallmark of NAFLD. However, the exploitation of peroxisome proliferator-activated receptor-γ (PPARγ) signaling in a therapeutic setting of NAFLD has been hampered by the limited understanding of its regulatory mechanisms and the lack of its precise function in liver-adipose tissue crosstalk.
The superfamily of peroxisome proliferator-activated receptors (PPARs) has three subtypes: PPARα, PPARγ, and PPAR-β/δ, which regulate many metabolic pathways in a tissue-specific manner [17]. PPARγ was identified previously by Tontonoz et al. [18] and the corresponding gene, Pparγ, was mapped on chromosome 3p25.2 in humans [19]. The human Pparγ gene is composed of 9 exons exon A1, exon A2, exon B, and exons 1–6. This gene generates four Pparγ splice variants (Pparγ 1–4), which differ at their 5-end due to differential promoter usage and alternative splicing, and encodes for two protein isoforms (Figure 1) [20,21]. The PPARγ1 isoform, which is a 477 amino acid protein, is produced by the mRNAs Pparγ1, Pparγ3, and Pparγ4. The PPARγ2 isoform is translated from the Pparγ2 mRNA transcript and has an extra 30 amino acids at its NH2-terminus [22].
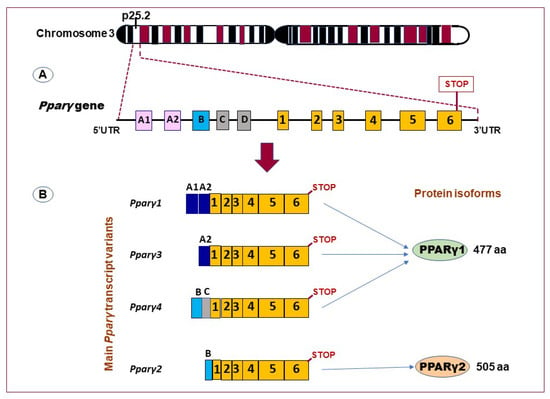
Figure 1.
Schematic representation of the human Pparγ gene and its main transcript variants. (A) Pparγ gene lies on chromosome 23, band 3p25, and composed of at least 11 exons (exon A1–2, exon B–D, and exons 1–6). (B) Alternative promoter and mRNA splicing generate several variants (the transcript variants Pparγ 1,3, and 4 encode PPARγ1 isoform (477 amino acids; aa). The transcript variants Pparγ2 encodes the PPARγ2 isoform (505 aa). (Adapted from reference [21]).
PPARγ is involved in multiple physiological and pathological processes [23]. Based on the tissue in which it is expressed, PPARγ has three different isoforms. PPARγ2 expression is mostly restricted to adipose tissue, whereas PPARγ1 is found in nearly all cells [18,24,25]. It is also worth noting that, while PPARγ2 expression is induced in fatty liver, it remains substantially lower than in adipose tissue. On the other hand, macrophages, adipose tissue, and the colon are where PPARγ3 is most abundantly expressed [26]. The primary function of PPARγ is believed to be in adipose tissue, where it is known to induce adipocyte differentiation and promote triglyceride storage, hence reducing liver lipotoxicity and improving steatosis [27]. Transcription levels of this nuclear factor have been found elevated in the steatotic livers of obese individuals and animal obesity models [28,29]. Consistent with this finding, a different study found that PPARγ2 inactivation reduced the severity of fatty liver that had been induced by a high saturated fat diet in mice [30]. Furthermore, rosiglitazone and pioglitazone significantly reduced hepatic steatosis, as shown by clinical human studies [16,31,32]. There is growing evidence that the pathophysiology of NAFLD is significantly influenced by epigenetic changes, including altered DNA methylation patterns, posttranslational modifications of histones, and ncRNAs. However, it is yet unclear how these changes along with PPARγ influence the onset and progression of the disease.
2. The Role of Epigenetic Mechanisms in Regulating PPARγ Regulatory Function in NAFLD
Contrary to genetic changes in heritability, epigenetics is the study of heritable modifications in gene activity that do not involve direct alteration of the underlying DNA sequences [33]. Epigenetics determines the architecture of chromatin in cell nucleus, and therefore affects specific genomic sequences accessible to cellular regulatory machineries. The epigenome is susceptible to dysregulation throughout life but is highly vulnerable to environmental factors during fetal life since this is a period of rapid DNA synthesis [34].
Cells’ transcriptomes can be modified in response to both internal signals and environmental cues via epigenetic modulators, primarily DNA methylation, histone modifications, and alterations in ncRNAs expression patterns. As was already indicated, alterations in the liver epigenetic mechanisms have been demonstrated to have a role in the emergence of NAFLD [35] in part through crosstalk with PPARγ. Given the crucial role that PPARγ plays in lipid metabolism and lipogenesis, the identification of epigenetic changes underlying its regulation in the development of NAFLD is of special relevance for study in this field. In light of this, the next sections provide an overview of the existing literature on altered reciprocal regulation between epigenetic modulators and the PPARγ signaling pathways in NAFLD.
2.1. DNA Methylation
The most prevalent epigenetic mark in the mammalian genome is DNA methylation, which occurs when a methyl group is added to the C5 position of cytosine to form 5-methylcytosine. DNA methylation affects the accessibility of the transcriptional machinery to a DNA region that regulates gene expression. When confined to gene promoters, DNA methylation is often a repressive epigenetic signal [36]. The methylation of DNA bases, which is important for controlling the expression of imprinted genes, has been linked to a variety of human diseases, including NAFLD [37,38,39]. In patients with NAFLD, both hepatic DNA methylation and insulin resistance play a key role in the disease progression from simple steatosis to severe fibrotic NASH [40].
When comparing the DNA methylation levels of numerous genes in liver samples from NAFLD patients to those from healthy subjects, differences have been observed [41,42]. Even though PPARγ is less abundant in the liver than PPARα, it is still crucial for liver function, and the DNA methylation state of the Pparγ gene has been identified as a marker of the progression of liver disease. In a case-control study of NAFLD patients, increased hepatic methylation of the promoter of the PPARγ coactivator one-alpha (PGC1-α) gene, a key transcriptional regulator of mitochondrial fatty acid oxidation, significantly correlated with peripheral IR status and fasting insulin levels [43]. Furthermore, in human NASH liver biopsies, it has been shown that the promoter region of Pparγ undergoes methylation remodeling and becomes hypermethylated as fibrosis severity increases [44], indicating that DNA methylation may be used as a non-invasive tool for stratifying the risk of fibrosis in NAFLD. In line with this finding, a different investigation on subjects with NAFLD revealed that DNA methylation at particular CpG dinucleotides within the human Pparα and Pparγ gene promoters can differentiate between patients with mild from those with severe fibrosis in NAFLD [45]. Later, a Turkish cohort study conducted by the same research team revealed a link between DNA methylation in the Pparγ promoter and fibrosis [46]. In a recent work, Hajri et al. showed that both HFD and palmitic acid alter global and Pparγ promoter DNA methylation, leading to significantly increased Pparγ expression and enhanced lipid retention in the liver, which causes NAFLD to develop [47]. Moreover, both in rat models and in NAFLD patients, it was found that Pparγ methylation levels significantly correlated with the severity of liver fibrosis [44,48]. It is of interest to note that Pparγ promoter methylation levels in plasma-free DNA were proposed as a non-invasive method to distinguish between patients with mild and severe fibrosis associated with NAFLD [48]. Another base alteration in DNA called 5-hydroxymethylcytosine (5hmC) has been found to affect DNA demethylation, which in turn affects both the activation and repression of gene transcription [49]. In this regard, an observational study by Pirola et al., suggested that the 5hmC might be involved in the pathogenesis of NAFLD by regulating liver mitochondrial biogenesis and PPARγ coactivator 1a (PGC-1α) expression [50].
Indirect effects of DNA methylation on Pparγ expression are also possible. In fact, a prior study found that, in diet-induced obese mice, methylation of hepatic interferon regulatory factor 6 (Irf6) reduces hepatic steatosis and metabolic abnormalities by transcriptionally repressing Pparγ [51]. It has been reported that the C-Maf inducing protein (Cmip) is associated with metabolic disorders such obesity, diabetes, and NAFLD. A further investigation demonstrated that hypomethylation of Cmip promotes its expression and facilitates the development and progression of NAFLD by activating the PPARγ-CD36 signaling pathway [52]. Furthermore, even though the findings reported here show interesting characteristics of Pparγ gene expression and methylation changes in relation to NAFLD, more research is required to clearly establish a causal link between the two events. Examples of DNA methylation patterns and the PPARγ pathway linked to the pathogenic feature of NAFLD are shown in Table 1.

Table 1.
Epigenetic regulation of PPARγ by DNA methylation in NAFLD.
2.2. Histone Modifications
An important component of the epigenetic changes that affect the transcriptional regulatory processes is the dynamic network of post-translational histone modifications. Much research has been conducted on histone methylation and acetylation as heritable epigenetic indicators for chromatin structure and function. Many enzymes control the posttranscriptional alterations of histones by interfering with particular DNA binding sites, which results in the dysregulation of specific gene expression [53]. In addition, to achieve the accurate regulation of gene expression, histone modifications frequently interact in a cooperative way with transcription factors (TFs). It has been demonstrated that an imbalance in histone modifications leads to an irregularity in transcriptional activity that is associated with the emergence of diseases such T2DM, obesity, and consequently MAFLD [54]. For instance, abnormal histone modifications have been shown to promote the development of insulin resistance and thus, NAFLD [55]. Hence, gaining a better knowledge of how cells connect histone changes to transcription factors (TFs) may open up new avenues for the identification of novel epigenetic targets and offer crucial hints for the design of functional investigations to come and prospective epigenetic treatments for NAFLD.
2.2.1. Histone Methylation/Demethylation
According to a study undertaken by Kim et al., the histone H3 lysine 4 (H3K4) methyltransferase myeloid/lymphoid or mixed-lineage leukemia 4 (MLL4/KMT2D) regulates overnutrition-induced steatosis by acting as a coactivator for PPARγ2 through H3K4 methylation [56]. Further studies suggested that H3K4 and H3K9 trimethylation may contribute to hepatic steatosis and disease progression [57]. In fact, Jun et al. demonstrated that, in HFD-fed mice, aberrant histone H3K4 and H3K9 trimethylation in Pparα and genes involved in lipid metabolism cause hepatic steatosis [57]. Moreover, both diet-induced obese mice and NAFLD patients have considerably higher levels of the histone-lysine N-methyltransferase suppressor of variegation 3-9 homologue 2 (Suv39h2), which represses the expression of the Sirt1 and Pparγ genes [58].
The process of histone demethylation is carried out by enzymes called histone demethylases (HDMs), which remove methyl groups from altered histones to activate or repress gene transcription. Many histone demethylases have been identified and classified into two classes: FAD-dependent amine oxidases (LSD demethylases) and Fe(II)- and α-ketoglutarate-dependent Jumonji C (JmjC) domain-containing demethylase (JMJD demethylase) [59]. PPARγ is also implicated in the regulation of adipogenic metabolism by certain demethylases. However, direct evidence that HDMs participate in the PPARγ pathway is scarce. The H3K9-specific Jumonji demethylase JMJD1A has been reported to bind to the Pparγ promoter, which then decrease the number of H3K9me2 marks in this region, causing modulation of hepatic stellate cells activation and liver fibrosis [60]. Inversely, increasing JMJD2B expression promoted adipogenesis and steatosis by increasing PPARγ2 expression, hepatic lipid uptake, and intracellular triglyceride accumulation [61]. Collectively, the reviewed phenotypic evidences, as summarized in t Table 2, demonstrate that histone methylation status/PPARγ axis plays important roles in the emergence of NAFLD. However, further studies are needed to comprehend the abnormalities in the histone system that may result in NAFLD through PPARγ signaling, which would greatly increase our understanding of the pathophysiology of this condition.

Table 2.
Epigenetic regulation of PPARγ through histone methylation mechanisms.
2.2.2. Histone Acetylation/Deacetylation
The balance between acetylation and deacetylation plays a role in the regulation of gene expression. Histone acetylation is catalyzed by histone acetyltransferases (HATs), which use acetyl-CoA as a co-substrate and acetylate lysine residues on histone tails. HATs modify chromatin histones and play an important role in the epigenetic modulation of gene transcription programs. Additionally, aberrant histone modifications have been shown to contribute to the onset of IR and consequently to fatty liver disease [55]. Indeed, numerous investigations have demonstrated an association between NAFLD and changes in histone acetylation [62,63]. Moreover, studies have carefully looked into how histone (de)acetylation in Pparγ locus influences its expression in NAFLD. For example, a prior study revealed that histone marks on histone H3 lysine 9 acetylation are increased at Pparγ binding sites during adipogenesis [64]. Chromatin profiling of H3K27ac revealed that this mark is highly induced at the Pparγ gene locus during the course of adipogenesis and correlates with Pparγ gene expression [65]. It is important to keep in mind that the process of adipogenesis is accompanied by the fat synthesis, which may contribute to the occurrence and progression of NAFLD. Unfortunately, little is known about the relationship between HATs and TFs in the development of NAFLD.
Histone deacetylases (HDACs) are known to repress gene expression by removing acetyl groups from lysine residues in the NH2 terminal tails of core histones and condensing chromatin, rendering the regions less accessible to transcription factors. A further study revealed that PPARγ deacetylation on two lysine residues (K268 and K293) induces brown remodeling of white adipose tissue and uncouples the adverse effects of TZDs from insulin sensitization [66]. Recent research has demonstrated that PPARγ deacetylation inhibits hypercholesterolemia and aging-associated atherosclerosis [67], confers the anti-atherogenic properties, and improves endothelial function in the treatment of diabetes [68]. Many aspects of mammalian development and physiology need HDAC3 [69,70]. HDAC3 genetic investigation indicates that it is a crucial regulatory component of molecular complexes that govern gene expression, which in turn affects metabolic function in the liver via numerous signaling pathways, and HDAC3 deletion in the liver affects normal metabolic homeostasis [70,71]. HDAC3 has also been demonstrated to modulate metabolism by increasing fatty acid oxidation and improving circadian histone deacetylation [72]. Interestingly, clinical studies revealed that HDAC3 expression levels in pediatric patients were correlated with overweight [73]. High levels of proinflammatory markers and insulin resistance are associated with enhanced expression of the deacetylase HDAC3 in the hepatocytes of fat-fed E3 rats that developed metabolic syndrome and in the peripheral blood mononuclear cells of T2DM patients [74]. Inhibition of HDAC3 may promote ligand-independent PPARγ activation by protein acetylation causing an increase in glucose uptake and improvement of insulin sensitivity in adipocytes [75].
A histone deacetylase known as Sirtuin 1 (SIRT1) has historically been associated with the control of hepatic metabolism, as well as glucose and lipid homeostasis [76]. A previous study has indicated that the deacetylating effect of SIRT1 on histone improves hepatic steatosis [77]. It was also demonstrated that better liver health is correlated with overexpression of SIRT1 in hepatocytes [78,79]. Fatty acid oxidation has been linked to SIRT1, and its deficiency negatively impacts PPARγ signaling. Interestingly, the interaction of PPARγ and SIRT1 is essential for the activation of PGC-1α. Moreover, hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and leads to hepatic steatosis and inflammation [80]. In fact, reduced levels of this protein have been observed in NAFLD patients as well as in animal models [81,82]. Parallel to this, SIRT1 suppression in the mouse liver is sufficient to cause hepatic steatosis [83], an effect that may be mediated via Ppar-γ and Pparα, the key regulators of glycolysis and lipolysis [80,84]. Adipose-specific deletion of Sirt1 generates a hyperacetylated PPARγ state and enhanced PPARγ activity, leading to higher insulin sensitivity [84]. Collectively, these preliminary findings highlight the significance of PPARγ epigenetic regulation and histone-modifying enzymes as possible pharmaceutical targets to treat NAFLD.
2.3. Noncoding RNAs
ncRNAs modulate various cell biological processes in cells, including metabolism, chromatin shaping, gene transcription and translation, and posttranslational modifications. Dysregulation of these transcripts has been implicated in a variety of pathologies including NAFLD. Therefore, understanding their underlying mechanisms of action and identifying factors with which they crosstalk will make them appealing non-invasive biomarkers and therapeutic targets in fatty liver disease. As previously highlighted, peroxisome proliferator-activated receptors (PPARs) regulate lipid homeostasis and have been proposed as important regulators in the development of NAFLD and its various stages. Furthermore, their cross-regulation with ncRNAs has emerged as an additional layer of complexity in the regulatory mechanisms of several diseases, including NAFLD [85,86,87]. Thus, expanding our knowledge of the ncRNAs/PPARγ regulatory axis may help us better understand how epigenetic mechanisms contribute to the physiopathology of NAFLD and advance the process of developing potential ncRNAs/PPARγ-based therapeutics for this condition.
2.3.1. miRNAs-PPARγ Axis
miRNAs, a type of small noncoding RNAs, have been associated with the epigenetic regulation of gene expression involved in the development of steatosis and its progression to NASH, fibrosis, and HCC [88,89,90]. There is currently mounting evidence that miRNAs influence the transcription of NAFLD-related genes including those involved in PPARγ pathway [91,92]. Further research revealed that the expression of a variety of miRNAs was induced in NASH and fibrosis, and that this induction was associated with PPARγ modulation. As will become apparent in the following discussion, the mutual crosstalk between PPARγ and specific miRNAs plays substantial roles in the development of NAFLD forms.
miR-21: It has been shown that miR-21 promotes hepatic lipid accumulation in part by interacting with multiple factors, including SREBP1 [93] and 3-hydroxy-3-methylglutaryl-co-enzyme A reductase [94]. Furthermore, by inhibiting the PPARα signaling pathway, miR-21 contributed to cell damage, inflammation, and fibrosis [95]. Several investigations have shown that the circulation and liver of NAFLD patients and mice models both have high amounts of miR-21 [96,97,98]. Rodrigues et al. showed that feeding miR-21 knockout animals an obeticholic acid-supplemented HFD causes a progressive decrease in steatosis, inflammation, and lipoapoptosis through PPARα upregulation and activation of the farnesoid X-activated receptor (FXR) [99]. Further research indicated that inhibition of miR-21 could alleviate steatosis by activating PPARα [95,100]. The relevance of the miR-21/PPARγ axis in NAFLD, however, remains poorly understood. According to a recent study, PPARγ regulates miR21-5p/Secreted Frizzled-related Protein 5 (SFRP5) to reduce inflammation and oxidative stress in mouse models and human tissue samples from NASH patients [101]. Mechanistically, PPARγ downregulates miR-21-5p by interacting with its promoter region, resulting in increased expression of SFRP5, an anti-inflammatory adipokine that regulates NASH progression [101]. The authors of this study suggested that the PPARγ/miR-21-5p//SFRP5 axis might be a promising target for NASH treatment.
miR-27: Early studies suggested that miR-27a decreases lipid accumulation in rat HSCs and human hepatoma cells by targeting the retinoid X receptor alpha [102,103] and impairs adipocyte differentiation by targeting PPARγ [103]. A different investigation revealed that miR-27a is essential for maintaining hepatic lipid homeostasis and for the pathogenesis of NAFLD [104]. miR-27b is part of a panel of miRNAs that has been proposed for highly accurate diagnosis of NAFLD [105] and NASH in the livers of rats and zebrafish [106]. The effects of three different Western diets, a low-fat diet, a high-fat diet, and a high-fat high-fructose diet exposure, on liver PPARγ/miRNA regulation were examined, and the results revealed that a high-fat, high-fructose diet induces an intermediate stage between fatty liver and fibrosis via miR-27b-5p-induced PPARγ downregulation [107]. The miR-27b targets 3′-UTR of Pparγ gene [108] and contributes to the destabilization of Pparγ mRNA by lipopolysaccharides [109]. Disruption of endogenous miR-27b activity by a transgenic miR-27b sponge in zebrafish enhances lipid accumulation and the expression of PPARγ in the liver, leading to early onset of NAFLD and NASH [110]. Moreover, miR-27a directly targets the 3′-UTR of the Pparγ gene to promote proliferation of hepatocellular carcinoma cells [111,112]. Taken together, these findings support the notion that the miR-27-Pparγ axis may be a target for NAFLD prevention.
miR-34: The expression of miR-34a is altered in patients with T2DM, steatosis, NASH, and NAFLD models [113,114,115]. Several studies [116,117] found that serum levels and hepatic expression of miR-34 were higher in NAFLD/NASH patients compared to controls [116,117]. Furthermore, in patients with coronary artery disease, miR-34a expression is upregulated, which is exacerbated when the patients also have NAFLD [118]. In a mouse model, inhibiting miR-34a improved hepatic steatosis by increasing PPAR levels, which promoted lipid oxidation [119]. In an in vitro and in vivo model, the antagomir circRNA 0046366 antagonized miR-34a and restored PPAR expression, alleviating NAFLD [120]. Derdak et al. showed that inhibiting p53 transcriptional activity partially reduced steatosis, associated oxidative stress, and apoptosis in a mouse model of NAFLD by downregulating miR-34a and activating the SIRT1/PGC1α/PPARα axis [121]. Furthermore, miNA-34a and miRNA-34c promoted HSCs activation by targeting PPARγ, implying that members of the miR-34 family may be involved in liver fibrosis via PPARγ pathways [122].
miR-132: Visceral adipose tissue from obese bariatric surgery patients with biopsy-proven NASH had considerably lower levels of a panel of miRNAs expression profiles, including miR-132 [123]. However, animal models of hepatic steatosis or NASH exhibited a substantial increase in hepatic miR-132 levels and a commensurate drop in selected miR-132 targets, while miR-132 repression attenuated the steatotic phenotype [124]. miR-132 was downregulated in stellate cells from CCl4-treated animals (to induce fibrosis), which resulted in the expression of one of its targets, methyl-CpG-binding protein 2 (MeCP2). In turn, MeCP2 binds to PPARγ and enhances the development of an epigenetic repressor complex, which inhibits PPARγ transcription and, in this mouse model, causes liver fibrosis [125].
miR-155: This transcript has been found to be implicated in several inflammatory processes that control innate immunity in both alcoholic and nonalcoholic fatty liver disease [126]. Additionally, miR-155 regulates various TFs involved in lipid metabolism such as LXRα [127], PPARα and PPARγ [128], and SIRT1 [129]. The expression of miR-155 is reduced during adipogenesis in vitro, while its overexpression inhibits PPARγ and cEBPα, clearly indicating that miR-155 acts as a negative regulator of adipogenesis [130]. By suppressing PPARγ, miR-155 contributes to insulin resistance, a glucose intolerance state conferred by obese adipose tissue macrophage (ATM) exosomes [131]. Tryggestad et al. found increased expression of miR-155 by ATM in obese individuals and predicted that this would have the same effect on PPARγ and GLUT4 [132]. In a mouse model of liver fibrosis, miR-155 is induced, and PPARγ is its direct target in both naive and alcohol-treated macrophages [128], implying that PPARγ also functions as an antifibrotic gene. More interesting, several miRNAs have been identified as key regulators of hepatic steatosis. miR-30a-3p and miR-3666, for example, protect hepatocytes from steatosis by targeting PPARα and PPARγ, respectively [133,134]. Last but not least, the studies reviewed here and compiled in Table 3 affirm the relevance and complexity of the regulation of the miRNAs/PPARγ network in NAFLD emphasizing the necessity of continuing research in this promising field.

Table 3.
Relevant miRNAs shown to be associated with PPARγ in NAFLD and its complications.
2.3.2. lncRNAs-PPARγ Axis
LncRNAs are a group of RNA molecules that have a length of more than 200 nucleotides but cannot be translated into functional proteins. These RNA transcripts have a variety of epigenetic regulatory functions in humans, including chromatin modification and remodeling, genomic imprinting, and transcriptional and translation processes [135]. lncRNAs alteration has been linked to the pathophysiology of a variety of diseases, including cancer, T2DM, CVD, and liver disease [136]. In fact, deregulation of lncRNAs has been proposed as a factor in NAFLD susceptibility [137]. There is ample evidence that lncRNAs crosstalk with nuclear receptors, including PPARs to play a pivotal role in triglyceride, cholesterol, and lipoprotein metabolism. For example, the hepatocyte-derived lncRNA (lnc-HC) regulates hepatic lipid droplets accumulation via the miR-130-3p/PPARγ pathway [138], implying that lnc-HC could be a potential therapeutic target to prevent excessive lipogenesis, lipid accumulation, and NAFLD phenotype. Through the expression of PPARγ, lncRNA AC096664.3 has a positive correlation with ATP Binding Cassette Subfamily G Member 1 expression [139]. In NAFLD animal and FFA-treated cell models, overexpression of lncRNA-H19 (H19) promotes steatosis and increases hepatic lipid accumulation and lipogenesis via the miR-130a/PPARγ axis [140]. Consistent with these results, Wang et al. revealed that the expression of H19 induces hepatic steatosis by activating the lipogenic transcription factor MLX interacting protein-like and the mammalian target of rapamycin complex 1 (mTORC1) transcriptional network in hepatocytes [141]. Another lncRNA that can affect the stage of adipogenesis, the lncRNA steroid receptor RNA activator (SRA), has also been shown to promote hepatic steatosis by repressing adipose triglyceride lipase expression [142]. SRA is known to act as a steroid receptor coactivator [143]. In adipogenesis, SRA associates with PPARγ and coactivates PPAR-dependent gene expression [144]. Aerobic exercise appears to improve lipid metabolism in obese mice via the LncSRA/p38/JNK/PPARγ signaling pathway [145]. In both humans and mice with NAFLD, the conserved Hedgehog (Hh) signaling pathway was activated [146,147]. According to one recent study, the Hh pathway plays an important role in the regulation of hepatic lipid metabolism and related diseases via direct regulation of a previously uncharacterized lncRNA termed Hedgehog signaling-induced long noncoding RNA (Hilnc) [148]. Hilnc was found to control the stability of PPARγ mRNA by directly interacting with IGF22BP2. The PPARγ signaling pathway was reduced in Hilnc knockout mice, which made them resistant to diet-induced obesity and hepatic steatosis [148]. It is interesting to note that the same research team also discovered h-Hilnc, the human homologue of Hilnc, which demonstrated a comparable role for Hilnc in lipid metabolism. These results collectively imply that understanding the epigenetic function of the lncRNA/PPARγ networks, with the most pertinent examples presented in Table 4, may improve lipid homeostasis and prevent NAFLD. Nonetheless, more research would be beneficial to pinpoint the precise role that these events played in the etiology of NAFLD.

Table 4.
Relevant lncRNAs shown to regulate PPARγ in NAFLD and associated complications.
2.3.3. circRNAs-PPARγ Axis
Circular RNAs (circRNAs) are endogenous noncoding RNA molecules that have recently attracted attention due to their stable structure [149]. Recently, it was found that circRNAs sponge miRNAs by binding to miRNA response elements (MREs) [150,151]. They have been implicated in numerous important physiological and pathological processes and may serve as potential biomarkers for a variety of diseases. For instance, some circRNAs can inhibit miRNA’s function that is associated with the progression and pathophysiology of chronic liver disease [151,152,153]. Insulin resistance, which is considered to be the “first hit” in NAFLD, has been attributed to the circHIPK3 and circANKRD36-mediated sponging of miR-192-5p and miR-145, respectively [154,155]. While there is a wealth of knowledge regarding the crosstalk between PPARγ and miRNAs or lncRNAs in NAFLD, very few studies have examined the function of the circRNAs/PPARγ network. In this respect, a prior study has revealed a tight association between circRNAs with hepatic steatosis and NASH [156] and circRNA_0046367 has been found to prevent hepatic steatosis by reversing the inhibitory effect of miR-34a on PPARα by blocking miRNA/mRNA interaction with MRE [153]. Likewise, circRNA_0046366 has been shown to inhibit hepatocellular steatosis by normalizing PPARα signaling [120]. The circRNA low-density lipoprotein receptor (circLDLR) acts as a sponge for miR-667-5p to regulate SIRT1 expression in NAFLD [157]. SIRT1 is the NAD-dependent deacetylase known as a PPARγ inhibitor [158]. Together, circLDLR and SIRT1 are common targets of miR-667-5p and contribute to the development of NALFD by regulating the autophagy pathway. Recently, Lin et al. identified a novel circRNA, circRNf111 (hsa_circ_0001982, generated from exon 2 of the RNF111 gene), whose expression is downregulated in metabolic syndrome, a risk factor for NAFLD [159]. Afterword, these authors demonstrated that circRNF111 protects against insulin resistance and fat deposition in the metabolic syndrome via controlling the miR-143-3p/IGF2R axis [159].
3. Limitations and Future Perspectives
Emerging evidence clearly indicates that alterations in PPARγ/epigenetic effectors network play a crucial role in the pathogenesis of NAFLD. These observations have sparked a renewed interest in investigating PPARγ epigenetic regulatory mechanisms as they may lead to the discovery of potential non-invasive biomarkers and therapeutic targets for NAFLD. Even though the concept is promising, there are still several limitations in the field, and these will be pointed out. (i) While the studies discussed above provided evidence that epigenetic factors contribute to the dysregulation of PPARγ in NAFLD, the impact of the genetic determinants of this TF was not considered in this work. This is challenging because variants in the Pparγ gene have been linked to metabolic disorders such as atherosclerosis, diabetes, obesity, and NAFLD. In fact, a missense Pro12Ala substitution in the Pparγ2 gene (rs1801282) has been investigated in relation to NAFLD risk in several ethnic groups, and those who carry the Pparγ2 Ala allele polymorphism showed a protective effect against NAFLD [160,161]. Additionally, the SNP rs3856806 (also referred to as C161T or C1431T) in the Pparγ gene increases NAFLD susceptibility through the adiponectin pathway [162,163]. Therefore, the fundamental question that remains unanswered is to what extent these genetic variations drive the potential interactions between PPARγ and epigenetic regulators. (ii) Another significant limitation in developing therapeutics for NAFLD/NASH is a lack of suitable and validated preclinical models that mimic the pathophysiology of human disease [164,165]. Despite the fact that molecular epigenetic modifications such as DNA methylation are apparent across species, there are no relevant preclinical models to investigate the role of epigenetic mechanisms in NAFLD. This drawback emphasizes the urgent need to develop newer and more pertinent epigenetic models that offer insightful information about the pathophysiology of NAFLD and promote their successful implementation in real-world human therapeutic settings. In this respect, epigenome editing is gaining ground as a promising strategy to reverse aberrant epigenetic drivers of disease. In fact, there have been recent attempts to enable gene expression reprogramming by targeting epigenetic editing of locus-specific sites using DNA technology to generate these models. For example, to attenuate fibrosis in a mouse model, Xu et al. successfully demonstrated targeted demethylation of the Rasal1 and Klotho genes by in vivo lentiviral delivery [166]. Similar to this, Horii et al. generated a mouse model for epigenetic disorders through targeted epigenome demethylation [167]. Consequently, using epigenome editing technologies in a preclinical model could lead to a better knowledge of how epigenetic expression is controlled and the development of new therapeutic tools. (iii) The tight bidirectional communication between adipose tissue and liver is known to involve multiple factors including lipids, adipokines, and secreted molecules that can affect the expression key TFs such as SREBP1c and PPARγ, thus leading to enhanced pathological process of NAFLD. In addition, inflammatory signals and immune mediators can interact with chromatin to influence changes in the epigenetic landscape. However, it is unclear how adipose tissue/liver axis dysfunction affects PPARγ/epigenetic mechanisms via immune and inflammatory factors. (iv) There is an ambiguity as to whether PPARγ is in favor or against the development of NAFLD. As was previously mentioned, PPARγ is abundantly expressed in adipose tissue, where it is crucial for the control of adipogenesis, fat storage, and glucose metabolism. Several clinical trials have examined the ability of TZDs to lower NAFLD/NASH by targeting PPARγ [168]. This decrease could be attributed to PPARγ activation in adipose tissue, which promotes adipogenesis while inhibiting lipolysis, lowering the amount of fatty acids entering the liver [169]. In fact, adipogenesis is known to be a physiological process that improves tissue’s ability to safely store lipids and avoid lipotoxicity in peripheral organs such as the liver. PPARγ expression is low in healthy liver, but once activated, it can provide several benefits such as protection from oxidation, inflammation, fibrosis, fatty liver [170], and insulin sensitizing activity [27]. Paradoxically, there is a strong correlation between the onset of NAFLD and hepatocyte-specific PPARγ expression. Furthermore, hepatic PPARγ expression is strongly induced in NAFLD patients and experimental models [28,171]. Therefore, this ambiguity must be resolved. Particular focus should be placed on the potential crosstalk between PPARγ pathways and epigenetic factors and how this might be fine-tuned in different tissues and contexts. (v) Another important pathway involved in the pathogenesis of NAFLD is the tripartite interaction of adipose tissue, gut, and liver [172]. It is unclear what role PPARγ and epigenetic mechanisms might play in regulating such an organ-organ connection. In the long run, exploring the gut-liver-adipose tissue axis, which integrates epigenetic and TFs mechanisms, may therefore lead to better understanding of the pathogenesis of liver disease as well as new treatment options.
4. Conclusions
The findings from numerous human and animal studies reviewed above and recapitulated in Figure 2 clearly support the notion that dysregulation in the crosstalk between PPARγ singling and epigenetic effectors plays a significant role in the pathophysiology of NAFLD and its progression stages. Because NAFLD is likely to affect more than one organ, this may be a compelling rationale to investigate the mechanisms governing the functional association and interplay of PPAR and epigenetic regulators, not only in the liver but also in other organs and systems implicated in NAFLD. It is hoped that this knowledge will shed more light on the pathogenesis of the condition and may lead to the identification of novel epigenetic targets and signaling pathways that may hold crucial information for the development of functional studies in the future and potential epigenetic treatments for NAFLD.
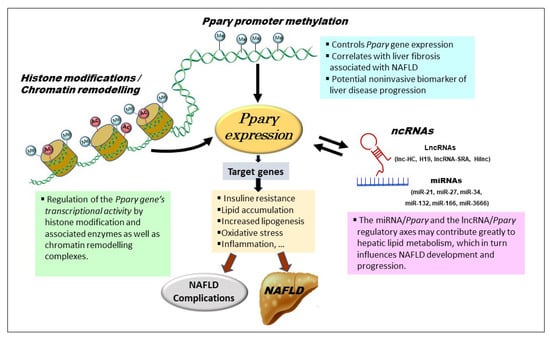
Figure 2.
Epigenetic regulatory mechanisms of PPARγ in NAFLD. Pparγ promoter methylation changes alter PPARγ expression and, as a result, contribute to NAFLD development. Histone modification in the vicinity of Pparγ promoter can regulate PPARγ expression and lipogenesis. The expression of PPARγ and its target genes is regulated by chromatin remodeling at the Pparγ locus. Many ncRNAs, primarily miRNAs and lncRNAs, can influence PPARγ expression via crosstalk regulation. Epigenetic modifiers are themselves subject to regulation by environmental stimuli and genetic factors. Ac—acetylation; Me—methylation; ncRNAs—noncoding RNAs.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Younossi, Z.M.; Koenig, A.B.; Abdelatif, D.; Fazel, Y.; Henry, L.; Wymer, M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016, 64, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Galoosian, A.; Kaswala, D.; Li, A.A.; Gadiparthi, C.; Cholankeril, G.; Kim, D.; Ahmed, A. Nonalcoholic Fatty Liver Disease: Epidemiology, Liver Transplantation Trends and Outcomes, and Risk of Recurrent Disease in the Graft. J. Clin. Transl. Hepatol. 2018, 6, 420–424. [Google Scholar] [CrossRef]
- Chalasani, N.; Younossi, Z.; Lavine, J.E.; Charlton, M.; Cusi, K.; Rinella, M.; Harrison, S.A.; Brunt, E.M.; Sanyal, A.J. The diagnosis and management of nonalcoholic fatty liver disease: Practice guidance from the American Association for the Study of Liver Diseases. Hepatology 2018, 67, 328–357. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Mingolla, L.; Rigolon, R.; Pichiri, I.; Cavalieri, V.; Zoppini, G.; Lippi, G.; Bonora, E.; Targher, G. Nonalcoholic fatty liver disease is independently associated with an increased incidence of cardiovascular disease in adult patients with type 1 diabetes. Int. J. Cardiol. 2016, 225, 387–391. [Google Scholar] [CrossRef] [PubMed]
- Tilg, H.; Moschen, A.R.; Roden, M. NAFLD and diabetes mellitus. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 32–42. [Google Scholar] [CrossRef]
- Romeo, S.; Kozlitina, J.; Xing, C.; Pertsemlidis, A.; Cox, D.; Pennacchio, L.A.; Boerwinkle, E.; Cohen, J.C.; Hobbs, H.H. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 2008, 40, 1461–1465. [Google Scholar] [CrossRef]
- Eslam, M.; Newsome, P.N.; Sarin, S.K.; Anstee, Q.M.; Targher, G.; Romero-Gomez, M.; Zelber-Sagi, S.; Wai-Sun Wong, V.; Dufour, J.F.; Schattenberg, J.M.; et al. A new definition for metabolic dysfunction-associated fatty liver disease: An international expert consensus statement. J. Hepatol. 2020, 73, 202–209. [Google Scholar] [CrossRef]
- Ratziu, V.; Rinella, M.; Beuers, U.; Loomba, R.; Anstee, Q.M.; Harrison, S.; Francque, S.; Sanyal, A.; Newsome, P.N.; Younossi, Z. The times they are a-changin’ (for NAFLD as well). J. Hepatol. 2020, 73, 1307–1309. [Google Scholar] [CrossRef]
- Kim, D.; Ahmed, A. Reply to: “NAFLD vs. MAFLD-It is not the name but the disease that decides the outcome in fatty liver”. J. Hepatol. 2022, 76, 477–478. [Google Scholar] [CrossRef]
- Day, C.P.; James, O.F. Steatohepatitis: A tale of two “hits”? Gastroenterology 1998, 114, 842–845. [Google Scholar] [CrossRef]
- Machado, M.V.; Diehl, A.M. Pathogenesis of Nonalcoholic Steatohepatitis. Gastroenterology 2016, 150, 1769–1777. [Google Scholar] [CrossRef]
- Buzzetti, E.; Pinzani, M.; Tsochatzis, E.A. The multiple-hit pathogenesis of non-alcoholic fatty liver disease (NAFLD). Metabolism 2016, 65, 1038–1048. [Google Scholar] [CrossRef]
- Tilg, H.; Moschen, A.R. Evolution of inflammation in nonalcoholic fatty liver disease: The multiple parallel hits hypothesis. Hepatology 2010, 52, 1836–1846. [Google Scholar] [CrossRef]
- Loomba, R.; Friedman, S.L.; Shulman, G.I. Mechanisms and disease consequences of nonalcoholic fatty liver disease. Cell 2021, 184, 2537–2564. [Google Scholar] [CrossRef]
- Ratziu, V. Starting the battle to control non-alcoholic steatohepatitis. Lancet 2015, 385, 922–924. [Google Scholar] [CrossRef]
- Sanyal, A.J.; Chalasani, N.; Kowdley, K.V.; McCullough, A.; Diehl, A.M.; Bass, N.M.; Neuschwander-Tetri, B.A.; Lavine, J.E.; Tonascia, J.; Unalp, A.; et al. Pioglitazone, vitamin E, or placebo for nonalcoholic steatohepatitis. N. Engl. J. Med. 2010, 362, 1675–1685. [Google Scholar] [CrossRef]
- Braissant, O.; Foufelle, F.; Scotto, C.; Dauça, M.; Wahli, W. Differential expression of peroxisome proliferator-activated receptors (PPARs): Tissue distribution of PPAR-alpha, -beta, and -gamma in the adult rat. Endocrinology 1996, 137, 354–366. [Google Scholar] [CrossRef]
- Tontonoz, P.; Hu, E.; Graves, R.A.; Budavari, A.I.; Spiegelman, B.M. mPPAR gamma 2: Tissue-specific regulator of an adipocyte enhancer. Genes Dev. 1994, 10, 1224–1234. [Google Scholar] [CrossRef]
- Greene, M.E.; Blumberg, B.; McBride, O.W.; Yi, H.F.; Kronquist, K.; Kwan, K.; Hsieh, L.; Greene, G.; Nimer, S.D. Isolation of the human peroxisome proliferator activated receptor gamma cDNA: Expression in hematopoietic cells and chromosomal mapping. Gene Expr. 1995, 4, 281–299. [Google Scholar]
- Fajas, L.; Auboeuf, D.; Raspé, E.; Schoonjans, K.; Lefebvre, A.M.; Saladin, R.; Najib, J.; Laville, M.; Fruchart, J.C.; Deeb, S.; et al. The organization, promoter analysis, and expression of the human PPARgamma gene. J. Biol. Chem. 1997, 272, 18779–18789. [Google Scholar] [CrossRef]
- Hernandez-Quiles, M.; Broekema, M.F.; Kalkhoven, E. PPARgamma in Metabolism, Immunity, and Cancer: Unified and Diverse Mechanisms of Action. Front. Endocrinol. 2021, 12, 624112. [Google Scholar] [CrossRef] [PubMed]
- Tontonoz, P.; Spiegelman, B.M. Fat and beyond: The diverse biology of PPARgamma. Annu. Rev. Biochem. 2008, 77, 289–312. [Google Scholar] [CrossRef] [PubMed]
- Lehrke, M.; Lazar, M.A. The many faces of PPARgamma. Cell 2005, 123, 993–999. [Google Scholar] [CrossRef] [PubMed]
- Chawla, A.; Schwarz, E.J.; Dimaculangan, D.D.; Lazar, M.A. Peroxisome proliferator-activated receptor (PPAR) gamma: Adipose-predominant expression and induction early in adipocyte differentiation. Endocrinology 1994, 135, 798–800. [Google Scholar] [CrossRef]
- Vidal-Puig, A.; Jimenez-Liñan, M.; Lowell, B.B.; Hamann, A.; Hu, E.; Spiegelman, B.; Flier, J.S.; Moller, D.E. Regulation of PPAR gamma gene expression by nutrition and obesity in rodents. J. Clin. Investig. 1996, 97, 2553–2561. [Google Scholar] [CrossRef]
- Chen, Y.; Jimenez, A.R.; Medh, J.D. Identification and regulation of novel PPAR-gamma splice variants in human THP-1 macrophages. Biochim. Biophys. Acta 2006, 1759, 32–43. [Google Scholar] [CrossRef]
- Wu, C.W.; Chu, E.S.; Lam, C.N.; Cheng, A.S.; Lee, C.W.; Wong, V.W.; Sung, J.J.; Yu, J. PPARgamma is essential for protection against nonalcoholic steatohepatitis. Gene Ther. 2010, 17, 790–798. [Google Scholar] [CrossRef]
- Pettinelli, P.; Videla, L.A. Up-Regulation of PPAR-γ mRNA Expression in the Liver of Obese Patients: An Additional Reinforcing Lipogenic Mechanism to SREBP-1c Induction. J. Clin. Endocrinol. Metab. 2011, 96, 1424–1430. [Google Scholar] [CrossRef]
- Lima-Cabello, E.; García-Mediavilla, M.V.; Miquilena-Colina, M.E.; Vargas-Castrillón, J.; Lozano-Rodríguez, T.; Fernández-Bermejo, M.; Olcoz, J.L.; González-Gallego, J.; García-Monzón, C.; Sánchez-Campos, S. Enhanced expression of pro-inflammatory mediators and liver X-receptor-regulated lipogenic genes in non-alcoholic fatty liver disease and hepatitis C. Clin. Sci. 2011, 120, 239–250. [Google Scholar] [CrossRef]
- Yamazaki, T.; Shiraishi, S.; Kishimoto, K.; Miura, S.; Ezaki, O. An increase in liver PPARγ2 is an initial event to induce fatty liver in response to a diet high in butter: PPARγ2 knockdown improves fatty liver induced by high-saturated fat. J. Nutr. Biochem. 2011, 22, 543–553. [Google Scholar] [CrossRef]
- Lutchman, G.; Promrat, K.; Kleiner, D.E.; Heller, T.; Ghany, M.G.; Yanovski, J.A.; Liang, T.J.; Hoofnagle, J.H. Changes in serum adipokine levels during pioglitazone treatment for nonalcoholic steatohepatitis: Relationship to histological improvement. Clin. Gastroenterol. Hepatol. 2006, 4, 1048–1052. [Google Scholar] [CrossRef]
- Ratziu, V.; Giral, P.; Jacqueminet, S.; Charlotte, F.; Hartemann-Heurtier, A.; Serfaty, L.; Podevin, P.; Lacorte, J.M.; Bernhardt, C.; Bruckert, E.; et al. Rosiglitazone for nonalcoholic steatohepatitis: One-year results of the randomized placebo-controlled Fatty Liver Improvement with Rosiglitazone Therapy (FLIRT) Trial. Gastroenterology 2008, 135, 100–110. [Google Scholar] [CrossRef]
- Berger, S.L.; Kouzarides, T.; Shiekhattar, R.; Shilatifard, A. An operational definition of epigenetics. Genes Dev. 2009, 23, 781–783. [Google Scholar] [CrossRef]
- Foley, D.L.; Craig, J.M.; Morley, R.; Olsson, C.A.; Dwyer, T.; Smith, K.; Saffery, R. Prospects for epigenetic epidemiology. Am. J. Epidemiol. 2009, 169, 389–400. [Google Scholar] [CrossRef]
- Eslam, M.; Valenti, L.; Romeo, S. Genetics and epigenetics of NAFLD and NASH: Clinical impact. J. Hepatol. 2018, 68, 268–279. [Google Scholar] [CrossRef]
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef]
- Baylin, S.B.; Jones, P.A. Epigenetic Determinants of Cancer. Cold Spring Harb. Perspect. Biol. 2016, 8, a019505. [Google Scholar] [CrossRef]
- Hyun, J.; Jung, Y. DNA Methylation in Nonalcoholic Fatty Liver Disease. Int. J. Mol. Sci. 2020, 21, 8138. [Google Scholar] [CrossRef]
- Zaiou, M.; Amrani, R.; Rihn, B.; Hajri, T. Dietary Patterns Influence Target Gene Expression through Emerging Epigenetic Mechanisms in Nonalcoholic Fatty Liver Disease. Biomedicines 2021, 9, 1256. [Google Scholar] [CrossRef]
- Tian, Y.; Wong, V.W.-S.; Chan, H.L.-Y.; Cheng, A.S.-L. Epigenetic regulation of hepatocellular carcinoma in non-alcoholic fatty liver disease. Semin. Cancer Biol. 2013, 23, 471–482. [Google Scholar] [CrossRef]
- Ahrens, M.; Ammerpohl, O.; von Schönfels, W.; Kolarova, J.; Bens, S.; Itzel, T.; Teufel, A.; Herrmann, A.; Brosch, M.; Hinrichsen, H.; et al. DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metab. 2013, 18, 296–302. [Google Scholar] [CrossRef] [PubMed]
- De Mello, V.D.; Matte, A.; Perfilyev, A.; Männistö, V.; Rönn, T.; Nilsson, E.; Käkelä, P.; Ling, C.; Pihlajamäki, J. Human liver epigenetic alterations in non-alcoholic steatohepatitis are related to insulin action. Epigenetics 2017, 12, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Sookoian, S.; Rosselli, M.S.; Gemma, C.; Burgueño, A.L.; Fernández Gianotti, T.; Castaño, G.O.; Pirola, C.J. Epigenetic regulation of insulin resistance in nonalcoholic fatty liver disease: Impact of liver methylation of the peroxisome proliferator–activated receptor γ coactivator 1α promoter. Hepatology 2010, 52, 1992–2000. [Google Scholar] [CrossRef] [PubMed]
- Zeybel, M.; Hardy, T.; Wong, Y.K.; Mathers, J.C.; Fox, C.R.; Gackowska, A.; Oakley, F.; Burt, A.D.; Wilson, C.L.; Anstee, Q.M.; et al. Multigenerational epigenetic adaptation of the hepatic wound-healing response. Nat. Med. 2012, 18, 1369–1377. [Google Scholar] [CrossRef]
- Zeybel, M.; Hardy, T.; Robinson, S.M.; Fox, C.; Anstee, Q.M.; Ness, T.; Masson, S.; Mathers, J.C.; French, J.; White, S.; et al. Differential DNA methylation of genes involved in fibrosis progression in non-alcoholic fatty liver disease and alcoholic liver disease. Clin. Epigenet. 2015, 7, 25. [Google Scholar] [CrossRef]
- Yigit, B.; Boyle, M.; Ozler, O.; Erden, N.; Tutucu, F.; Hardy, T.; Bergmann, C.; Distler, J.H.W.; Adali, G.; Dayangac, M.; et al. Plasma cell-free DNA methylation: A liquid biomarker of hepatic fibrosis. Gut 2018, 67, 1907–1908. [Google Scholar] [CrossRef]
- Hajri, T.; Zaiou, M.; Fungwe, T.V.; Ouguerram, K.; Besong, S. Epigenetic Regulation of Peroxisome Proliferator-Activated Receptor Gamma Mediates High-Fat Diet-Induced Non-Alcoholic Fatty Liver Disease. Cells 2021, 10, 1355. [Google Scholar] [CrossRef]
- Hardy, T.; Zeybel, M.; Day, C.P.; Dipper, C.; Masson, S.; McPherson, S.; Henderson, E.; Tiniakos, D.; White, S.; French, J.; et al. Plasma DNA methylation: A potential biomarker for stratification of liver fibrosis in non-alcoholic fatty liver disease. Gut 2017, 66, 1321–1328. [Google Scholar] [CrossRef]
- Laird, A.; Thomson, J.P.; Harrison, D.J.; Meehan, R.R. 5-hydroxymethylcytosine profiling as an indicator of cellular state. Epigenomics 2013, 5, 655–669. [Google Scholar] [CrossRef]
- Pirola, C.J.; Scian, R.; Gianotti, T.F.; Dopazo, H.; Rohr, C.; Martino, J.S.; Castaño, G.O.; Sookoian, S. Epigenetic Modifications in the Biology of Nonalcoholic Fatty Liver Disease: The Role of DNA Hydroxymethylation and TET Proteins. Medicine 2015, 94, e1480. [Google Scholar] [CrossRef]
- Tong, J.; Han, C.J.; Zhang, J.Z.; He, W.Z.; Zhao, G.J.; Cheng, X.; Zhang, L.; Deng, K.Q.; Liu, Y.; Fan, H.F.; et al. Hepatic Interferon Regulatory Factor 6 Alleviates Liver Steatosis and Metabolic Disorder by Transcriptionally Suppressing Peroxisome Proliferator-Activated Receptor γ in Mice. Hepatology 2019, 69, 2471–2488. [Google Scholar] [CrossRef]
- Lee, J.; Song, J.H.; Park, J.H.; Chung, M.Y.; Lee, S.H.; Jeon, S.B.; Park, S.H.; Hwang, J.T.; Choi, H.K. Dnmt1/Tet2-mediated changes in Cmip methylation regulate the development of nonalcoholic fatty liver disease by controlling the Gbp2-Pparγ-CD36 axis. Exp. Mol. Med. 2023, 55, 143–157. [Google Scholar] [CrossRef]
- Jenuwein, T.; Allis, C.D. Translating the Histone Code. Science 2001, 293, 1074–1080. [Google Scholar] [CrossRef]
- Lee, J.; Kim, Y.; Friso, S.; Choi, S.W. Epigenetics in non-alcoholic fatty liver disease. Mol. Asp. Med. 2017, 54, 78–88. [Google Scholar] [CrossRef]
- Ling, C.; Groop, L. Epigenetics: A molecular link between environmental factors and type 2 diabetes. Diabetes 2009, 58, 2718–2725. [Google Scholar] [CrossRef]
- Kim, D.H.; Kim, J.; Kwon, J.S.; Sandhu, J.; Tontonoz, P.; Lee, S.K.; Lee, S.; Lee, J.W. Critical Roles of the Histone Methyltransferase MLL4/KMT2D in Murine Hepatic Steatosis Directed by ABL1 and PPARγ2. Cell Rep. 2016, 17, 1671–1682. [Google Scholar] [CrossRef]
- Jun, H.J.; Kim, J.; Hoang, M.H.; Lee, S.J. Hepatic lipid accumulation alters global histone h3 lysine 9 and 4 trimethylation in the peroxisome proliferator-activated receptor alpha network. PLoS ONE 2012, 7, e44345. [Google Scholar] [CrossRef]
- Fan, Z.; Li, L.; Li, M.; Zhang, X.; Hao, C.; Yu, L.; Zeng, S.; Xu, H.; Fang, M.; Shen, A.; et al. The histone methyltransferase Suv39h2 contributes to nonalcoholic steatohepatitis in mice. Hepatology 2017, 65, 1904–1919. [Google Scholar] [CrossRef]
- Mosammaparast, N.; Shi, Y. Reversal of histone methylation: Biochemical and molecular mechanisms of histone demethylases. Annu. Rev. Biochem. 2010, 79, 155–179. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, S.; Zhao, Y.; Lin, C.; Zhong, F.; Jin, L.; He, F.; Wang, H. Histone H3K9 demethylase JMJD1A modulates hepatic stellate cells activation and liver fibrosis by epigenetically regulating peroxisome proliferator-activated receptor γ. FASEB J. 2015, 29, 1830–1841. [Google Scholar] [CrossRef]
- Kim, J.H.; Jung, D.Y.; Nagappan, A.; Jung, M.H. Histone H3K9 demethylase JMJD2B induces hepatic steatosis through upregulation of PPARγ2. Sci. Rep. 2018, 8, 13734. [Google Scholar] [CrossRef]
- Colak, Y.; Yesil, A.; Mutlu, H.H.; Caklili, O.T.; Ulasoglu, C.; Senates, E.; Takir, M.; Kostek, O.; Yilmaz, Y.; Yilmaz Enc, F.; et al. A potential treatment of non-alcoholic fatty liver disease with SIRT1 activators. J. Gastrointestin. Liver Dis. 2014, 23, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Bricambert, J.; Miranda, J.; Benhamed, F.; Girard, J.; Postic, C.; Dentin, R. Salt-inducible kinase 2 links transcriptional coactivator p300 phosphorylation to the prevention of ChREBP-dependent hepatic steatosis in mice. Clin. Investig. 2010, 120, 4316–4331. [Google Scholar] [CrossRef] [PubMed]
- Lefterova, M.I.; Zhang, Y.; Steger, D.J.; Schupp, M.; Schug, J.; Cristancho, A.; Feng, D.; Zhuo, D.; Stoeckert, C.J.; Liu, X.S.; et al. PPARγ and C/EBP Factors Orchestrate Adipocyte Biology via Adjacent Binding on a Genome-Wide Scale. Genes Dev. 2008, 22, 2941–2952. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, T.S.; Xu, Z.; Zhang, X.; Wang, L.; Gimble, J.M.; Lander, E.S.; Rosen, E.D. Comparative epigenomic analysis of murine and human adipogenesis. Cell 2010, 143, 156–169. [Google Scholar] [CrossRef]
- Kraakman, M.J.; Liu, Q.; Postigo-Fernandez, J.; Ji, R.; Kon, N.; Larrea, D.; Namwanje, M.; Fan, L.; Chan, M.; Area-Gomez, E.; et al. PPARγ deacetylation dissociates thiazolidinedione’s metabolic benefits from its adverse effects. J. Clin. Investig. 2018, 128, 2600–2612. [Google Scholar] [CrossRef]
- Zahr, T.; Liu, L.; Chan, M.; Zhou, Q.; Cai, B.; He, Y.; Aaron, N.; Accili, D.; Sun, L.; Qiang, L. PPARγ (Peroxisome Proliferator-Activated Receptor γ) Deacetylation Suppresses Aging-Associated Atherosclerosis and Hypercholesterolemia. Arterioscler. Thromb. Vasc. Biol. 2023, 43, 30–44. [Google Scholar] [CrossRef]
- Liu, L.; Fan, L.; Chan, M.; Kraakman, M.J.; Yang, J.; Fan, Y.; Aaron, N.; Wan, Q.; Carrillo-Sepulveda, M.A.; Tall, A.R.; et al. PPARγ Deacetylation Confers the Antiatherogenic Effect and Improves Endothelial Function in Diabetes Treatment. Diabetes 2020, 69, 1793–1803. [Google Scholar] [CrossRef]
- Rodrigues, B.; McNeill, J.H. Cardiac dysfunction in isolated perfused hearts from spontaneously diabetic BB rats. Can. J. Physiol. Pharmacol. 1990, 68, 514–518. [Google Scholar] [CrossRef]
- Emmett, M.J.; Lazar, M.A. Integrative regulation of physiology by histone deacetylase 3. Nat. Rev. Mol. Cell. Biol. 2019, 20, 102–115. [Google Scholar] [CrossRef]
- Knutson, S.K.; Chyla, B.J.; Amann, J.M.; Bhaskara, S.; Huppert, S.S.; Hiebert, S.W. Liver-specific deletion of histone deacetylase 3 disrupts metabolic transcriptional networks. EMBO J. 2008, 27, 1017–1028. [Google Scholar] [CrossRef]
- Ferrari, A.; Longo, R.; Fiorino, E.; Silva, R.; Mitro, N.; Cermenati, G.; Gilardi, F.; Desvergne, B.; Andolfo, A.; Magagnotti, C.; et al. HDAC3 is a molecular brake of the metabolic switch supporting white adipose tissue browning. Nat. Commun. 2017, 8, 93. [Google Scholar] [CrossRef]
- Whitt, J.; Woo, V.; Lee, P.; Moncivaiz, J.; Haberman, Y.; Denson, L.; Tso, P.; Alenghat, T. Disruption of Epithelial HDAC3 in Intestine Prevents Diet-Induced Obesity in Mice. Gastroenterology 2018, 155, 501–513. [Google Scholar] [CrossRef]
- Li, D.; Wang, X.; Ren, W.; Ren, J.; Lan, X.; Wang, F.; Li, H.; Zhang, F.; Han, Y.; Song, T.; et al. High expression of liver histone deacetylase 3 contributes to high-fat-diet-induced metabolic syndrome by suppressing the PPAR-γ and LXR-α-pathways in E3 rats. Mol. Cell. Endocrinol. 2011, 344, 69–80. [Google Scholar] [CrossRef]
- Jiang, X.; Ye, X.; Guo, W.; Lu, H.; Gao, Z. Inhibition of HDAC3 promotes ligand-independent PPARγ activation by protein acetylation. J. Mol. Endocrinol. 2014, 53, 191–200. [Google Scholar] [CrossRef]
- Ding, R.-B.; Bao, J.; Deng, C.-X. Emerging roles of SIRT1 in fatty liver diseases. Int. J. Biol. Sci. 2017, 13, 852–867. [Google Scholar] [CrossRef]
- Cao, Y.; Xue, Y.; Xue, L.; Jiang, X.; Wang, X.; Zhang, Z.; Yang, J.; Lu, J.; Zhang, C.; Wang, W.; et al. Hepatic menin recruits SIRT1 to control liver steatosis through histone deacetylation. J. Hepatol. 2013, 59, 1299–1306. [Google Scholar] [CrossRef]
- de Gregorio, E.; Colell, A.; Morales, A.; Marí, M. Relevance of SIRT1-NF-κB Axis as Therapeutic Target to Ameliorate Inflammation in Liver Disease. Int. J. Mol. Sci. 2020, 21, 3858. [Google Scholar] [CrossRef]
- Vilà, L.; Elias, I.; Roca, C.; Ribera, A.; Ferré, T.; Casellas, A.; Lage, R.; Franckhauser, S.; Bosch, F. AAV8-mediated Sirt1 gene transfer to the liver prevents high carbohydrate diet-induced nonalcoholic fatty liver disease. Molecular therapy. Mol. Ther. Methods Clin. Dev. 2014, 1, 14039. [Google Scholar] [CrossRef]
- Purushotham, A.; Schug, T.T.; Xu, Q.; Surapureddi, S.; Guo, X.; Li, X. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 2009, 9, 327–338. [Google Scholar] [CrossRef]
- Deng, X.Q.; Chen, L.L.; Li, N.X. The expression of SIRT1 in nonalcoholic fatty liver disease induced by high-fat diet in rats. Liver Int. 2007, 27, 708–715. [Google Scholar] [CrossRef] [PubMed]
- Mariani, S.; Fiore, D.; Basciani, S.; Persichetti, A.; Contini, S.; Lubrano, C.; Salvatori, L.; Lenzi, A.; Gnessi, L. Plasma levels of SIRT1 associate with non-alcoholic fatty liver disease in obese patients. Endocrine 2015, 49, 711–716. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Hu, M.; Liang, X.; Ajmo, J.M.; Li, X.; Bataller, R.; Odena, G.; Stevens, S.M., Jr.; You, M. Deletion of SIRT1 from hepatocytes in mice disrupts lipin-1 signaling and aggravates alcoholic fatty liver. Gastroenterology 2014, 146, 801–811. [Google Scholar] [CrossRef] [PubMed]
- Mayoral, R.; Osborn, O.; McNelis, J.; Johnson, A.M.; Oh, D.Y.; Izquierdo, C.L.; Chung, H.; Li, P.; Traves, P.G.; Bandyopadhyay, G.; et al. Adipocyte SIRT1 knockout promotes PPARγ activity, adipogenesis and insulin sensitivity in chronic-HFD and obesity. Mol. Metab. 2015, 4, 378–391. [Google Scholar] [CrossRef]
- Li, P.; Ruan, X.; Yang, L.; Kiesewetter, K.; Zhao, Y.; Luo, H.; Chen, Y.; Gucek, M.; Zhu, J.; Cao, H. A Liver-Enriched Long Non-Coding RNA, lncLSTR, Regulates Systemic Lipid Metabolism in Mice. Cell Metab. 2015, 21, 455–467. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, C.; Barbier, O.; Smalling, R.; Tsuchiya, H.; Lee, S.; Delker, D.; Zou, A.; Hagedorn, C.H.; Wang, L. Bcl2 is a critical regulator of bile acid homeostasis by dictating Shp and lncRNA H19 function. Sci. Rep. 2016, 6, 20559. [Google Scholar] [CrossRef]
- Zaiou, M. Noncoding RNAs as additional mediators of epigenetic regulation in nonalcoholic fatty liver disease. World J. Gastroenterol. 2022, 28, 5111–5128. [Google Scholar] [CrossRef]
- Liu, C.H.; Ampuero, J.; Gil-Gómez, A.; Montero-Vallejo, R.; Rojas, Á.; Muñoz-Hernández, R.; Gallego-Durán, R.; Romero-Gómez, M. miRNAs in patients with non-alcoholic fatty liver disease: A systematic review and meta-analysis. J. Hepatol. 2018, 69, 1335–1348. [Google Scholar] [CrossRef]
- Nobili, V.; Alisi, A.; Valenti, L.; Miele, L.; Feldstein, A.E.; Alkhouri, N. NAFLD in children: New genes, new diagnostic modalities and new drugs. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 517–530. [Google Scholar] [CrossRef]
- Zhu, M.; Wang, Q.; Zhou, W.; Liu, T.; Yang, L.; Zheng, P.; Zhang, L.; Ji, G. Integrated analysis of hepatic mRNA and miRNA profiles identified molecular networks and potential biomarkers of NAFLD. Sci. Rep. 2018, 8, 7628. [Google Scholar] [CrossRef]
- Lv, X.; Yan, J.; Jiang, J.; Zhou, X.; Lu, Y.; Jiang, H. MicroRNA-27a-3p suppression of peroxisome proliferator-activated receptor-γ contributes to cognitive impairments resulting from sevoflurane treatment. J. Neurochem. 2017, 143, 306–319. [Google Scholar] [CrossRef]
- Baffy, G. MicroRNAs in Nonalcoholic Fatty Liver Disease. J. Clin. Med. 2015, 4, 1977–1988. [Google Scholar] [CrossRef]
- Wu, H.; Ng, R.; Chen, X.; Steer, C.J.; Song, G. MicroRNA-21 is a potential link between non-alcoholic fatty liver disease and hepatocellular carcinoma via modulation of the HBP1-p53-Srebp1c pathway. Gut 2016, 65, 1850–1860. [Google Scholar] [CrossRef]
- Sun, C.; Huang, F.; Liu, X.; Xiao, X.; Yang, M.; Hu, G.; Liu, H.; Liao, L. miR-21 regulates triglyceride and cholesterol metabolism in non-alcoholic fatty liver disease by targeting HMGCR. Int. J. Mol. Med. 2015, 35, 847–853. [Google Scholar] [CrossRef]
- Loyer, X.; Paradis, V.; Hénique, C.; Vion, A.-C.; Colnot, N.; Guerin, C.L.; Devue, C.; On, S.; Scetbun, J.; Romain, M.; et al. Liver microRNA-21 is overexpressed in non-alcoholic steatohepatitis and contributes to the disease in experimental models by inhibiting PPARα expression. Gut 2015, 65, 1882–1894. [Google Scholar] [CrossRef]
- Benhamouche-Trouillet, S.; Postic, C. Emerging role of miR-21 in non-alcoholic fatty liver disease. Gut 2016, 65, 1781–1783. [Google Scholar] [CrossRef]
- Pillai, S.S.; Lakhani, H.V.; Zehra, M.; Wang, J.; Dilip, A.; Puri, N.; O’Hanlon, K.; Sodhi, K. Predicting Nonalcoholic Fatty Liver Disease through a Panel of Plasma Biomarkers and MicroRNAs in Female West Virginia Population. Int. J. Mol. Sci. 2020, 21, 6698. [Google Scholar] [CrossRef]
- Liu, J.; Xiao, Y.; Wu, X.; Jiang, L.; Yang, S.; Ding, Z.; Fang, Z.; Hua, H.; Kirby, M.S.; Shou, J. A circulating microRNA signature as noninvasive diagnostic and prognostic biomarkers for nonalcoholic steatohepatitis. BMC Genom. 2018, 19, 188. [Google Scholar] [CrossRef]
- Rodrigues, P.M.; Afonso, M.B.; Simão, A.L.; Carvalho, C.C.; Trindade, A.; Duarte, A.; Borralho, P.M.; Machado, M.V.; Cortez-Pinto, H.; Rodrigues, C.M.; et al. miR-21 ablation and obeticholic acid ameliorate nonalcoholic steatohepatitis in mice. Cell Death Dis. 2017, 8, e2825. [Google Scholar] [CrossRef]
- Almohawes, Z.N.; El-Kott, A.; Morsy, K.; Shati, A.A.; El-Kenawy, A.E.; Khalifa, H.S.; Elsaid, F.G.; Abd-Lateif, A.M.; Abu-Zaiton, A.; Ebealy, E.R.; et al. Salidroside inhibits insulin resistance and hepatic steatosis by downregulating miR-21 and subsequent activation of AMPK and upregulation of PPARα in the liver and muscles of high fat diet-fed rats. Arch. Physiol. Biochem. 2022, 1–18. [Google Scholar] [CrossRef]
- Zhang, X.; Deng, F.; Zhang, Y.; Zhang, X.; Chen, J.; Jiang, Y. PPARγ attenuates hepatic inflammation and oxidative stress of non-alcoholic steatohepatitis via modulating the miR-21-5p/SFRP5 pathway. Mol. Med. Rep. 2021, 24, 823. [Google Scholar] [CrossRef] [PubMed]
- Shirasaki, T.; Honda, M.; Shimakami, T.; Horii, R.; Yamashita, T.; Sakai, Y.; Sakai, A.; Okada, H.; Watanabe, R.; Murakami, S.; et al. MicroRNA-27a Regulates Lipid Metabolism and Inhibits Hepatitis C Virus Replication in Human Hepatoma Cells. J. Virol. 2013, 87, 5270–5286. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Kim, A.Y.; Lee, H.W.; Son, Y.H.; Lee, G.Y.; Lee, J.W.; Lee, Y.K.; Kim, J.B. miR-27a is a negative regulator of adipocyte differentiation via suppressing PPARgamma expression. Biochem. Biophys. Res. Commun. 2010, 392, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Sun, W.; Zhou, M.; Tang, Y. MicroRNA-27a regulates hepatic lipid metabolism and alleviates NAFLD via repressing FAS and SCD1. Sci. Rep. 2017, 7, 14493. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Ge, G.; Pan, T.; Wen, D.; Gan, J. A pilot study of serum microRNAs panel as potential biomarkers for diagnosis of nonalcoholic fatty liver disease. PLoS ONE 2014, 9, e105192. [Google Scholar] [CrossRef]
- Gerhard, G.S. Micro RNAs in the development of non-alcoholic fatty liver disease. World J. Hepatol. 2015, 7, 226–234. [Google Scholar] [CrossRef]
- Zhang, J.; Powell, C.; Kay, M.; Sonkar, R.; Meruvu, S.; Choudhury, M. Effect of Chronic Western Diets on Non-Alcoholic Fatty Liver of Male Mice Modifying the PPAR-γ Pathway via miR-27b-5p Regulation. Int. J. Mol. Sci. 2021, 22, 1822. [Google Scholar] [CrossRef]
- Zhu, D.; Lyu, L.; Shen, P.; Wang, J.; Chen, J.; Sun, X.; Chen, L.; Zhang, L.; Zhou, Q.; Duan, Y. rSjP40 protein promotes PPARγ expression in LX-2 cells through microRNA-27b. FASEB J. 2018, 32, 4798–4803. [Google Scholar] [CrossRef]
- Jennewein, C.; von Knethen, A.; Schmid, T.; Brüne, B. MicroRNA-27b contributes to lipopolysaccharide-mediated peroxisome proliferator-activated receptor gamma (PPARgamma) mRNA destabilization. J. Biol. Chem. 2010, 285, 11846–11853. [Google Scholar] [CrossRef]
- Hsu, C.C.; Lai, C.Y.; Lin, C.Y.; Yeh, K.Y.; Her, G.M. MicroRNA-27b Depletion Enhances Endotrophic and Intravascular Lipid Accumulation and Induces Adipocyte Hyperplasia in Zebrafish. Int. J. Mol. Sci. 2017, 19, 93. [Google Scholar] [CrossRef]
- Li, S.; Li, J.; Fei, B.Y.; Shao, D.; Pan, Y.; Mo, Z.H.; Sun, B.Z.; Zhang, D.; Zheng, X.; Zhang, M.; et al. MiR-27a promotes hepatocellular carcinoma cell proliferation through suppression of its target gene peroxisome proliferator-activated receptor γ Chin. Med. J. 2015, 128, 941–947. [Google Scholar] [CrossRef]
- Zheng, X.; Zhang, F.; Zhao, Y.; Zhang, J.; Dawulieti, J.; Pan, Y.; Cui, L.; Sun, M.; Shao, D.; Li, M.; et al. Self-assembled dual fluorescence nanoparticles for CD44-targeted delivery of anti-miR-27a in liver cancer theranostics. Theranostics 2018, 8, 3808–3823. [Google Scholar] [CrossRef]
- Zarfeshani, A.; Ngo, S.; Sheppard, A.M. MicroRNA Expression Relating to Dietary-Induced Liver Steatosis and NASH. J. Clin. Med. 2015, 4, 1938–1950. [Google Scholar] [CrossRef]
- Castro, R.E.; Ferreira, D.M.; Afonso, M.B.; Borralho, P.M.; Machado, M.V.; Cortez-Pinto, H.; Rodrigues, C.M. miR-34a/SIRT1/p53 is suppressed by ursodeoxycholic acid in the rat liver and activated by disease severity in human non-alcoholic fatty liver disease. J. Hepatol. 2013, 58, 119–125. [Google Scholar] [CrossRef]
- Meng, F.; Glaser, S.S.; Francis, H.; Yang, F.; Han, Y.; Stokes, A.; Staloch, D.; McCarra, J.; Liu, J.; Venter, J.; et al. Epigenetic regulation of miR-34a expression in alcoholic liver injury. Am. J. Pathol. 2012, 181, 804–817. [Google Scholar] [CrossRef]
- Cermelli, S.; Ruggieri, A.; Marrero, J.A.; Ioannou, G.N.; Beretta, L. Circulating MicroRNAs in Patients with Chronic Hepatitis C and Non-Alcoholic Fatty Liver Disease. PLoS ONE 2011, 6, e23937. [Google Scholar] [CrossRef]
- Salvoza, N.C.; Klinzing, D.C.; Gopez-Cervantes, J.; Baclig, M.O. Association of Circulating Serum miR-34a and miR-122 with Dyslipidemia among Patients with Non-Alcoholic Fatty Liver Disease. PLoS ONE 2016, 11, e0153497. [Google Scholar] [CrossRef]
- Braza-Boïls, A.; Marí-Alexandre, J.; Molina, P.; Arnau, M.A.; Barceló-Molina, M.; Domingo, D.; Girbes, J.; Giner, J.; Martínez-Dolz, L.; Zorio, E. Deregulated hepatic microRNAs underlie the association between non-alcoholic fatty liver disease and coronary artery disease. Liver Int. 2016, 36, 1221–1229. [Google Scholar] [CrossRef]
- Ding, J.; Li, M.; Wan, X.; Jin, X.; Chen, S.; Yu, C.; Li, Y. Effect of miR-34a in regulating steatosis by targeting PPARα expression in nonalcoholic fatty liver disease. Sci. Rep. 2015, 5, 13729. [Google Scholar] [CrossRef]
- Guo, X.-Y.; Sun, F.; Chen, J.-N.; Wang, Y.-Q.; Pan, Q.; Fan, J.-G. circRNA_0046366 inhibits hepatocellular steatosis by normalization of PPAR signaling. World J. Gastroenterol. 2018, 24, 323–337. [Google Scholar] [CrossRef]
- Derdak, Z.; Villegas, K.A.; Harb, R.; Wu, A.M.; Sousa, A.; Wands, J.R. Inhibition of p53 attenuates steatosis and liver injury in a mouse model of non-alcoholic fatty liver disease. J. Hepatol. 2013, 58, 785–791. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, Y.; Wu, S.; He, J.; Lou, L.; Ye, W.; Wang, J. microRNA-34a and microRNA-34c promote the activation of human hepatic stellate cells by targeting peroxisome proliferator-activated receptor γ. Mol. Med. Rep. 2015, 11, 1017–1024. [Google Scholar] [CrossRef] [PubMed]
- Estep, M.; Armistead, D.; Hossain, N.; Elarainy, H.; Goodman, Z.; Baranova, A.; Chandhoke, V.; Younossi, Z.M. Differential expression of miRNAs in the visceral adipose tissue of patients with non-alcoholic fatty liver disease. Aliment. Pharmacol. Ther. 2010, 32, 487–497. [Google Scholar] [CrossRef] [PubMed]
- Hanin, G.; Yayon, N.; Tzur, Y.; Haviv, R.; Bennett, E.R.; Udi, S.; Krishnamoorthy, Y.R.; Kotsiliti, E.; Zangen, R.; Efron, B.; et al. miRNA-132 induces hepatic steatosis and hyperlipidaemia by synergistic multitarget suppression. Gut 2018, 67, 1124–1134. [Google Scholar] [CrossRef] [PubMed]
- Mann, J.; Chu, D.C.; Maxwell, A.; Oakley, F.; Zhu, N.L.; Tsukamoto, H.; Mann, D.A. MeCP2 controls an epigenetic pathway that promotes myofibroblast transdifferentiation and fibrosis. Gastroenterology 2010, 138, 705–714. [Google Scholar] [CrossRef]
- Csak, T.; Bala, S.; Lippai, D.; Kodys, K.; Catalano, D.; Iracheta-Vellve, A.; Szabo, G. MicroRNA-155 Deficiency Attenuates Liver Steatosis and Fibrosis without Reducing Inflammation in a Mouse Model of Steatohepatitis. PLoS ONE 2015, 10, e0129251. [Google Scholar] [CrossRef]
- Miller, A.M.; Gilchrist, D.S.; Nijjar, J.; Araldi, E.; Ramírez, C.M.; Lavery, C.A.; Fernandez-Hernando, C.; McInnes, I.B.; Kurowska-Stolarska, M. MiR-155 Has a Protective Role in the Development of Non-Alcoholic Hepatosteatosis in Mice. PLoS ONE 2013, 8, e72324. [Google Scholar] [CrossRef]
- Bala, S.; Csak, T.; Saha, B.; Zatsiorsky, J.; Kodys, K.; Catalano, D.; Satishchandran, A.; Szabo, G. The pro-inflammatory effects of miR-155 promote liver fibrosis and alcohol-induced steatohepatitis. J. Hepatol. 2016, 64, 1378–1387. [Google Scholar] [CrossRef]
- Wang, Y.; Zheng, Z.J.; Jia, Y.J.; Yang, Y.L.; Xue, Y.M. Role of p53/miR-155-5p/sirt1 loop in renal tubular injury of diabetic kidney disease. J. Transl. Med. 2018, 16, 146. [Google Scholar] [CrossRef]
- Skårn, M.; Namløs, H.M.; Noordhuis, P.; Wang, M.-Y.; Meza-Zepeda, L.A.; Myklebost, O. Adipocyte Differentiation of Human Bone Marrow-Derived Stromal Cells Is Modulated by MicroRNA-155, MicroRNA-221, and MicroRNA-222. Stem Cells Dev. 2012, 21, 873–883. [Google Scholar] [CrossRef]
- Ying, W.; Riopel, M.; Bandyopadhyay, G.; Dong, Y.; Birmingham, A.; Seo, J.B.; Ofrecio, J.M.; Wollam, J.; Hernandez-Carretero, A.; Fu, W.; et al. Adipose Tissue Macrophage-Derived Exosomal miRNAs Can Modulate In Vivo and In Vitro Insulin Sensitivity. Cell 2017, 171, 372–384.e12. [Google Scholar] [CrossRef]
- Tryggestad, J.B.; Teague, A.M.; Sparling, D.P.; Jiang, S.; Chernausek, S.D. Macrophage-Derived MicroRNA-155 Increases in Obesity and Influences Adipocyte Metabolism by Targeting Peroxisome Proliferator-Activated Receptor Gamma. Obesity 2019, 27, 1856–1864. [Google Scholar] [CrossRef]
- Wang, D.R.; Wang, B.; Yang, M.; Liu, Z.L.; Sun, J.; Wang, Y.; Sun, H.; Xie, L.J. Suppression of miR-30a-3p Attenuates Hepatic Steatosis in Non-alcoholic Fatty Liver Disease. Biochem. Genet. 2020, 58, 691–704. [Google Scholar] [CrossRef]
- Mittal, S.; Inamdar, S.; Acharya, J.; Pekhale, K.; Kalamkar, S.; Boppana, R.; Ghaskadbi, S. miR-3666 inhibits development of hepatic steatosis by negatively regulating PPARγ. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2020, 1865, 158777. [Google Scholar] [CrossRef]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 1–17. [Google Scholar] [CrossRef]
- Huarte, M. The emerging role of lncRNAs in cancer. Nat. Med. 2015, 21, 1253–1261. [Google Scholar] [CrossRef]
- Sookoian, S.; Rohr, C.; Salatino, A.; Dopazo, H.; Fernandez Gianotti, T.; Castaño, G.O.; Pirola, C.J. Genetic variation in long noncoding RNAs and the risk of nonalcoholic fatty liver disease. Oncotarget 2017, 8, 22917–22926. [Google Scholar] [CrossRef]
- Lan, X.; Wu, L.; Wu, N.; Chen, Q.; Li, Y.; Du, X.; Wei, C.; Feng, L.; Li, Y.; Osoro, E.K.; et al. Long Noncoding RNA lnc-HC Regulates PPARγ-Mediated Hepatic Lipid Metabolism through miR-130b-3p. Mol. Ther. Nucleic Acids 2019, 18, 954–965. [Google Scholar] [CrossRef]
- Xu, B.M.; Xiao, L.; Kang, C.M.; Ding, L.; Guo, F.X.; Li, P.; Lu, Z.F.; Wu, Q.; Xu, Y.J.; Bai, H.L.; et al. LncRNA AC096664.3/PPAR-γ/ABCG1-dependent signal transduction pathway contributes to the regulation of cholesterol homeostasis. J. Cell. Biochem. 2019, 120, 13775–13782. [Google Scholar] [CrossRef]
- Liu, J.; Tang, T.; Wang, G.D.; Liu, B. LncRNA-H19 promotes hepatic lipogenesis by directly regulating miR-130a/PPARγ axis in non-alcoholic fatty liver disease. Biosci. Rep. 2019, 39, BSR20181722. [Google Scholar] [CrossRef]
- Wang, H.; Cao, Y.; Shu, L.; Zhu, Y.; Peng, Q.; Ran, L.; Wu, J.; Luo, Y.; Zuo, G.; Luo, J.; et al. Long non-coding RNA (lncRNA) H19 induces hepatic steatosis through activating MLXIPL and mTORC1 networks in hepatocytes. J. Cell. Mol. Med. 2020, 24, 1399–1412. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Yu, D.; Nian, X.; Liu, J.; Koenig, R.J.; Xu, B.; Sheng, L. LncRNA SRA promotes hepatic steatosis through repressing the expression of adipose triglyceride lipase (ATGL). Sci. Rep. 2016, 6, 35531. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.X.; Li, B.Z.; Mitchell, L.A.; Wu, Y.; Qi, X.; Jin, Z.; Jia, B.; Wang, X.; Zeng, B.X.; Liu, H.M.; et al. “Perfect” designer chromosome V and behavior of a ring derivative. Science 2017, 355, eaaf4704. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Gerin, I.; Miao, H.; Vu-Phan, D.; Johnson, C.N.; Xu, R.; Chen, X.W.; Cawthorn, W.P.; MacDougald, O.A.; Koenig, R.J. Multiple roles for the non-coding RNA SRA in regulation of adipogenesis and insulin sensitivity. PLoS ONE 2010, 5, e14199. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Ding, J.; Chen, A.; Song, Y.; Xu, C.; Tian, F.; Zhao, J. Aerobic exercise improves adipogenesis in diet-induced obese mice via the lncSRA/p38/JNK/PPARγ pathway. Nutr. Res. 2022, 105, 20–32. [Google Scholar] [CrossRef]
- Guy, C.D.; Suzuki, A.; Zdanowicz, M.; Abdelmalek, M.F.; Burchette, J.; Unalp, A.; Diehl, A.M.; NASH CRN. Hedgehog pathway activation parallels histologic severity of injury and fibrosis in human nonalcoholic fatty liver disease. Hepatology 2012, 55, 1711–1721. [Google Scholar] [CrossRef]
- Kwon, H.; Song, K.; Han, C.; Chen, W.; Wang, Y.; Dash, S.; Lim, K.; Wu, T. Inhibition of hedgehog signaling ameliorates hepatic inflammation in mice with nonalcoholic fatty liver disease. Hepatology 2016, 63, 1155–1169. [Google Scholar] [CrossRef]
- Jiang, Y.; Peng, J.; Song, J.; He, J.; Jiang, M.; Wang, J.; Ma, L.; Wang, Y.; Lin, M.; Wu, H.; et al. Loss of Hilnc prevents diet-induced hepatic steatosis through binding of IGF2BP2. Nat. Metab. 2021, 3, 1569–1584. [Google Scholar] [CrossRef]
- Barrett, S.P.; Salzman, J. Circular RNAs: Analysis, expression and potential functions. Development 2016, 143, 1838–1847. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Zaiou, M. The Emerging Role and Promise of Circular RNAs in Obesity and Related Metabolic Disorders. Cells 2020, 9, 1473. [Google Scholar] [CrossRef]
- Jin, X.; Feng, C.Y.; Xiang, Z.; Chen, Y.P.; Li, Y.M. CircRNA expression pattern and circRNA-miRNA-mRNA network in the pathogenesis of nonalcoholic steatohepatitis. Oncotarget 2016, 7, 66455–66467. [Google Scholar] [CrossRef]
- Guo, X.Y.; Chen, J.N.; Sun, F.; Wang, Y.Q.; Pan, Q.; Fan, J.G. circRNA_0046367 Prevents Hepatoxicity of Lipid Peroxidation: An Inhibitory Role against Hepatic Steatosis. Oxid. Med. Cell. Longev. 2017, 2017, 3960197. [Google Scholar] [CrossRef]
- Cai, H.; Jiang, Z.; Yang, X.; Lin, J.; Cai, Q.; Li, X. Circular RNA HIPK3 contributes to hyperglycemia and insulin homeostasis by sponging miR-192-5p and upregulating transcription factor forkhead box O1. Endocr. J. 2020, 67, 397–408. [Google Scholar] [CrossRef]
- Lu, J.; Pang, L.; Zhang, B.; Gong, Z.; Song, C. Silencing circANKRD36 inhibits streptozotocin-induced insulin resistance and inflammation in diabetic rats by targeting miR-145 via XBP1. Inflamm. Res. 2021, 70, 695–704. [Google Scholar] [CrossRef]
- Guo, X.Y.; He, C.X.; Wang, Y.Q.; Sun, C.; Li, G.M.; Su, Q.; Pan, Q.; Fan, J.G. Circular RNA Profiling and Bioinformatic Modeling Identify Its Regulatory Role in Hepatic Steatosis. BioMed Res. Int. 2017, 2017, 5936171. [Google Scholar] [CrossRef]
- Yuan, X.; Li, Y.; Wen, S.; Xu, C.; Wang, C.; He, Y.; Zhou, L. CircLDLR acts as a sponge for miR-667-5p to regulate SIRT1 expression in non-alcoholic fatty liver disease. Lipids Health Dis. 2022, 21, 127. [Google Scholar] [CrossRef]
- Picard, F.; Kurtev, M.; Chung, N.; Topark-Ngarm, A.; Senawong, T.; Machado De Oliveira, R.; Leid, M.; McBurney, M.W.; Guarente, L. Sirt1 Promotes Fat Mobilization in White Adipocytes by Repressing PPAR-Gamma. Nature 2004, 429, 771–776. [Google Scholar] [CrossRef]
- Lin, X.; Du, Y.; Lu, W.; Gui, W.; Sun, S.; Zhu, Y.; Wang, G.; Eserberg, D.T.; Zheng, F.; Zhou, J.; et al. CircRNF111 Protects Against Insulin Resistance and Lipid Deposition via Regulating miR-143-3p/IGF2R Axis in Metabolic Syndrome. Front. Cell Dev. Biol. 2021, 9, 663148. [Google Scholar] [CrossRef]
- Lee, Y.H.; Bae, S.C.; Song, G.G. Meta-analysis of associations between the peroxisome proliferator-activated receptor-γ Pro12Ala polymorphism and susceptibility to nonalcoholic fatty liver disease, rheumatoid arthritis, and psoriatic arthritis. Genet. Test. Mol. Biomark. 2014, 18, 341–348. [Google Scholar] [CrossRef]
- Xie, R.; Tang, S.; Yang, Y. Associations of peroxisome proliferator-activated receptor-γ Pro12Ala polymorphism with non-alcoholic fatty liver disease: A meta-analysis. J. Diabetes Complicat. 2022, 36, 108261. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Li, Y.-Y.; Nie, Y.-Q.; Sha, W.-H.; Du, Y.-L.; Lai, X.-B.; Zhou, Y.-J. Effect of peroxisome proliferator-activated receptors-gamma and co-activator-1alpha genetic polymorphisms on plasma adiponectin levels and susceptibility of non-alcoholic fatty liver disease in Chinese people. Liver Int. 2008, 28, 385–392. [Google Scholar] [CrossRef]
- Zhou, Y.J.; Li, Y.Y.; Nie, Y.Q.; Yang, H.; Zhan, Q.; Huang, J.; Shi, S.L.; Lai, X.B.; Huang, H.L. Influence of polygenetic polymorphisms on the susceptibility to non-alcoholic fatty liver disease of Chinese people. J. Gastroenterol. Hepatol. 2010, 25, 772–777. [Google Scholar] [CrossRef] [PubMed]
- Flessa, C.M.; Nasiri-Ansari, N.; Kyrou, I.; Leca, B.M.; Lianou, M.; Chatzigeorgiou, A.; Kaltsas, G.; Kassi, E.; Randeva, H.S. Genetic and Diet-Induced Animal Models for Non-Alcoholic Fatty Liver Disease (NAFLD) Research. Int. J. Mol. Sci. 2022, 23, 15791. [Google Scholar] [CrossRef]
- Santhekadur, P.K.; Kumar, D.P.; Sanyal, A.J. Preclinical models of non-alcoholic fatty liver disease. J. Hepatol. 2018, 68, 230–237. [Google Scholar] [CrossRef]
- Xu, X.; Tan, X.; Tampe, B.; Wilhelmi, T.; Hulshoff, M.S.; Saito, S.; Moser, T.; Kalluri, R.; Hasenfuss, G.; Zeisberg, E.M.; et al. High-fidelity CRISPR/Cas9- based gene-specific hydroxymethylation rescues gene expression and attenuates renal fibrosis. Nat. Commun. 2018, 9, 3509. [Google Scholar] [CrossRef]
- Horii, T.; Morita, S.; Hatada, I. Generation of Epigenetic Disease Model Mice by Targeted Demethylation of the Epigenome. Methods Mol. Biol. 2023, 2577, 255–268. [Google Scholar] [CrossRef]
- Ratziu, V.; Charlotte, F.; Bernhardt, C.; Giral, P.; Halbron, M.; Lenaour, G.; Hartmann-Heurtier, A.; Bruckert, E.; Poynard, T.; LIDO Study Group. Long-term efficacy of rosiglitazone in nonalcoholic steatohepatitis: Results of the fatty liver improvement by rosiglitazone therapy (FLIRT 2) extension trial. Hepatology 2010, 51, 445–453. [Google Scholar] [CrossRef]
- Skat-Rørdam, J.; Højland Ipsen, D.; Lykkesfeldt, J.; Tveden-Nyborg, P. A role of peroxisome proliferator-activated receptor γ in non-alcoholic fatty liver disease. Basic Clin. Pharmacol. Toxicol. 2019, 124, 528–537. [Google Scholar] [CrossRef]
- Yang, X.; Frank, J.; Gonzalez, F.J.; Huang, M.; Bi, H. Nuclear receptors and non-alcoholic fatty liver disease: An update. Liver Res. 2020, 4, 88–93. [Google Scholar] [CrossRef]
- Ahmadian, M.; Myoung Suh, J.; Hah, N.; Liddle, C.; Atkins, A.R.; Downes, M.; Evans, R.M. PPARgamma signaling and metabolism: The good, the bad and the future. Nat. Med. 2013, 19, 557–566. [Google Scholar] [CrossRef]
- Romualdo, G.R.; Valente, L.C.; Sprocatti, A.C.; Bacil, G.P.; de Souza, I.P.; Rodrigues, J.; Rodrigues, M.A.M.; Vinken, M.; Cogliati, B.; Barbisan, L.F. Western diet-induced mouse model of non-alcoholic fatty liver disease associated with metabolic outcomes: Features of gut microbiome-liver-adipose tissue axis. Nutrition 2022, 103–104, 111836. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).