Gene Fusion Detection in NSCLC Routine Clinical Practice: Targeted-NGS or FISH?
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Cohort
2.2. FISH Analysis
2.3. ALK IHC
2.4. RNA Extraction
2.5. Targeted RNA-Based NGS
2.6. Real-Time PCR
3. Results
3.1. Fusion Variant Detection by FISH and IHC: In Situ Approach
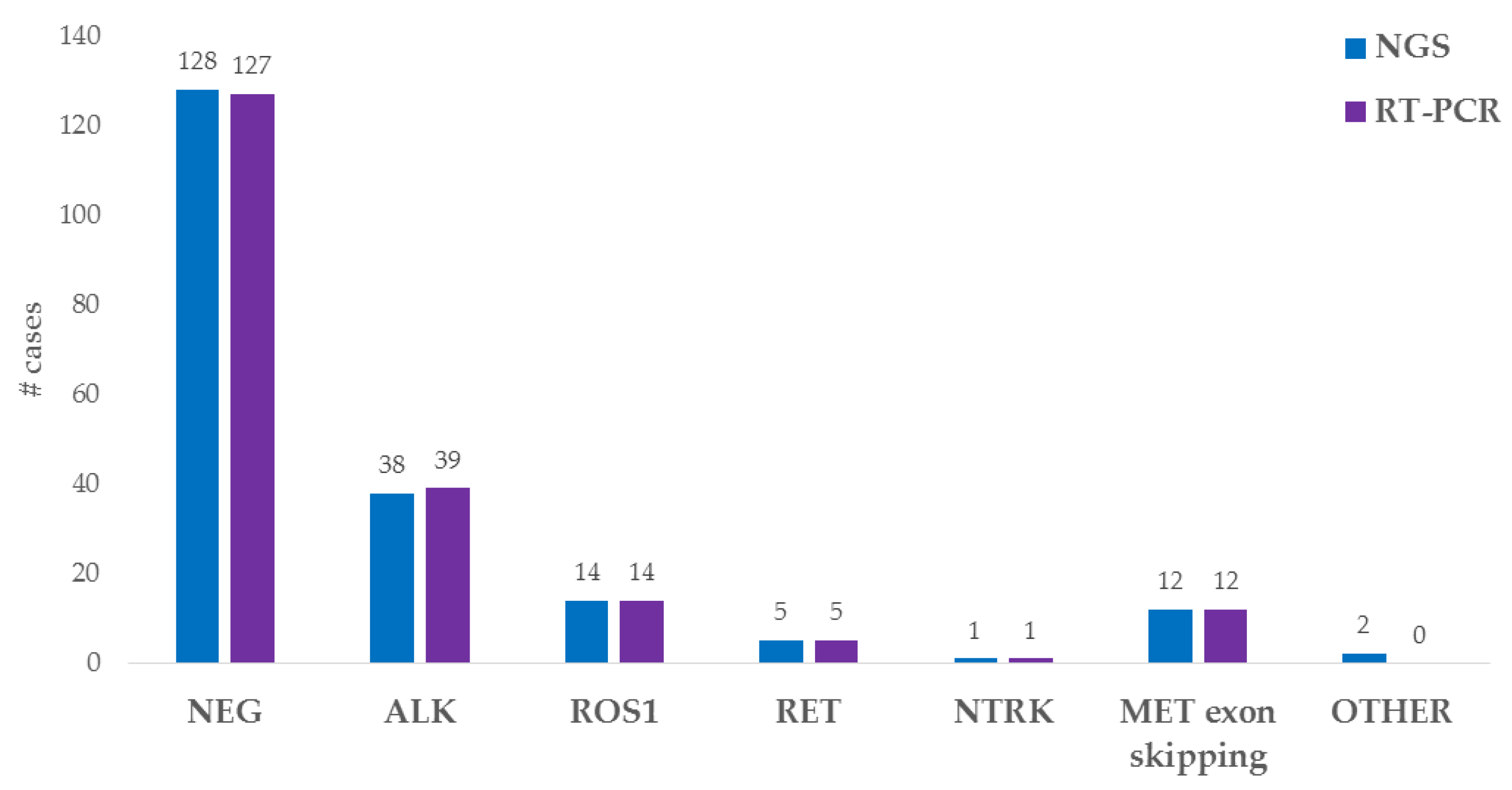
3.2. Fusion Variant Detection by NGS and Real-Time PCR: Molecular Approach
3.3. Comparison of the In Situ and Molecular Approaches
3.4. Selected Patients’ Clinical Follow-Up
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mosele, F.; Remon, J.; Mateo, J.; Westphalen, C.B.; Barlesi, F.; Lolkema, M.P.; Normanno, N.; Scarpa, A.; Robson, M.; Meric-Bernstam, F.; et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: A report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2020, 31, 1491–1505. [Google Scholar] [CrossRef] [PubMed]
- Lindeman, N.I.; Cagle, P.T.; Aisner, D.L.; Arcila, M.E.; Beasley, B.; Bernicker, E.H.; Colasacco, C.; Dacic, S.; Hirsch, F.R.; Kerr, K.; et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment With Targeted Tyrosine Kinase Inhibitors Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch. Pathol. Lab. Med. 2018, 13, 323–358. [Google Scholar]
- Kazdal, D.; Hofman, V.; Christopoulos, P.; Ilié, M.; Stenzinger, A.; Hofman, P. Fusion-positive non-small cell lung carcinoma: Biological principles, clinical practice, and diagnostic implications. Genes Chromosom. Cancer 2022, 61, 244–260. [Google Scholar] [CrossRef]
- Saigí, M.; Carcereny, E.; Morán, T.; Cucurull, M.; Domènech, M.; Hernandez, A.; Martinez-Cardús, A.; Pros, E.; Sanchez-Cespedes, M. Biological and clinical perspectives of the actionable gene fusions and amplifications involving tyrosine kinase receptors in lung cancer. Cancer Treat. Rev. 2022, 109, 102430. [Google Scholar] [CrossRef] [PubMed]
- Westphalen, C.B.; Krebs, M.G.; Le Tourneau, C.; Sokol, E.S.; Maund, S.L.; Wilson, T.R.; Jin, D.X.; Newberg, J.Y.; Fabrizio, D.; Veronese, L.; et al. Genomic context of NTRK1/2/3 fusion-positive tumours from a large real-world population. NPJ Precis. Oncol. 2021, 5, 69. [Google Scholar] [CrossRef] [PubMed]
- Lindeman, N.I.; Cagle, P.T.; Beasley, M.B.; Chitale, D.A.; Dacic, S.; Giaccone, G.; Jenkins, R.B.; Kwiatkowski, D.J.; Saldivar, J.S.; Squire, J.; et al. Molecular testing guidelines for selection of lung cancer patients for ALK tyrosine kinase inhibitors. Guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J. Mol. Diagn. 2013, 15, 415–453. [Google Scholar] [CrossRef]
- Tuononen, K.; Sarhadi, V.K.; Wirtanen, A.; Rönty, M.; Salmenkivi, K.; Knuuttila, A.; Remes, S.; Telaranta-Keerie, A.I.; Bloor, S.; Ellonen, P.; et al. Targeted resequencing reveals ALK fusions in non-small cell lung carcinomas detected by FISH, immunohistochemistry, and real-time RT-PCR: A comparison of four methods. Biomed. Res. Int. 2013, 2013, 757490. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Shu, W.; Li, P. Fluorescence in situ hybridization: Cell-based genetic diagnostic and research applications. Front. Cell Dev. Biol. 2016, 4, 89. [Google Scholar] [CrossRef]
- Lazzari, C.; Bulotta, A.; Cangi, M.G.; Bucci, G.; Pecciarini, L.; Bonfiglio, S.; Lorusso, V.; Ippati, S.; Arrigoni, G.; Grassini, G.; et al. Next generation sequencing in non-small cell lung cancer: Pitfalls and opportunities. Diagnostics 2020, 10, 1092. [Google Scholar] [CrossRef]
- Gregorc, V.; Mazzarella, L.; Lazzari, C.; Graziano, P.; Vigneri, P.; Genova, C.; Toschi, L.; Ciliberto, G.; Bonanno, L.; Delmonte, A.; et al. Prospective Validation of the Italian Alliance against Cancer Lung Panel in Patients with Advanced Non–Small-Cell Lung Cancer. Clin. Lung Cancer 2021, 22, e637–e641. [Google Scholar] [CrossRef]
- Magliacane, G.; Grassini, G.; Bartocci, P.; Francaviglia, I.; Cin, E.D.; Barbieri, G.; Arrigoni, G.; Pecciarini, L.; Doglioni, C.; Cangi, M.G. Rapid targeted somatic mutation analysis of solid tumors in routine clinical diagnostics. Oncotarget 2015, 6, 30592–30603. [Google Scholar] [CrossRef]
- Redegalli, M.; Grassini, G.; Magliacane, G.; Pecciarini, L.; Lena, M.S.; Smart, C.E.; Johnston, R.L.; Waddell, N.; Maestro, R.; Macchini, M.; et al. Routine molecular profiling in both resectable and unresectable pancreatic adenocarcinoma: Relevance of cytological samples. Clin. Gastroenterol. Hepatol. 2022. online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Francaviglia, I.; Magliacane, G.; Lazzari, C.; Grassini, G.; Brunetto, E.; Cin, E.D.; Girlando, S.; Medicina, D.; Smart, C.E.; Bulotta, A.; et al. Identification and monitoring of somatic mutations in circulating cell-free tumor DNA in lung cancer patients. Lung Cancer 2019, 134, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Belli, C.; Penault-Llorca, F.; Ladanyi, M.; Normanno, N.; Scoazec, J.-Y.; Lacroix, L.; Reis-Filho, J.; Subbiah, V.; Gainor, J.; Endris, V.; et al. ESMO recommendations on the standard methods to detect RET fusions and mutations in daily practice and clinical research. Ann. Oncol. 2021, 32, 337–350. [Google Scholar] [CrossRef] [PubMed]
- Lazzari, C.; Pecciarini, L.; Doglioni, C.; Pedica, F.; Gajate, A.M.S.; Bulotta, A.; Gregorc, V.; Cangi, M.G. Case report: EML4::NTRK3 gene fusion in a patient with metastatic lung adenocarcinoma successfully treated with entrectinib. Front. Oncol. 2022, 12, 1038774. [Google Scholar] [CrossRef] [PubMed]
- Postmus, P.E.; Kerr, K.M.; Oudkerk, M.; Senan, S.; Waller, D.A.; Vansteenkiste, J.; Escriu, C.; Peters, S.; ESMO Guidelines Committee. Early and locally advanced non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2017, 28 (Suppl. S4), iv1–iv21. [Google Scholar] [CrossRef] [PubMed]
- Kirchner, M.; Neumann, O.; Volckmar, A.-L.; Stögbauer, F.; Allgäuer, M.; Kazdal, D.; Budczies, J.; Rempel, E.; Brandt, R.; Talla, S.B.; et al. RNA-Based Detection of Gene Fusions in. Cancers 2019, 11, 1309. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; McNulty, S.N.; Evenson, M.J.; Zhu, X.; Robinson, J.A.; Mann, P.R.; Duncavage, E.J.; Pfeifer, J.D. Clinical Implications of a Targeted RNA-Sequencing Panel in the Detection of Gene Fusions in Solid Tumors. J. Mol. Diagn. 2021, 23, 1749–1760. [Google Scholar] [CrossRef]
- Tabbò, F.; Muscarella, L.A.; Gobbini, E.; Trombetta, D.; Castellana, S.; Rigutto, A.; Galetta, D.; Maiello, E.; Martelli, O.; Tiseo, M.; et al. Detection of ALK fusion variants by RNA-based NGS and clinical outcome correlation in NSCLC patients treated with ALK-TKI sequences. Eur. J. Cancer 2022, 174, 200–211. [Google Scholar] [CrossRef]
- Luca, C.; Pepe, F.; Pisapia, P.; Iaccarino, A.; Righi, L.; Listì, A.; Russo, G.; Campione, S.; Pagni, F.; Nacchio, M.; et al. RNA-based next-generation sequencing in non-small-cell lung cancer in a routine setting: An experience from an Italian referral center. Per. Med. 2022, 19, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Liebers, M.; Zhelyazkova, B.; Cao, Y.; Panditi, D.; Lynch, K.D.; Chen, J.; Robinson, H.E.; Shim, H.S.; Chmielecki, J.; et al. Technical Reports Anchored multiplex PCR for targeted next-generation sequencing. Nat. Med. 2014, 20, 1479–1484. [Google Scholar] [CrossRef] [PubMed]
- Pfarr, N.; Kirchner, M.; Lehmann, U.; Leichsenring, J.; Merkelbach-Bruse, S.; Glade, J.; Hummel, M.; Stögbauer, F.; Lehmann, A.; Trautmann, M.; et al. Testing NTRK testing: Wet-lab and in silico comparison of RNA-based targeted sequencing assays. Genes Chromosom. Cancer 2020, 59, 178–188. [Google Scholar] [CrossRef] [PubMed]
- Racanelli, D.; Brenca, M.; Baldazzi, D.; Goeman, F.; Casini, B.; De Angelis, B.; Guercio, M.; Milano, G.M.; Tamborini, E.; Busico, A.; et al. Next-Generation Sequencing Approaches for the Identification of Pathognomonic Fusion Transcripts in Sarcomas: The Experience of the Italian ACC Sarcoma Working Group. Front. Oncol. 2020, 10, 489. [Google Scholar] [CrossRef] [PubMed]
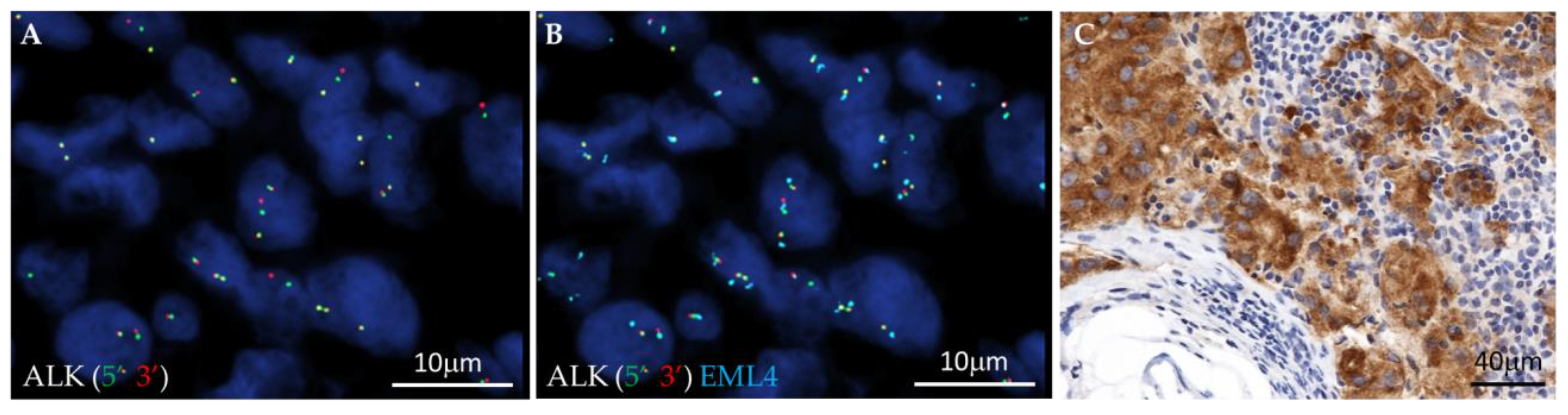
| No | Pt_ID | FISH | NGS | RT-PCR | ALK IHC | FISH_Data | NGS_Data | RT-PCR_Data |
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | + | + | + | + | ALK split >50%, no EML4 | KIF5B(24)::ALK(20) | ALK POS |
| 2 | 3 | + | + | + | + | ALK split >50%, no EML4 | KIF5B(17)::ALK(20) | ALK POS |
| 3 | 4 | + | + | + | + | ALK split >50%, no EML4 | KIF5B(17)::ALK(20) | ALK POS |
| 4 | 5 | + | + | + | + | EML4::ALK > 50% | EML4(20)::ALK(20) | ALK POS |
| 5 | 6 | + | + | + | + | EML4::ALK > 50% | EML4(6)::ALK(20) | ALK POS |
| 6 | 7 | + | + | + | + | EML4::ALK > 50% | EML4(6)::ALK(20) | ALK POS |
| 7 | 8 | + | + | + | + | EML4::ALK > 50% | EML4(6)::ALK(20) | ALK POS |
| 8 | 9 | + | + | + | + | EML4::ALK > 50% | EML4(6)::ALK(20) | ALK POS |
| 9 | 10 | + | + | + | + | EML4::ALK > 50% | EML4(2)::ALK(20) | ALK POS |
| 10 | 11 | + | + | + | + | EML4::ALK > 50% | EML4(13)::ALK(20) | ALK POS |
| 11 | 12 | + | + | + | + | EML4::ALK > 50% | EML4(13)::ALK(20) | ALK POS |
| 12 | 13 | + | + | + | + | EML4::ALK > 50% | EML4(13)::ALK(20) | ALK POS |
| 13 | 14 | + | + | - | + | EML4::ALK > 50% | EML4(6)::ALK(18) | NEG |
| 14 | 15 | + | + | + | + | EML4::ALK > 50% | EML4(13)::ALK(20) | ALK POS |
| 15 | 16 | + | + | + | + | EML4::ALK > 50% | EML4(13)::ALK(20) | ALK POS |
| 16 | 17 | + | + | + | + | EML4::ALK > 50% | EML4(20)::ALK(20) | ALK POS |
| 17 | 18 | + | + | + | + | EML4::ALK > 50% | EML4(13)::ALK(20) | ALK POS |
| 18 | 19 | + | + | + | + | EML4::ALK > 50% | EML4(20)::ALK(20) | ALK POS |
| 19 | 20 | + | + | + | + | EML4::ALK > 50% | EML4(6)::ALK(20) | ALK POS |
| 20 | 21 | + | + | + | + | EML4::ALK > 50% | EML4(13)::ALK(20) | ALK POS |
| 21 | 22 | + | + | + | + | EML4::ALK > 50% | EML4(6)::ALK(20) | ALK POS |
| 22 | 23 | + | + | + | + | EML4::ALK >50% | EML4(6)::ALK(20) | ALK POS |
| 23 | 24 | + | + | + | + | EML4::ALK > 50% | EML4(13)::ALK(20) | ALK POS |
| 24 | 25 | + | + | + | + | EML4::ALK > 50% | EML4(13)::ALK(20) | ALK POS |
| 25 | 26 | + | + | + | + | EML4::ALK > 50% | EML4(6)::ALK(20) | ALK POS |
| 26 | 27 | + | + | + | + | EML4::ALK > 50% | EML4(13)::ALK(20) | ALK POS |
| 27 | 28 | + | + | + | + | EML4::ALK > 50% | EML4(13)::ALK(20) | ALK POS |
| 28 | 29 | + | + | + | + | EML4::ALK > 50% | EML4(13)::ALK(20) | ALK POS |
| 29 | 30 | + | + | + | + | EML4::ALK > 50% | EML4(13)::ALK(20) | ALK POS |
| 30 | 31 | + | + | + | + | EML4::ALK > 50% | EML4(2)::ALK(20) | ALK POS |
| 31 | 32 | + | + | + | + | EML4::ALK > 50% | EML4(20)::ALK(20) | ALK POS |
| 32 | 33 | + | + | + | + | EML4::ALK > 50% | EML4(6)::ALK(20) | ALK POS |
| 33 | 34 | + | + | + | + | EML4::ALK > 50% | EML4(6)::ALK(20) | ALK POS |
| 34 | 35 | + | + | + | + | EML4::ALK > 50% | EML4(13)::ALK(20) | ALK POS |
| 35 | 212 | + | + | + | + | EML4::ALK > 50% | EML4(6)::ALK(20) | ALK POS |
| 36 | 36 | + | + | + | + | EML4::ALK = 12% | EML4(6)::ALK(20) | ALK POS |
| 37 | 37 | + | + | + | + | EML4::ALK = 40% | EML4(13)::ALK(20) | ALK POS |
| 38 | 162 | + | + | + | - | EML4::NTRK3 > 50% | EML4(2)::NTRK3(14) | NTRK POS |
| 39 | 166 | + | + | + | - | ROS1 split > 50% | SDC4(2)::ROS1(32) | ROS1 POS |
| 40 | 167 | + | + | + | - | ROS1 split > 50% | SLC34A2(13)::ROS1(34) | ROS1 POS |
| 41 | 168 | + | + | + | - | ROS1 split > 50% | EZR(10)::ROS1(34) | ROS1 POS |
| 42 | 169 | + | + | + | - | ROS1 split > 50% | CD74(6)::ROS1(34) | ROS1 POS |
| 43 | 170 | + | + | + | - | ROS1 split > 50% | SLC34A2(13)::ROS1(32) | ROS1 POS |
| 44 | 171 | + | + | + | - | ROS1 split > 50% | SDC4(2)::ROS1(32) | ROS1 POS |
| 45 | 172 | + | + | + | - | ROS1 split > 50% | CD74(6)::ROS1(34) | ROS1 POS |
| 46 | 173 | + | + | + | - | ROS1 split > 50% | CD74(6)::ROS1(34) | ROS1 POS |
| 47 | 174 | + | + | + | - | ROS1 split > 50% | SLC34A2(13)::ROS1(32) | ROS1 POS |
| 48 | 175 | + | + | + | - | ROS1 split > 50% | SLC34A2(13)::ROS1(32) | ROS1 POS |
| 49 | 176 | + | + | + | - | ROS1 split > 50% | SDC4(2)::ROS1(32) | ROS1 POS |
| 50 | 177 | + | + | + | - | ROS1 split > 50% | SDC4(2)::ROS1(32) | ROS1 POS |
| 51 | 209 | + | + | + | - | ROS1 split > 50% | CD74(6)::ROS1(34) | ROS1 POS |
| 52 | 210 | + | + | + | - | ROS1 split > 50% | CD74(6)::ROS1(34) | ROS1 POS |
| 53 | 163 | + | + | + | - | RET split > 50% | KIF5B(15)::RET(12) | RET POS |
| 54 | 164 | + | + | + | - | RET split > 50% | KIF5B(15)::RET(12) | RET POS |
| 55 | 165 | + | + | + | - | RET split > 50% | KIF5B(15)::RET(12) | RET POS |
| 56 | 207 | + | + | + | - | RET split > 50% | KIF5B(23)::RET(12) | RET POS |
| 57 | 208 | + | + | + | - | RET split > 50% | KIF5B(15)::RET(12) | RET POS |
| No | Pt_ID | FISH | NGS | RT-PCR | ALK IHC | FISH_Data | NGS_Data | RT-PCR_Data |
|---|---|---|---|---|---|---|---|---|
| 1 | 178 | + | - | - | + | ALK split = 25%, no EML4 | No fusions | NEG |
| 2 | 179 | + | - | - | + | ALK split = 25%, no EML4 | No fusions | NEG |
| 3 | 180 | + | - | - | + | ALK split = 50%, no EML4 | No fusions | NEG |
| 4 | 181 | + | - | - | + | ALK split = 50%, no EML4 | No fusions | NEG |
| 5 | 182 | + | - | + | + | EML4::ALK > 50% | No fusions | ALK POS |
| 6 | 183 | + | - | - | + | EML4::ALK > 50% | No fusions | NEG |
| 7 | 184 | + | - | - | + | EML4::ALK > 50% | No fusions | NEG |
| 8 | 185 | + | - | + | + | EML4::ALK > 50% | No fusions | ALK POS |
| 9 | 192 | + | - | - | - | ROS1 split > 50% | No fusions | NEG |
| 10 | 193 | + | - | - | - | ROS1 split > 50% | No fusions | NEG |
| 11 | 194 | + | - | - | - | ROS1 split > 50% | No fusions | NEG |
| 12 | 195 | + | - | - | - | ROS1 split = 25% | No fusions | NEG |
| 13 | 196 | + | - | - | - | ROS1 split = 25% | No fusions | NEG |
| 14 | 189 | + | - | - | - | RET split > 50% | No fusions | NEG |
| 15 | 190 | + | - | - | - | RET split > 50% | No fusions | NEG |
| 16 | 191 | + | - | - | - | RET split > 50% | No fusions | NEG |
| 17 | 188 | + | - | - | - | RET single 3′ > 50% | No fusions | NEG |
| 18 | 186 | - | + | + | - | ALK split in rare cell | EML4(6)::ALK(20) | ALK POS |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pecciarini, L.; Brunetto, E.; Grassini, G.; De Pascali, V.; Ogliari, F.R.; Talarico, A.; Marra, G.; Magliacane, G.; Redegalli, M.; Arrigoni, G.; et al. Gene Fusion Detection in NSCLC Routine Clinical Practice: Targeted-NGS or FISH? Cells 2023, 12, 1135. https://doi.org/10.3390/cells12081135
Pecciarini L, Brunetto E, Grassini G, De Pascali V, Ogliari FR, Talarico A, Marra G, Magliacane G, Redegalli M, Arrigoni G, et al. Gene Fusion Detection in NSCLC Routine Clinical Practice: Targeted-NGS or FISH? Cells. 2023; 12(8):1135. https://doi.org/10.3390/cells12081135
Chicago/Turabian StylePecciarini, Lorenza, Emanuela Brunetto, Greta Grassini, Valeria De Pascali, Francesca Rita Ogliari, Anna Talarico, Giovanna Marra, Gilda Magliacane, Miriam Redegalli, Gianluigi Arrigoni, and et al. 2023. "Gene Fusion Detection in NSCLC Routine Clinical Practice: Targeted-NGS or FISH?" Cells 12, no. 8: 1135. https://doi.org/10.3390/cells12081135
APA StylePecciarini, L., Brunetto, E., Grassini, G., De Pascali, V., Ogliari, F. R., Talarico, A., Marra, G., Magliacane, G., Redegalli, M., Arrigoni, G., Lazzari, C., Gregorc, V., Bulotta, A., Doglioni, C., & Cangi, M. G. (2023). Gene Fusion Detection in NSCLC Routine Clinical Practice: Targeted-NGS or FISH? Cells, 12(8), 1135. https://doi.org/10.3390/cells12081135







