Signal-Retaining Autophagy Indicator as a Quantitative Imaging Method for ER-Phagy
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Cell Lines
2.2. Plasmids and Molecular Cloning
2.3. Viruses, Transduction and Stable Cell Lines
2.4. Generation of ΔAtg7 PDAC Cell Line Using CRISPR/Cas9 Gene Editing
2.5. Generation of SRAI-Expressing PDAC Cell Lines Using FACS Sorting
2.6. Cell Imaging by Wide-Field Microscopy and Automated Acquisition
2.7. Imaging Quantification of the TOLLES:YPet Index
2.8. Immunocytochemistry and Confocal Imaging
2.9. Immunobloting
2.10. Image Analysis and Statistical Analysis
3. Results
3.1. Characterization of SRAI as a Probe to Observe Bulk Autophagic Flux, Extending the Utility of This Probe beyond Mitophagy
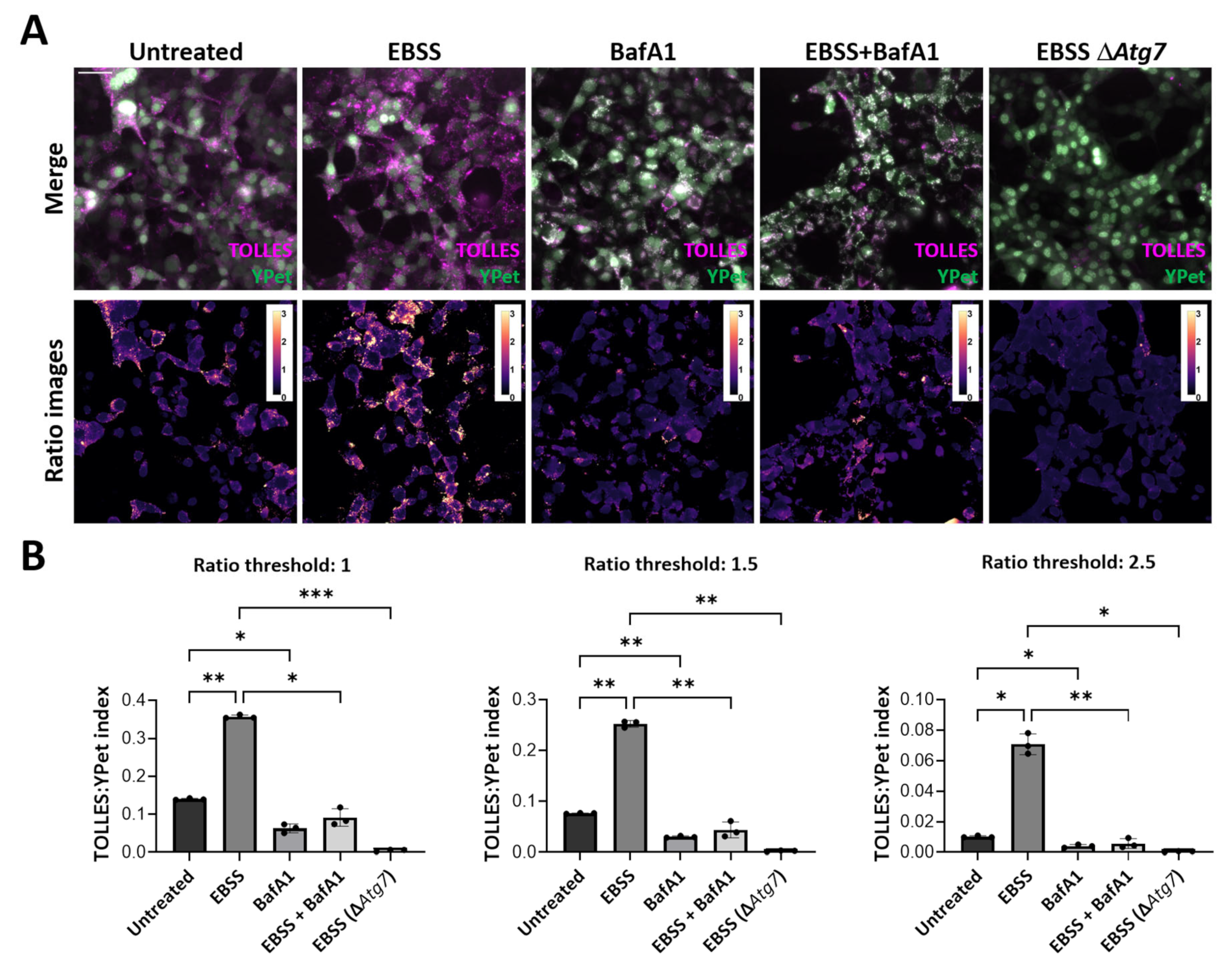
3.2. Establishing a Methodology for Semi-Automated Quantification of Autophagic Flux
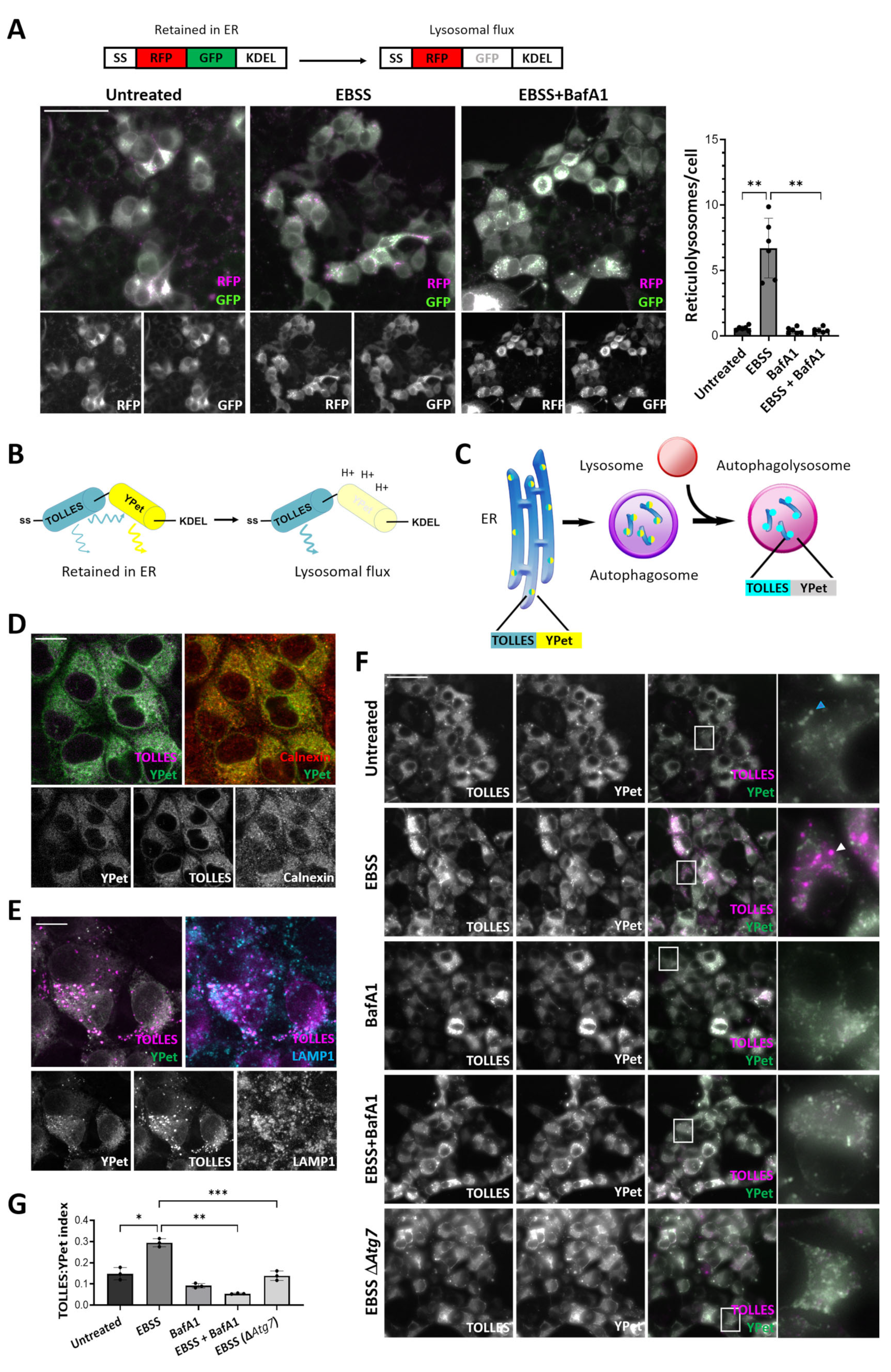
3.3. Development of ss-SRAI-KDEL for Reliable Measurement of General ER-Phagy
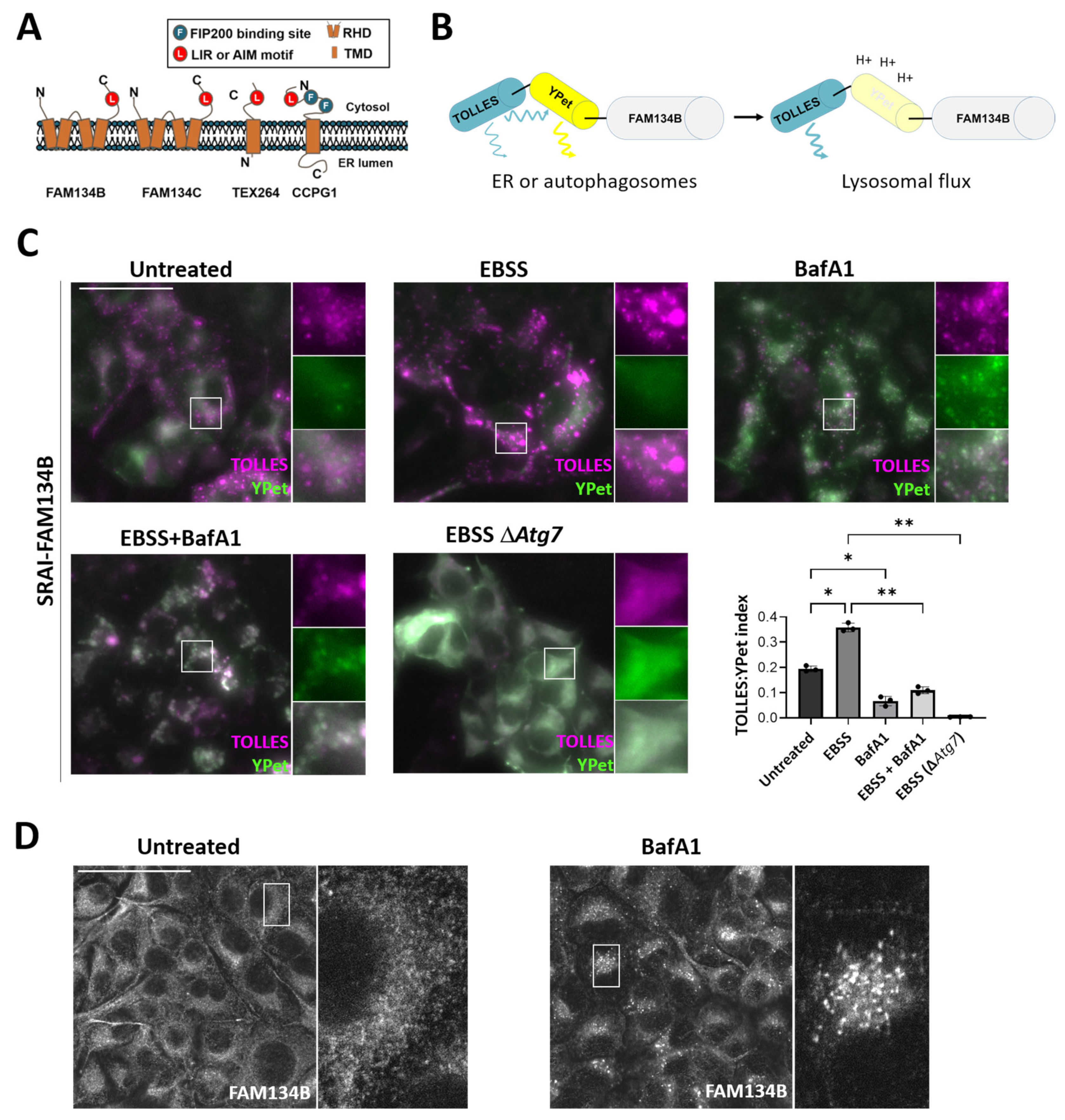
3.4. ER-Phagy Flux Associated with Specific ER-Phagy Receptors Can Be Detected Using the SRAI Probe
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Dikic, I.; Elazar, Z. Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 2018, 19, 349–364. [Google Scholar] [CrossRef]
- Yamamoto, H.; Zhang, S.; Mizushima, N. Autophagy genes in biology and disease. Nat. Rev. Genet. 2023, 22, 1–19. [Google Scholar] [CrossRef]
- Zhao, Y.G.; Codogno, P.; Zhang, H. Machinery, regulation and pathophysiological implications of autophagosome maturation. Nat. Rev. Mol. Cell Biol. 2021, 22, 733–750. [Google Scholar] [CrossRef] [PubMed]
- Kirkin, V. History of the Selective Autophagy Research: How Did It Begin and Where Does It Stand Today? J. Mol. Biol. 2020, 432, 3–27. [Google Scholar] [CrossRef]
- Hernandez, G.A.; Perera, R.M. Autophagy in cancer cell remodeling and quality control. Mol. Cell 2022, 82, 1514–1527. [Google Scholar] [CrossRef]
- Aman, Y.; Schmauck-Medina, T.; Hansen, M.; Morimoto, R.I.; Simon, A.K.; Bjedov, I.; Palikaras, K.; Simonsen, A.; Johansen, T.; Tavernarakis, N.; et al. Autophagy in healthy aging and disease. Nat. Aging 2021, 1, 634–650. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Petroni, G.; Amaravadi, R.K.; Baehrecke, E.H.; Ballabio, A.; Boya, P.; Pedro, J.M.B.; Cadwell, K.; Cecconi, F.; Choi, A.M.K.; et al. Autophagy in major human diseases. EMBO J. 2021, 40, e108863. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; White, E.; Rubinsztein, D.C. Breakthroughs and bottlenecks in autophagy research. Trends Mol. Med. 2021, 27, 835–838. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N. The exponential growth of autophagy-related research: From the humble yeast to the Nobel Prize. FEBS Lett. 2017, 591, 681–689. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition) (1). Autophagy 2021, 17, 1–382. [Google Scholar] [CrossRef]
- Mizushima, N.; Murphy, L.O. Autophagy Assays for Biological Discovery and Therapeutic Development. Trends Biochem. Sci. 2020, 45, 1080–1093. [Google Scholar] [CrossRef]
- Yoshii, S.R.; Mizushima, N. Monitoring and Measuring Autophagy. Int. J. Mol. Sci. 2017, 18, 1865. [Google Scholar] [CrossRef]
- Kaizuka, T.; Morishita, H.; Hama, Y.; Tsukamoto, S.; Matsui, T.; Toyota, Y.; Kodama, A.; Ishihara, T.; Mizushima, T.; Mizushima, N. An Autophagic Flux Probe that Releases an Internal Control. Mol. Cell 2016, 64, 835–849. [Google Scholar] [CrossRef]
- McWilliams, T.G.; Prescott, A.R.; Allen, G.F.; Tamjar, J.; Munson, M.J.; Thomson, C.; Muqit, M.M.; Ganley, I.G. Mito-QC illuminates mitophagy and mitochondrial architecture in vivo. J. Cell Biol. 2016, 214, 333–345. [Google Scholar] [CrossRef] [PubMed]
- Kimura, S.; Noda, T.; Yoshimori, T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 2007, 3, 452–460. [Google Scholar] [CrossRef]
- Katayama, H.; Yamamoto, A.; Mizushima, N.; Yoshimori, T.; Miyawaki, A. GFP-like proteins stably accumulate in lysosomes. Cell Struct. Funct. 2008, 33, 1–12. [Google Scholar] [CrossRef]
- Katayama, H.; Hama, H.; Nagasawa, K.; Kurokawa, H.; Sugiyama, M.; Ando, R.; Funata, M.; Yoshida, N.; Homma, M.; Nishimura, T.; et al. Visualizing and Modulating Mitophagy for Therapeutic Studies of Neurodegeneration. Cell 2020, 181, 1176–1187.e16. [Google Scholar] [CrossRef] [PubMed]
- Katayama, H.; Kogure, T.; Mizushima, N.; Yoshimori, T.; Miyawaki, A. A sensitive and quantitative technique for detecting autophagic events based on lysosomal delivery. Chem. Biol. 2011, 18, 1042–1052. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.W.; Daugherty, P.S. Evolutionary optimization of fluorescent proteins for intracellular FRET. Nat. Biotechnol. 2005, 23, 355–360. [Google Scholar] [CrossRef]
- Stringer, B.W.; Day, B.W.; D’Souza, R.C.J.; Jamieson, P.R.; Ensbey, K.S.; Bruce, Z.C.; Lim, Y.C.; Goasdoué, K.; Offenhäuser, C.; Akgül, S.; et al. A reference collection of patient-derived cell line and xenograft models of proneural, classical and mesenchymal glioblastoma. Sci. Rep. 2019, 9, 4902. [Google Scholar] [CrossRef]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Shimozono, S.; Miyawaki, A. Engineering FRET constructs using CFP and YFP. Methods Cell Biol. 2008, 85, 381–393. [Google Scholar] [PubMed]
- Yang, X.; Boehm, J.S.; Yang, X.; Salehi-Ashtiani, K.; Hao, T.; Shen, Y.; Lubonja, R.; Thomas, S.R.; Alkan, O.; Bhimdi, T.; et al. A public genome-scale lentiviral expression library of human ORFs. Nat. Methods 2011, 8, 659–661. [Google Scholar] [CrossRef] [PubMed]
- Chino, H.; Hatta, T.; Natsume, T.; Mizushima, N. Intrinsically Disordered Protein TEX264 Mediates ER-phagy. Mol. Cell 2019, 74, 909–921.e6. [Google Scholar] [CrossRef]
- Behrends, C.; Sowa, M.E.; Gygi, S.P.; Harper, J.W. Network organization of the human autophagy system. Nature 2010, 466, 68–76. [Google Scholar] [CrossRef]
- Fraser, J.; Simpson, J.; Fontana, R.; Kishi-Itakura, C.; Ktistakis, N.T.; Gammoh, N. Targeting of early endosomes by autophagy facilitates EGFR recycling and signalling. EMBO Rep. 2019, 20, e47734. [Google Scholar] [CrossRef]
- Gao, Z.; Herrera-Carrillo, E.; Berkhout, B. A Single H1 Promoter Can Drive Both Guide RNA and Endonuclease Expression in the CRISPR-Cas9 System. Mol. Ther. Nucleic Acids 2019, 14, 32–40. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef]
- Murphy, L.C.; Pearson, M.; Jimenez-Moreno, N.; Wilkinson, S. Tools Used for Tolles: YPet Reporter Paper. 2023. Available online: https://github.com/IGC-Advanced-Imaging-Resource/JimenezMoreno_2022 (accessed on 5 April 2023).
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Smith, M.D.; Harley, M.E.; Kemp, A.J.; Wills, J.; Lee, M.; Arends, M.; von Kriegsheim, A.; Behrends, C.; Wilkinson, S. CCPG1 Is a Non-canonical Autophagy Cargo Receptor Essential for ER-Phagy and Pancreatic ER Proteostasis. Dev. Cell 2018, 44, 217–232.e11. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Yoshimori, T.; Levine, B. Methods in mammalian autophagy research. Cell 2010, 140, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Reggiori, F.; Molinari, M. ER-phagy: Mechanisms, regulation, and diseases connected to the lysosomal clearance of the endoplasmic reticulum. Physiol. Rev. 2022, 102, 1393–1448. [Google Scholar] [CrossRef] [PubMed]
- Gubas, A.; Dikic, I. ER remodeling via ER-phagy. Mol. Cell 2022, 82, 1492–1500. [Google Scholar] [CrossRef]
- Chino, H.; Mizushima, N. ER-Phagy: Quality Control and Turnover of Endoplasmic Reticulum. Trends Cell Biol. 2020, 30, 384–398. [Google Scholar] [CrossRef]
- An, H.; Ordureau, A.; Paulo, J.A.; Shoemaker, C.J.; Denic, V.; Harper, J.W. TEX264 Is an Endoplasmic Reticulum-Resident ATG8-Interacting Protein Critical for ER Remodeling during Nutrient Stress. Mol. Cell 2019, 74, 891–908.e10. [Google Scholar] [CrossRef] [PubMed]
- Johansen, T.; Lamark, T. Selective Autophagy: ATG8 Family Proteins, LIR Motifs and Cargo Receptors. J. Mol. Biol. 2020, 432, 80–103. [Google Scholar] [CrossRef]
- Wilkinson, S. Emerging Principles of Selective ER Autophagy. J. Mol. Biol. 2020, 432, 185–205. [Google Scholar] [CrossRef]
- Nthiga, T.M.; Kumar Shrestha, B.; Sjøttem, E.; Bruun, J.A.; Bowitz Larsen, K.; Bhujabal, Z.; Lamark, T.; Johansen, T. CALCOCO1 acts with VAMP-associated proteins to mediate ER-phagy. EMBO J. 2020, 39, e103649. [Google Scholar] [CrossRef] [PubMed]
- Molinari, M. ER-phagy responses in yeast, plants, and mammalian cells and their crosstalk with UPR and ERAD. Dev. Cell 2021, 56, 949–966. [Google Scholar] [CrossRef]
- Reggio, A.; Buonomo, V.; Berkane, R.; Bhaskara, R.M.; Tellechea, M.; Peluso, I.; Polishchuk, E.; Di Lorenzo, G.; Cirillo, C.; Esposito, M.; et al. Role of FAM134 paralogues in endoplasmic reticulum remodeling, ER-phagy, and Collagen quality control. EMBO Rep. 2021, 22, e52289. [Google Scholar] [CrossRef] [PubMed]
- Khaminets, A.; Heinrich, T.; Mari, M.; Grumati, P.; Huebner, A.K.; Akutsu, M.; Liebmann, L.; Stolz, A.; Nietzsche, S.; Koch, N.; et al. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature 2015, 522, 354–358. [Google Scholar] [CrossRef] [PubMed]
- Di Lorenzo, G.; Iavarone, F.; Maddaluno, M.; Plata-Gómez, A.B.; Aureli, S.; Meza, C.P.Q.; Cinque, L.; Palma, A.; Reggio, A.; Cirillo, C.; et al. Phosphorylation of FAM134C by CK2 controls starvation-induced ER-phagy. Sci. Adv. 2022, 8, eabo1215. [Google Scholar] [CrossRef]
- Cinque, L.; De Leonibus, C.; Iavazzo, M.; Krahmer, N.; Intartaglia, D.; Salierno, F.G.; De Cegli, R.; Di Malta, C.; Svelto, M.; Lanzara, C.; et al. MiT/TFE factors control ER-phagy via transcriptional regulation of FAM134B. EMBO J. 2020, 39, e105696. [Google Scholar] [CrossRef] [PubMed]
- Yim, W.W.; Yamamoto, H.; Mizushima, N. A pulse-chasable reporter processing assay for mammalian autophagic flux with HaloTag. eLife 2022, 11, e78923. [Google Scholar] [CrossRef] [PubMed]
- Rudinskiy, M.; Bergmann, T.J.; Molinari, M. Quantitative and time-resolved monitoring of organelle and protein delivery to the lysosome with a tandem fluorescent Halo-GFP reporter. Mol. Biol. Cell 2022, 33, ar57. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Zhao, J.; Qiu, M.; Mi, Z.; Meng, M.; Guo, Y.; Wang, H.; Yuan, Z. CRISPR/Cas9 Mediated GFP Knock-in at the MAP1LC3B Locus in 293FT Cells Is Better for Bona Fide Monitoring Cellular Autophagy. Biotechnol. J. 2018, 13, e1700674. [Google Scholar] [CrossRef] [PubMed]
- Bräuer, M.; Zich, M.T.; Önder, K.; Müller, N. The influence of commonly used tags on structural propensities and internal dynamics of peptides. Mon. Chem.-Chem. Mon. 2019, 150, 913–925. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jimenez-Moreno, N.; Salomo-Coll, C.; Murphy, L.C.; Wilkinson, S. Signal-Retaining Autophagy Indicator as a Quantitative Imaging Method for ER-Phagy. Cells 2023, 12, 1134. https://doi.org/10.3390/cells12081134
Jimenez-Moreno N, Salomo-Coll C, Murphy LC, Wilkinson S. Signal-Retaining Autophagy Indicator as a Quantitative Imaging Method for ER-Phagy. Cells. 2023; 12(8):1134. https://doi.org/10.3390/cells12081134
Chicago/Turabian StyleJimenez-Moreno, Natalia, Carla Salomo-Coll, Laura C. Murphy, and Simon Wilkinson. 2023. "Signal-Retaining Autophagy Indicator as a Quantitative Imaging Method for ER-Phagy" Cells 12, no. 8: 1134. https://doi.org/10.3390/cells12081134
APA StyleJimenez-Moreno, N., Salomo-Coll, C., Murphy, L. C., & Wilkinson, S. (2023). Signal-Retaining Autophagy Indicator as a Quantitative Imaging Method for ER-Phagy. Cells, 12(8), 1134. https://doi.org/10.3390/cells12081134





