Zinc Supplementation Induced Transcriptional Changes in Primary Human Retinal Pigment Epithelium: A Single-Cell RNA Sequencing Study to Understand Age-Related Macular Degeneration
Abstract
1. Introduction
2. Materials and Methods
2.1. Retinal Pigment Epithelial (RPE) Cell Culture
2.2. Experiment Overview
2.3. scRNA-Seq
2.4. Bioinformatics
2.5. Functional Classification Pathway and Network Analysis
2.6. Immunofluorescence
3. Results
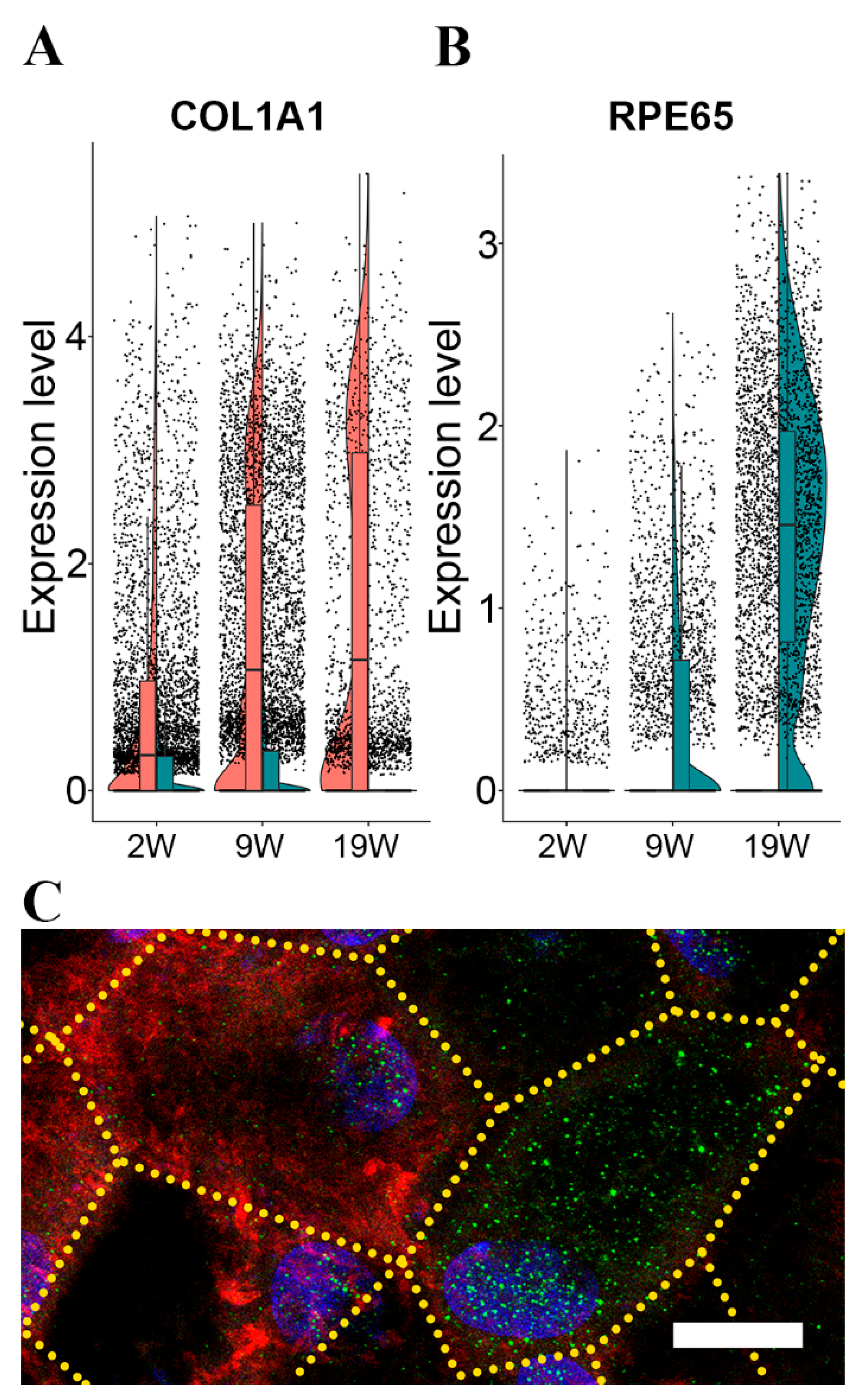
3.1. Maturation of RPE Cells in Culture
3.2. Cluster Analysis of the scRNA-Seq Data Identifies Significant Heterogeneity of RPE Cells
3.2.1. Unsupervised Clustering Analysis
3.2.2. Hierarchical Clustering Analysis Using Markers of Mature RPE Cells
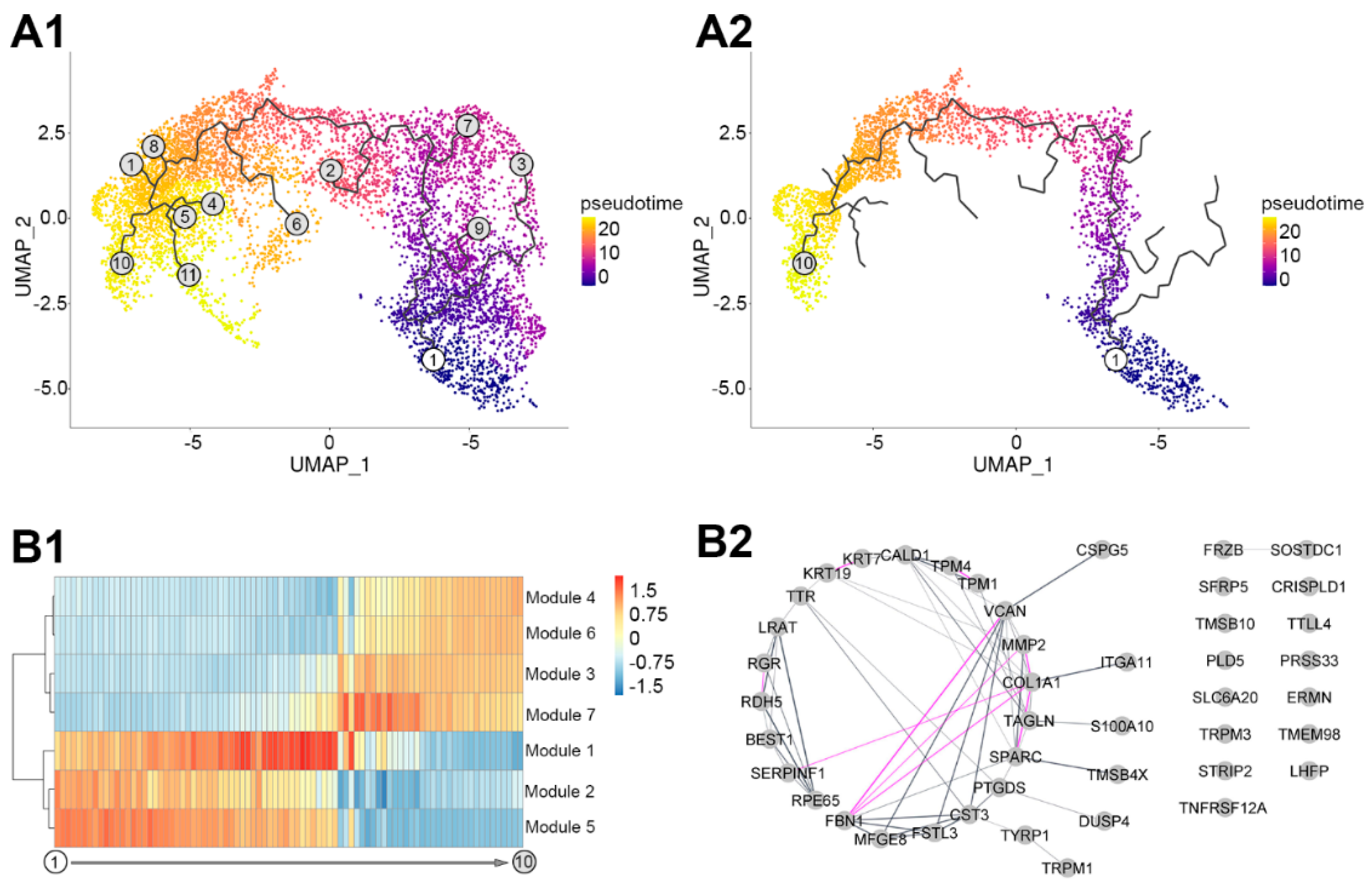
3.3. Pseudotemporal Ordering of the Expressed RPE Genes
3.4. Acute Zinc Supplementation Has a Multitude of Effects on Transcription in RPE Cells
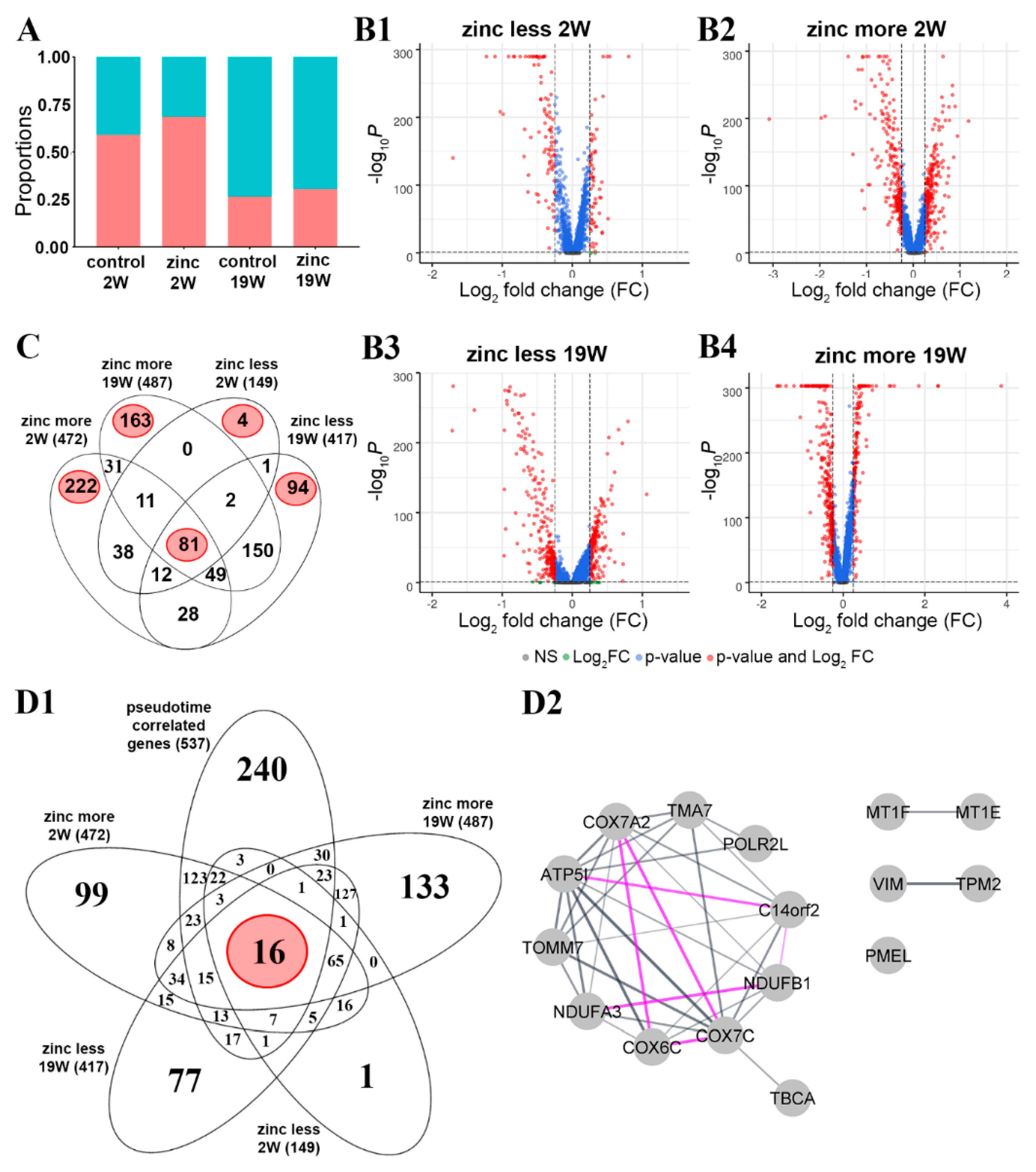
3.4.1. Transcriptional Changes in Response to Acute Zinc Supplementation
3.4.2. Influence of Zinc on Transcription Dynamics
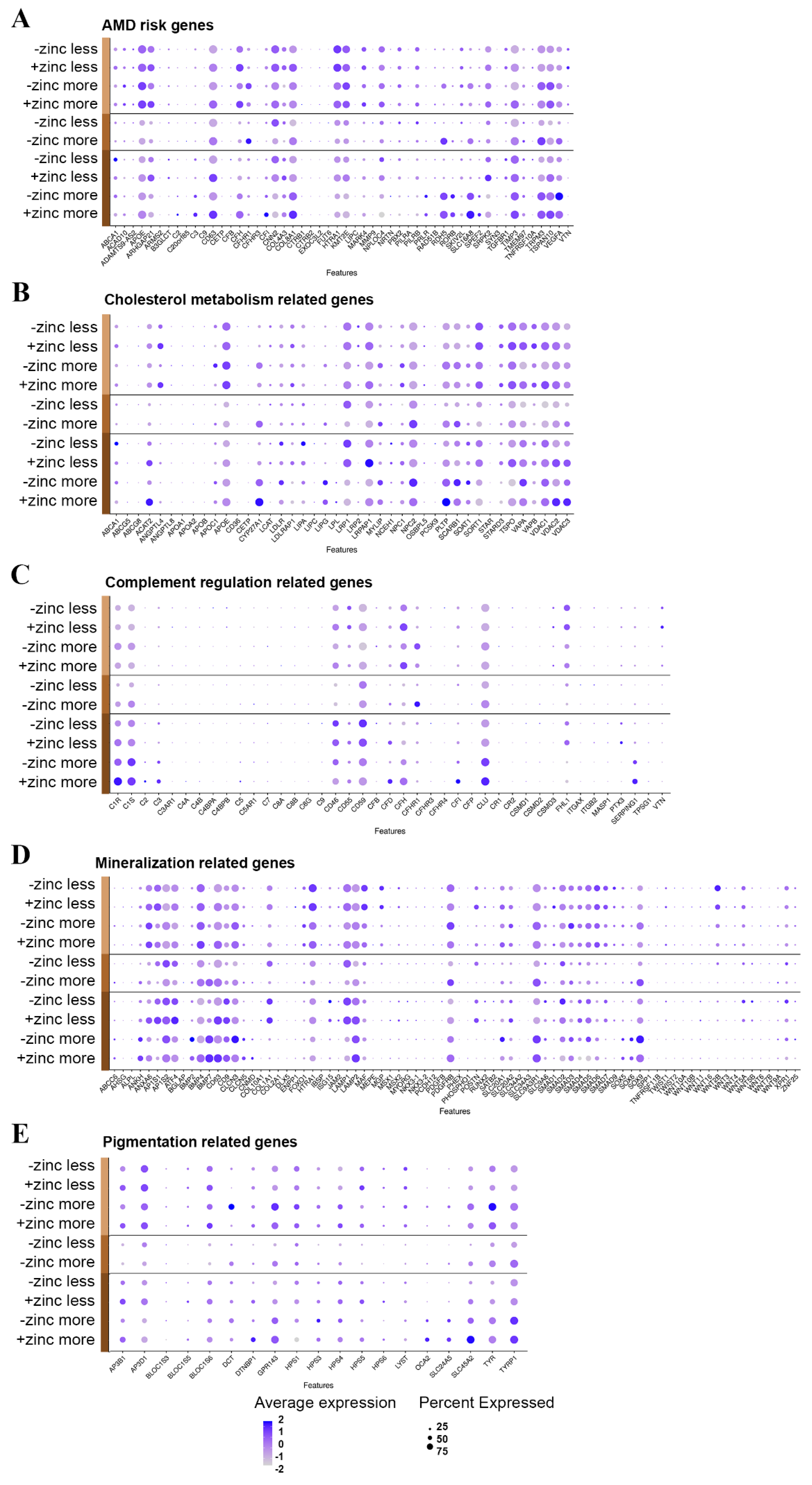
3.5. Sub-RPE Deposition-Related Gene Expression Pattern Depends on Maturation State and Zinc Supplementation
3.5.1. Genelist 01
3.5.2. Genelist 02
3.5.3. Genelist 03
3.5.4. Genelist 04
3.5.5. Genelist 05
4. Discussion
4.1. Study Rationale
4.2. Heterogeneity of RPE Cells
4.3. Transition from Less to More Differentiated RPE
4.4. Genes Involved in Transitioning RPE from Less to More Differentiated Cells
4.5. Response to Acute Zinc Supplementation
4.6. The Effects of Zinc on the Genes in the Pseudotemporal Trajectory
4.7. AMD-Specific Gene Expression Changes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Boulton, M.; Dayhaw-Barker, P. The role of the retinal pigment epithelium: Topographical variation and ageing changes. Eye 2001, 15, 384–389. [Google Scholar] [CrossRef] [PubMed]
- Bonilha, V.L. Age and disease-related structural changes in the retinal pigment epithelium. Clin. Ophthalmol. 2008, 2, 413–424. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.X.; Germer, C.J.; La Cunza, N.; Lakkaraju, A. Complement activation, lipid metabolism, and mitochondrial injury: Converging pathways in age-related macular degeneration. Redox Biol. 2020, 101781. [Google Scholar] [CrossRef] [PubMed]
- Bergen, A.A.; Arya, S.; Koster, C.; Pilgrim, M.G.; Wiatrek-Moumoulidis, D.; van der Spek, P.J.; Hauck, S.M.; Boon, C.J.F.; Emri, E.; Stewart, A.J.; et al. On the origin of proteins in human drusen: The meet, greet and stick hypothesis. Prog. Retin. Eye. Res. 2019, 70, 55–84. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.C.S.; Pilgrim, M.G.; Fearn, S.; Bertazzo, S.; Tsolaki, E.; Morrell, A.P.; Li, M.; Messinger, J.D.; Dolz-Marco, R.; Lei, J.; et al. Calcified nodules in retinal drusen are associated with disease progression in age-related macular degeneration. Sci. Transl. Med. 2018, 10. [Google Scholar] [CrossRef]
- Curcio, C.A.; Johnson, M.; Rudolf, M.; Huang, J.D. The oil spill in ageing Bruch membrane. Br. J. Ophthalmol. 2011, 95, 1638–1645. [Google Scholar] [CrossRef]
- Flinn, J.M.; Kakalec, P.; Tappero, R.; Jones, B.; Lengyel, I. Correlations in distribution and concentration of calcium, copper and iron with zinc in isolated extracellular deposits associated with age-related macular degeneration. Metallomics 2014, 6, 1223–1228. [Google Scholar] [CrossRef]
- Jhingan, M.; Singh, S.R.; Samanta, A.; Arora, S.; Tucci, D.; Amarasekera, S.; Cagini, C.; Lupidi, M.; Chhablani, J. Drusen ooze: Predictor for progression of dry age-related macular degeneration. Graefes. Arch. Clin. Exp. Ophthalmol. 2021, 259, 2687–2694. [Google Scholar] [CrossRef]
- Ferris, F.L., III; Wilkinson, C.P.; Bird, A.; Chakravarthy, U.; Chew, E.; Csaky, K.; Sadda, S.R. Clinical classification of age-related macular degeneration. Ophthalmology 2013, 120, 844–851. [Google Scholar] [CrossRef]
- Lee, J.; Kang, H.G.; Kim, H.R.; Lee, C.S.; Kim, M.; Kim, S.S.; Byeon, S.H. Pigmentary abnormality without significant drusen as a risk factor for late age-related macular degeneration. Sci. Rep. 2022, 12, 769. [Google Scholar] [CrossRef]
- Freund, K.B.; Staurenghi, G.; Jung, J.J.; Zweifel, S.A.; Cozzi, M.; Hill, L.; Blotner, S.; Tsuboi, M.; Gune, S. Macular neovascularization lesion type and vision outcomes in neovascular age-related macular degeneration: Post hoc analysis of HARBOR. Graefes. Arch. Clin. Exp. Ophthalmol. 2022, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Spaide, R.F.; Jaffe, G.J.; Sarraf, D.; Freund, K.B.; Sadda, S.R.; Staurenghi, G.; Waheed, N.K.; Chakravarthy, U.; Rosenfeld, P.J.; Holz, F.G.; et al. Consensus Nomenclature for Reporting Neovascular Age-Related Macular Degeneration Data: Consensus on Neovascular Age-Related Macular Degeneration Nomenclature Study Group. Ophthalmology 2020, 127, 616–636. [Google Scholar] [CrossRef] [PubMed]
- Age-Related Eye Disease Study Research Group. A randomized, placebo-controlled, clinical trial of high-dose supplementation with vitamins C and E, beta carotene, and zinc for age-related macular degeneration and vision loss: AREDS report no. 8. Arch. Ophthalmol. 2001, 119, 1417–1436. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, B.A.; Philp, N.J. Cell culture of retinal pigment epithelium: Special Issue. Exp. Eye. Res. 2014, 126, 1–4. [Google Scholar] [CrossRef]
- Pao, P.J.; Emri, E.; Abdirahman, S.B.; Soorma, T.; Zeng, H.H.; Hauck, S.M.; Thompson, R.B.; Lengyel, I. The effects of zinc supplementation on primary human retinal pigment epithelium. J. Trace. Elem. Med. Biol. 2018, 49, 184–191. [Google Scholar] [CrossRef]
- Pilgrim, M.G.; Lengyel, I.; Lanzirotti, A.; Newville, M.; Fearn, S.; Emri, E.; Knowles, J.C.; Messinger, J.D.; Read, R.W.; Guidry, C.; et al. Subretinal Pigment Epithelial Deposition of Drusen Components Including Hydroxyapatite in a Primary Cell Culture Model. Investig. Ophthalmol. Vis. Sci. 2017, 58, 708–719. [Google Scholar] [CrossRef]
- Emri, E.; Kortvely, E.; Dammeier, S.; Klose, F.; Simpson, D.; Consortium, E.R.; Den Hollander, A.I.; Ueffing, M.; Lengyel, I. A Multi-Omics Approach Identifies Key Regulatory Pathways Induced by Long-Term Zinc Supplementation in Human Primary Retinal Pigment Epithelium. Nutrients 2020, 12, 3051. [Google Scholar] [CrossRef]
- Maminishkis, A.; Chen, S.; Jalickee, S.; Banzon, T.; Shi, G.; Wang, F.E.; Ehalt, T.; Hammer, J.A.; Miller, S.S. Confluent monolayers of cultured human fetal retinal pigment epithelium exhibit morphology and physiology of native tissue. Investig. Ophthalmol. Vis. Sci. 2006, 47, 3612–3624. [Google Scholar] [CrossRef]
- Sonoda, S.; Spee, C.; Barron, E.; Ryan, S.J.; Kannan, R.; Hinton, D.R. A protocol for the culture and differentiation of highly polarized human retinal pigment epithelial cells. Nat. Protoc. 2009, 4, 662–673. [Google Scholar] [CrossRef]
- Bozym, R.; Hurst, T.K.; Westerberg, N.; Stoddard, A.; Fierke, C.A.; Frederickson, C.J.; Thompson, R.B. Determination of zinc using carbonic anhydrase-based fluorescence biosensors. Methods. Enzymol. 2008, 450, 287–309. [Google Scholar] [CrossRef]
- Booij, J.C.; Baas, D.C.; Beisekeeva, J.; Gorgels, T.G.; Bergen, A.A. The dynamic nature of Bruch’s membrane. Prog. Retin. Eye. Res. 2010, 29, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Bennis, A.; Gorgels, T.G.; Ten Brink, J.B.; van der Spek, P.J.; Bossers, K.; Heine, V.M.; Bergen, A.A. Comparison of Mouse and Human Retinal Pigment Epithelium Gene Expression Profiles: Potential Implications for Age-Related Macular Degeneration. PLoS ONE 2015, 10, e0141597. [Google Scholar] [CrossRef] [PubMed]
- Strunnikova, N.V.; Maminishkis, A.; Barb, J.J.; Wang, F.; Zhi, C.; Sergeev, Y.; Chen, W.; Edwards, A.O.; Stambolian, D.; Abecasis, G.; et al. Transcriptome analysis and molecular signature of human retinal pigment epithelium. Hum. Mol. Genet. 2010, 19, 2468–2486. [Google Scholar] [CrossRef]
- Liao, J.L.; Yu, J.; Huang, K.; Hu, J.; Diemer, T.; Ma, Z.; Dvash, T.; Yang, X.J.; Travis, G.H.; Williams, D.S.; et al. Molecular signature of primary retinal pigment epithelium and stem-cell-derived RPE cells. Hum. Mol. Genet. 2010, 19, 4229–4238. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Spielmann, M.; Qiu, X.; Huang, X.; Ibrahim, D.M.; Hill, A.J.; Zhang, F.; Mundlos, S.; Christiansen, L.; Steemers, F.J.; et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 2019, 566, 496–502. [Google Scholar] [CrossRef] [PubMed]
- Franceschini, A.; Szklarczyk, D.; Frankild, S.; Kuhn, M.; Simonovic, M.; Roth, A.; Lin, J.; Minguez, P.; Bork, P.; von Mering, C.; et al. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013, 41, D808–D815. [Google Scholar] [CrossRef] [PubMed]
- Bandettini, W.P.; Kellman, P.; Mancini, C.; Booker, O.J.; Vasu, S.; Leung, S.W.; Wilson, J.R.; Shanbhag, S.M.; Chen, M.Y.; Arai, A.E. MultiContrast Delayed Enhancement (MCODE) improves detection of subendocardial myocardial infarction by late gadolinium enhancement cardiovascular magnetic resonance: A clinical validation study. J. Cardiovasc. Magn. Reson. 2012, 14, 83. [Google Scholar] [CrossRef]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M., III; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive Integration of Single-Cell Data. Cell 2019, 177, 1888–1902.e21. [Google Scholar] [CrossRef]
- Özdemir, V. OMICS 2.0: An Accelerator for Global Science, Systems Medicine and Responsible Innovation. OMICS J. Integr. Biol. 2015, 19, 579–580. [Google Scholar] [CrossRef]
- An, E.; Lu, X.; Flippin, J.; Devaney, J.M.; Halligan, B.; Hoffman, E.P.; Strunnikova, N.; Csaky, K.; Hathout, Y. Secreted proteome profiling in human RPE cell cultures derived from donors with age related macular degeneration and age matched healthy donors. J. Proteome. Res. 2006, 5, 2599–2610. [Google Scholar] [CrossRef]
- Hu, Y.; Wang, X.; Hu, B.; Mao, Y.; Chen, Y.; Yan, L.; Yong, J.; Dong, J.; Wei, Y.; Wang, W.; et al. Dissecting the transcriptome landscape of the human fetal neural retina and retinal pigment epithelium by single-cell RNA-seq analysis. PLoS. Biol. 2019, 17, e3000365. [Google Scholar] [CrossRef] [PubMed]
- Voigt, A.P.; Mulfaul, K.; Mullin, N.K.; Flamme-Wiese, M.J.; Giacalone, J.C.; Stone, E.M.; Tucker, B.A.; Scheetz, T.E.; Mullins, R.F. Single-cell transcriptomics of the human retinal pigment epithelium and choroid in health and macular degeneration. Proc. Natl. Acad. Sci. USA 2019, 116, 24100–24107. [Google Scholar] [CrossRef] [PubMed]
- Ho, L.; van Leeuwen, R.; Witteman, J.C.; van Duijn, C.M.; Uitterlinden, A.G.; Hofman, A.; de Jong, P.T.; Vingerling, J.R.; Klaver, C.C. Reducing the genetic risk of age-related macular degeneration with dietary antioxidants, zinc, and omega-3 fatty acids: The Rotterdam study. Arch. Ophthalmol. 2011, 129, 758–766. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.S.; Wang, J.J.; Flood, V.; Rochtchina, E.; Smith, W.; Mitchell, P. Dietary antioxidants and the long-term incidence of age-related macular degeneration: The Blue Mountains Eye Study. Ophthalmology 2008, 115, 334–341. [Google Scholar] [CrossRef] [PubMed]
- King, J.C.; Shames, D.M.; Woodhouse, L.R. Zinc homeostasis in humans. J. Nutr. 2000, 130, 1360S–1366S. [Google Scholar] [CrossRef] [PubMed]
- Fritsche, L.G.; Igl, W.; Bailey, J.N.; Grassmann, F.; Sengupta, S.; Bragg-Gresham, J.L.; Burdon, K.P.; Hebbring, S.J.; Wen, C.; Gorski, M.; et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat. Genet. 2016, 48, 134–143. [Google Scholar] [CrossRef]
- Mollnes, T.E.; Jokiranta, T.S.; Truedsson, L.; Nilsson, B.; Rodriguez de Cordoba, S.; Kirschfink, M. Complement analysis in the 21st century. Mol. Immunol. 2007, 44, 3838–3849. [Google Scholar] [CrossRef]
- Skattum, L. Clinical Complement Analysis-An Overview. Transfus. Med. Rev. 2019, 33, 207–216. [Google Scholar] [CrossRef]
- Johnson, L.V.; Leitner, W.P.; Staples, M.K.; Anderson, D.H. Complement activation and inflammatory processes in Drusen formation and age related macular degeneration. Exp. Eye. Res. 2001, 73, 887–896. [Google Scholar] [CrossRef]
- database, K.P. KEGG Database. Available online: https://www.genome.jp/dbget-bin/www_bget?pathway+hsa04979 (accessed on 14 March 2021).
- Wang, L.; Clark, M.E.; Crossman, D.K.; Kojima, K.; Messinger, J.D.; Mobley, J.A.; Curcio, C.A. Abundant lipid and protein components of drusen. PLoS ONE 2010, 5, e10329. [Google Scholar] [CrossRef]
- Hofmann Bowman, M.A.; McNally, E.M. Genetic pathways of vascular calcification. Trends. Cardiovasc. Med. 2012, 22, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Rai, M.F.; Schmidt, E.J.; Hashimoto, S.; Cheverud, J.M.; Sandell, L.J. Genetic loci that regulate ectopic calcification in response to knee trauma in LG/J by SM/J advanced intercross mice. J. Orthop. Res. 2015, 33, 1412–1423. [Google Scholar] [CrossRef] [PubMed]
- Tsang, H.G.; Clark, E.L.; Markby, G.R.; Bush, S.J.; Hume, D.A.; Corcoran, B.M.; MacRae, V.E.; Summers, K.M. Expression of Calcification and Extracellular Matrix Genes in the Cardiovascular System of the Healthy Domestic Sheep (Ovis aries). Front. Genet. 2020, 11, 919. [Google Scholar] [CrossRef] [PubMed]
- Thompson, R.B.; Reffatto, V.; Bundy, J.G.; Kortvely, E.; Flinn, J.M.; Lanzirotti, A.; Jones, E.A.; McPhail, D.S.; Fearn, S.; Boldt, K.; et al. Identification of hydroxyapatite spherules provides new insight into subretinal pigment epithelial deposit formation in the aging eye. Proc. Natl. Acad. Sci. USA 2015, 112, 1565–1570. [Google Scholar] [CrossRef]
- McKay, B.S. Pigmentation and vision: Is GPR143 in control? J. Neurosci. Res. 2019, 97, 77–87. [Google Scholar] [CrossRef]
- Bakker, R.; Wagstaff, P.E.; Kruijt, C.C.; Emri, E.; van Karnebeek, C.D.M.; Hoffmann, M.B.; Brooks, B.P.; Booij, J.C.; Montoliu, L.; van Genderen, M.M.; et al. The retinal pigmentation pathway in human albinism: Not so black and white. Progress in Retinal and Eye Research 2022. [Google Scholar] [CrossRef]
- Cao, D.; Leong, B.; Messinger, J.D.; Kar, D.; Ach, T.; Yannuzzi, L.A.; Freund, K.B.; Curcio, C.A. Hyperreflective Foci, Optical Coherence Tomography Progression Indicators in Age-Related Macular Degeneration, Include Transdifferentiated Retinal Pigment Epithelium. Investig. Ophthalmol. Vis. Sci. 2021, 62, 34. [Google Scholar] [CrossRef]
- Ortolan, D.; Sharma, R.; Volkov, A.; Maminishkis, A.; Hotaling, N.A.; Huryn, L.A.; Cukras, C.; Di Marco, S.; Bisti, S.; Bharti, K. Single-cell-resolution map of human retinal pigment epithelium helps discover subpopulations with differential disease sensitivity. Proc. Natl. Acad. Sci. USA 2022, 119, e2117553119. [Google Scholar] [CrossRef]
- Miceli, M.V.; Tate, D.J., Jr.; Alcock, N.W.; Newsome, D.A. Zinc deficiency and oxidative stress in the retina of pigmented rats. Investig. Ophthalmol. Vis. Sci. 1999, 40, 1238–1244. [Google Scholar]
- Age-Related Eye Disease Study Research Group. Lutein + zeaxanthin and omega-3 fatty acids for age-related macular degeneration: The Age-Related Eye Disease Study 2 (AREDS2) randomized clinical trial. JAMA 2013, 309, 2005–2015. [Google Scholar] [CrossRef]
- Newsome, D.A. A randomized, prospective, placebo-controlled clinical trial of a novel zinc-monocysteine compound in age-related macular degeneration. Curr. Eye. Res. 2008, 33, 591–598. [Google Scholar] [CrossRef] [PubMed]
- Barzegar-Befroei, N.; Cahyadi, S.; Gango, A.; Peto, T.; Lengyel, I. Zinc and eye diseases. In Zinc in Human Health; Rink, L., Ed.; IOS Press: Amsterdam, The Netherlands, 2011. [Google Scholar]
- Bennis, A.; Jacobs, J.G.; Catsburg, L.A.E.; Ten Brink, J.B.; Koster, C.; Schlingemann, R.O.; van Meurs, J.; Gorgels, T.; Moerland, P.D.; Heine, V.M.; et al. Stem Cell Derived Retinal Pigment Epithelium: The Role of Pigmentation as Maturation Marker and Gene Expression Profile Comparison with Human Endogenous Retinal Pigment Epithelium. Stem Cell. Rev. Rep. 2017, 13, 659–669. [Google Scholar] [CrossRef] [PubMed]
- da Cruz, L.; Fynes, K.; Georgiadis, O.; Kerby, J.; Luo, Y.H.; Ahmado, A.; Vernon, A.; Daniels, J.T.; Nommiste, B.; Hasan, S.M.; et al. Phase 1 clinical study of an embryonic stem cell-derived retinal pigment epithelium patch in age-related macular degeneration. Nat. Biotechnol. 2018, 36, 328–337. [Google Scholar] [CrossRef] [PubMed]
- Ahmado, A.; Carr, A.J.; Vugler, A.A.; Semo, M.; Gias, C.; Lawrence, J.M.; Chen, L.L.; Chen, F.K.; Turowski, P.; da Cruz, L.; et al. Induction of differentiation by pyruvate and DMEM in the human retinal pigment epithelium cell line ARPE-19. Investig. Ophthalmol. Vis. Sci. 2011, 52, 7148–7159. [Google Scholar] [CrossRef] [PubMed]
- Lidgerwood, G.E.; Senabouth, A.; Smith-Anttila, C.J.A.; Gnanasambandapillai, V.; Kaczorowski, D.C.; Amann-Zalcenstein, D.; Fletcher, E.L.; Naik, S.H.; Hewitt, A.W.; Powell, J.E.; et al. Transcriptomic Profiling of Human Pluripotent Stem Cell-derived Retinal Pigment Epithelium over Time. Genom. Proteom. Bioinform. 2021, 19, 223–242. [Google Scholar] [CrossRef]
- Ferrington, D.A.; Ebeling, M.C.; Kapphahn, R.J.; Terluk, M.R.; Fisher, C.R.; Polanco, J.R.; Roehrich, H.; Leary, M.M.; Geng, Z.; Dutton, J.R.; et al. Altered bioenergetics and enhanced resistance to oxidative stress in human retinal pigment epithelial cells from donors with age-related macular degeneration. Redox Biol. 2017, 13, 255–265. [Google Scholar] [CrossRef]
- Cai, H.; Fields, M.A.; Hoshino, R.; Priore, L.V. Effects of aging and anatomic location on gene expression in human retina. Front. Aging Neurosci. 2012, 4, 8. [Google Scholar] [CrossRef]
- Orozco, L.D.; Chen, H.H.; Cox, C.; Katschke, K.J., Jr.; Arceo, R.; Espiritu, C.; Caplazi, P.; Nghiem, S.S.; Chen, Y.J.; Modrusan, Z.; et al. Integration of eQTL and a Single-Cell Atlas in the Human Eye Identifies Causal Genes for Age-Related Macular Degeneration. Cell. Rep. 2020, 30, 1246–1259.e6. [Google Scholar] [CrossRef]
- Senabouth, A.; Daniszewski, M.; Lidgerwood, G.E.; Liang, H.H.; Hernández, D.; Mirzaei, M.; Zhang, R.; Han, X.; Neavin, D.; Rooney, L.; et al. Transcriptomic and proteomic retinal pigment epithelium signatures of age-related macular degeneration. BioRxiv 2021. [Google Scholar] [CrossRef]
- Burke, J.M.; Skumatz, C.M.; Irving, P.E.; McKay, B.S. Phenotypic heterogeneity of retinal pigment epithelial cells in vitro and in situ. Exp. Eye. Res. 1996, 62, 63–73. [Google Scholar] [CrossRef]
- Jung, H.; Liu, J.; Liu, T.; George, A.; Smelkinson, M.G.; Cohen, S.; Sharma, R.; Schwartz, O.; Maminishkis, A.; Bharti, K.; et al. Longitudinal adaptive optics fluorescence microscopy reveals cellular mosaicism in patients. JCI. Insight 2019, 4. [Google Scholar] [CrossRef] [PubMed]
- Burke, J.M.; Hjelmeland, L.M. Mosaicism of the retinal pigment epithelium: Seeing the small picture. Mol. Interv. 2005, 5, 241–249. [Google Scholar] [CrossRef]
- McKay, B.S.; Burke, J.M. Separation of phenotypically distinct subpopulations of cultured human retinal pigment epithelial cells. Exp. Cell. Res. 1994, 213, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Berson, J.F.; Harper, D.C.; Tenza, D.; Raposo, G.; Marks, M.S. Pmel17 initiates premelanosome morphogenesis within multivesicular bodies. Mol. Biol. Cell. 2001, 12, 3451–3464. [Google Scholar] [CrossRef]
- Galloway, C.A.; Dalvi, S.; Shadforth, A.M.A.; Suzuki, S.; Wilson, M.; Kuai, D.; Hashim, A.; MacDonald, L.A.; Gamm, D.M.; Harkin, D.G.; et al. Characterization of Human iPSC-RPE on a Prosthetic Bruch’s Membrane Manufactured From Silk Fibroin. Investig. Ophthalmol. Vis. Sci. 2018, 59, 2792–2800. [Google Scholar] [CrossRef] [PubMed]
- Eamegdool, S.S.; Sitiwin, E.I.; Cioanca, A.V.; Madigan, M.C. Extracellular matrix and oxidative stress regulate human retinal pigment epithelium growth. Free Radic. Biol. Med. 2020, 146, 357–371. [Google Scholar] [CrossRef]
- Tamura, Y.; Konomi, H.; Sawada, H.; Takashima, S.; Nakajima, A. Tissue distribution of type VIII collagen in human adult and fetal eyes. Investig. Ophthalmol. Vis. Sci. 1991, 32, 2636–2644. [Google Scholar]
- Corominas, J.; Colijn, J.M.; Geerlings, M.J.; Pauper, M.; Bakker, B.; Amin, N.; Lores Motta, L.; Kersten, E.; Garanto, A.; Verlouw, J.A.M.; et al. Whole-Exome Sequencing in Age-Related Macular Degeneration Identifies Rare Variants in COL8A1, a Component of Bruch’s Membrane. Ophthalmology 2018, 125, 1433–1443. [Google Scholar] [CrossRef]
- Butler, J.M.; Supharattanasitthi, W.; Yang, Y.C.; Paraoan, L. RNA-seq analysis of ageing human retinal pigment epithelium: Unexpected up-regulation of visual cycle gene transcription. J. Cell. Mol. Med. 2021, 25, 5572–5585. [Google Scholar] [CrossRef] [PubMed]
- Ueda, K.; Kim, H.J.; Zhao, J.; Sparrow, J.R. Bisretinoid Photodegradation Is Likely Not a Good Thing. Adv. Exp. Med. Biol. 2018, 1074, 395–401. [Google Scholar]
- Sparrow, J.R.; Gregory-Roberts, E.; Yamamoto, K.; Blonska, A.; Ghosh, S.K.; Ueda, K.; Zhou, J. The bisretinoids of retinal pigment epithelium. Prog. Retin. Eye. Res. 2012, 31, 121–135. [Google Scholar] [CrossRef]
- Yeong, J.L.; Loveman, E.; Colquitt, J.L.; Royle, P.; Waugh, N.; Lois, N. Visual cycle modulators versus placebo or observation for the prevention and treatment of geographic atrophy due to age-related macular degeneration. Cochrane Database Syst. Rev. 2020, 12, CD013154. [Google Scholar] [CrossRef]
- Wang, Z.; Paik, D.C.; Del Priore, L.V.; Burch, R.L.; Gaillard, E.R. Nitrite-modified extracellular matrix proteins deleteriously affect retinal pigment epithelial cell function and viability: A comparison study with nonenzymatic glycation mechanisms. Curr. Eye. Res. 2005, 30, 691–702. [Google Scholar] [CrossRef]
- Fernandez-Godino, R.; Bujakowska, K.M.; Pierce, E.A. Changes in extracellular matrix cause RPE cells to make basal deposits and activate the alternative complement pathway. Hum. Mol. Genet. 2018, 27, 147–159. [Google Scholar] [CrossRef]
- Fernandez-Godino, R.; Pierce, E.A. C3a triggers formation of sub-retinal pigment epithelium deposits via the ubiquitin proteasome pathway. Sci. Rep. 2018, 8, 9679. [Google Scholar] [CrossRef]
- Curcio, C.A.; Johnson, M. Structure, Function, and Pathology of Bruch’s Membrane. Retina 2013, 1, 465–481. [Google Scholar]
- Knupp, C.; Amin, S.Z.; Munro, P.M.; Luthert, P.J.; Squire, J.M. Collagen VI assemblies in age-related macular degeneration. J. Struct. Biol. 2002, 139, 181–189. [Google Scholar] [CrossRef]
- Clark, S.J.; McHarg, S.; Tilakaratna, V.; Brace, N.; Bishop, P.N. Bruch’s Membrane Compartmentalizes Complement Regulation in the Eye with Implications for Therapeutic Design in Age-Related Macular Degeneration. Front Immunol. 2017, 8, 1778. [Google Scholar] [CrossRef]
- Wojnarowicz, P.M.; Lima, E.S.R.; Ohnaka, M.; Lee, S.B.; Chin, Y.; Kulukian, A.; Chang, S.H.; Desai, B.; Garcia Escolano, M.; Shah, R.; et al. A Small-Molecule Pan-Id Antagonist Inhibits Pathologic Ocular Neovascularization. Cell. Rep. 2019, 29, 62–75.e7. [Google Scholar] [CrossRef]
- Perk, J.; Iavarone, A.; Benezra, R. Id family of helix-loop-helix proteins in cancer. Nat. Rev. Cancer 2005, 5, 603–614. [Google Scholar] [CrossRef]
- Ling, F.; Kang, B.; Sun, X.H. Id proteins: Small molecules, mighty regulators. Curr. Top. Dev. Biol. 2014, 110, 189–216. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Yip, H.K. The expression and roles of inhibitor of DNA binding helix-loop-helix proteins in the developing and adult mouse retina. Neuroscience 2011, 175, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Lasorella, A.; Benezra, R.; Iavarone, A. The ID proteins: Master regulators of cancer stem cells and tumour aggressiveness. Nat. Rev. Cancer 2014, 14, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Kjørholt, C.; Akerfeldt, M.C.; Biden, T.J.; Laybutt, D.R. Chronic hyperglycemia, independent of plasma lipid levels, is sufficient for the loss of beta-cell differentiation and secretory function in the db/db mouse model of diabetes. Diabetes 2005, 54, 2755–2763. [Google Scholar] [CrossRef] [PubMed]
- Sakurai, D.; Tsuchiya, N.; Yamaguchi, A.; Okaji, Y.; Tsuno, N.H.; Kobata, T.; Takahashi, K.; Tokunaga, K. Crucial role of inhibitor of DNA binding/differentiation in the vascular endothelial growth factor-induced activation and angiogenic processes of human endothelial cells. J. Immunol. 2004, 173, 5801–5809. [Google Scholar] [CrossRef]
- Vishwanathan, R.; Chung, M.; Johnson, E.J. A systematic review on zinc for the prevention and treatment of age-related macular degeneration. Investig. Ophthalmol. Vis. Sci. 2013, 54, 3985–3998. [Google Scholar] [CrossRef]
- Rajapakse, D.; Curtis, T.; Chen, M.; Xu, H. Zinc Protects Oxidative Stress-Induced RPE Death by Reducing Mitochondrial Damage and Preventing Lysosome Rupture. Oxid. Med. Cell. Longev. 2017, 2017, 6926485. [Google Scholar] [CrossRef]
- Rodríguez-Menéndez, S.; García, M.; Fernández, B.; Álvarez, L.; Fernández-Vega-Cueto, A.; Coca-Prados, M.; Pereiro, R.; González-Iglesias, H. The Zinc-Metallothionein Redox System Reduces Oxidative Stress in Retinal Pigment Epithelial Cells. Nutrients 2018, 10, 1874. [Google Scholar] [CrossRef]
- Mocchegiani, E.; Giacconi, R.; Cipriano, C.; Costarelli, L.; Muti, E.; Tesei, S.; Giuli, C.; Papa, R.; Marcellini, F.; Mariani, E.; et al. Zinc, metallothioneins, and longevity--effect of zinc supplementation: Zincage study. Ann. N. Y. Acad. Sci. 2007, 1119, 129–146. [Google Scholar] [CrossRef]
- Liu, Z.; Yu, S.; Ye, S.; Shen, Z.; Gao, L.; Han, Z.; Zhang, P.; Luo, F.; Chen, S.; Kang, M. Keratin 17 activates AKT signalling and induces epithelial-mesenchymal transition in oesophageal squamous cell carcinoma. J. Proteomics 2020, 211, 103557. [Google Scholar] [CrossRef]
- Inana, G.; Murat, C.; An, W.; Yao, X.; Harris, I.R.; Cao, J. RPE phagocytic function declines in age-related macular degeneration and is rescued by human umbilical tissue derived cells. J. Transl. Med. 2018, 16, 63. [Google Scholar] [CrossRef]
- Nandrot, E.F.; Anand, M.; Almeida, D.; Atabai, K.; Sheppard, D.; Finnemann, S.C. Essential role for MFG-E8 as ligand for alphavbeta5 integrin in diurnal retinal phagocytosis. Proc. Natl. Acad. Sci. USA 2007, 104, 12005–12010. [Google Scholar] [CrossRef] [PubMed]
- Tarau, I.S.; Berlin, A.; Curcio, C.A.; Ach, T. The Cytoskeleton of the Retinal Pigment Epithelium: From Normal Aging to Age-Related Macular Degeneration. Int. J. Mol. Sci. 2019, 20, 3578. [Google Scholar] [CrossRef] [PubMed]
- Jarrett, S.G.; Boulton, M.E. Consequences of oxidative stress in age-related macular degeneration. Mol. Aspects. Med. 2012, 33, 399–417. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Jiang, N.; Chu, Y.; Postnikova, O.; Varghese, R.; Horvath, A.; Cheema, A.K.; Golestaneh, N. Dysregulated metabolic pathways in age-related macular degeneration. Sci. Rep. 2020, 10, 2464. [Google Scholar] [CrossRef]
- Chew, E.Y.; Clemons, T.E.; Agrón, E.; Sperduto, R.D.; Sangiovanni, J.P.; Kurinij, N.; Davis, M.D. Long-term effects of vitamins C and E, β-carotene, and zinc on age-related macular degeneration: AREDS report no. 35. Ophthalmology 2013, 120, 1604–1611.e4. [Google Scholar] [CrossRef]
- van Leeuwen, R.; Boekhoorn, S.; Vingerling, J.R.; Witteman, J.C.; Klaver, C.C.; Hofman, A.; de Jong, P.T. Dietary intake of antioxidants and risk of age-related macular degeneration. JAMA 2005, 294, 3101–3107. [Google Scholar] [CrossRef]
- Oliver, P.D.; Tate, D.J., Jr.; Newsome, D.A. Metallothionein in human retinal pigment epithelial cells: Expression, induction and zinc uptake. Curr. Eye. Res. 1992, 11, 183–188. [Google Scholar] [CrossRef]
- Del Priore, L.V.; Geng, L.; Tezel, T.H.; Kaplan, H.J. Extracellular matrix ligands promote RPE attachment to inner Bruch’s membrane. Curr. Eye. Res. 2002, 25, 79–89. [Google Scholar] [CrossRef]
- Ling, M.T.; Wang, X.; Tsao, S.W.; Wong, Y.C. Down-regulation of Id-1 expression is associated with TGF beta 1-induced growth arrest in prostate epithelial cells. Biochim. Biophys. Acta 2002, 1570, 145–152. [Google Scholar] [CrossRef]
- Emilsson, V.; Gudmundsson, E.F.; Jonmundsson, T.; Jonsson, B.G.; Twarog, M.; Gudmundsdottir, V.; Li, Z.; Finkel, N.; Poor, S.; Liu, X.; et al. A proteogenomic signature of age-related macular degeneration in blood. Nat. Commun. 2022, 13, 3401. [Google Scholar] [CrossRef]
- Smailhodzic, D.; van Asten, F.; Blom, A.M.; Mohlin, F.C.; den Hollander, A.I.; van de Ven, J.P.; van Huet, R.A.; Groenewoud, J.M.; Tian, Y.; Berendschot, T.T.; et al. Zinc supplementation inhibits complement activation in age-related macular degeneration. PLoS. One. 2014, 9, e112682. [Google Scholar] [CrossRef]
- Arya, S.; Emri, E.; Synowsky, S.A.; Shirran, S.L.; Barzegar-Befroei, N.; Peto, T.; Botting, C.H.; Lengyel, I.; Stewart, A.J. Quantitative analysis of hydroxyapatite-binding plasma proteins in genotyped individuals with late-stage age-related macular degeneration. Exp. Eye. Res. 2018, 172, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Nan, R.; Tetchner, S.; Rodriguez, E.; Pao, P.J.; Gor, J.; Lengyel, I.; Perkins, S.J. Zinc-induced self-association of complement C3b and Factor H: Implications for inflammation and age-related macular degeneration. J. Biol. Chem. 2013, 288, 19197–19210. [Google Scholar] [CrossRef] [PubMed]
- Volanakis, J.E. Transcriptional regulation of complement genes. Annu. Rev. Immunol. 1995, 13, 277–305. [Google Scholar] [CrossRef] [PubMed]
- Rodenkirchen, V.; Schettgen, T.; Rink, L. Zinc deficiency impairs interferon-γ production on post-transcriptional level. J. Trace. Elem. Med. Biol. 2020, 62, 126598. [Google Scholar] [CrossRef] [PubMed]
- Blom, A.M.; Kask, L.; Ramesh, B.; Hillarp, A. Effects of zinc on factor I cofactor activity of C4b-binding protein and factor H. Arch. Biochem. Biophys. 2003, 418, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Tsiftsoglou, S.A.; Sim, R.B. Human complement factor I does not require cofactors for cleavage of synthetic substrates. J. Immunol. 2004, 173, 367–375. [Google Scholar] [CrossRef]
- Leoni, G.; Rosato, A.; Perozzi, G.; Murgia, C. Zinc proteome interaction network as a model to identify nutrient-affected pathways in human pathologies. Genes Nutr. 2014, 9, 436. [Google Scholar] [CrossRef]
- Curcio, C.A. Soft Drusen in Age-Related Macular Degeneration: Biology and Targeting Via the Oil Spill Strategies. Investig. Ophthalmol. Vis. Sci. 2018, 59, AMD160–AMD181. [Google Scholar] [CrossRef]
- Chen, L.; Messinger, J.D.; Sloan, K.R.; Wong, J.; Roorda, A.; Duncan, J.L.; Curcio, C.A. Abundance and multimodal visibility of soft drusen in early age-related macular degeneration: A Clinicopathologic Correlation. Retina 2020, 40, 1644–1648. [Google Scholar] [CrossRef]
- van Leeuwen, E.M.; Emri, E.; Merle, B.M.J.; Colijn, J.M.; Kersten, E.; Cougnard-Gregoire, A.; Dammeier, S.; Meester-Smoor, M.; Pool, F.M.; de Jong, E.K.; et al. A new perspective on lipid research in age-related macular degeneration. Prog. Retin. Eye. Res. 2018, 67, 56–86. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Cheng, Y.; Su, G. A review of the multifunctionality of angiopoietin-like 4 in eye disease. Biosci. Rep. 2018, 38. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Shin, J.P.; Kim, I.T.; Park, D.H. Angiopoietin-like 4 correlates with response to intravitreal ranibizumab injections in neovascular age-related macular degeneration. Retina 2018, 38, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Cao, J.; Du, Y.; Gong, Q.; Cheng, Y.; Su, G. Angiopoietin-Like Protein 4 (ANGPTL4) Induces Retinal Pigment Epithelial Barrier Breakdown by Activating Signal Transducer and Activator of Transcription 3 (STAT3): Evidence from ARPE-19 Cells Under Hypoxic Condition and Diabetic Rats. Med. Sci. Monit. 2019, 25, 6742–6754. [Google Scholar] [CrossRef]
- Willnow, T.E.; Armstrong, S.A.; Hammer, R.E.; Herz, J. Functional expression of low density lipoprotein receptor-related protein is controlled by receptor-associated protein in vivo. Proc. Natl. Acad. Sci. USA 1995, 92, 4537–4541. [Google Scholar] [CrossRef]
- Dadsena, S.; Bockelmann, S.; Mina, J.G.M.; Hassan, D.G.; Korneev, S.; Razzera, G.; Jahn, H.; Niekamp, P.; Müller, D.; Schneider, M.; et al. Ceramides bind VDAC2 to trigger mitochondrial apoptosis. Nat. Commun. 2019, 10, 1832. [Google Scholar] [CrossRef]
- Martel, C.; Wang, Z.; Brenner, C. VDAC phosphorylation, a lipid sensor influencing the cell fate. Mitochondrion 2014, 19, 69–77. [Google Scholar] [CrossRef]
- Zhu, D.; Sreekumar, P.G.; Hinton, D.R.; Kannan, R. Expression and regulation of enzymes in the ceramide metabolic pathway in human retinal pigment epithelial cells and their relevance to retinal degeneration. Vision. Res. 2010, 50, 643–651. [Google Scholar] [CrossRef]
- McKay, G.J.; Patterson, C.C.; Chakravarthy, U.; Dasari, S.; Klaver, C.C.; Vingerling, J.R.; Ho, L.; de Jong, P.T.; Fletcher, A.E.; Young, I.S.; et al. Evidence of association of APOE with age-related macular degeneration: A pooled analysis of 15 studies. Hum. Mutat. 2011, 32, 1407–1416. [Google Scholar] [CrossRef]
- Anderson, D.H.; Ozaki, S.; Nealon, M.; Neitz, J.; Mullins, R.F.; Hageman, G.S.; Johnson, L.V. Local cellular sources of apolipoprotein E in the human retina and retinal pigmented epithelium: Implications for the process of drusen formation. Am. J. Ophthalmol. 2001, 131, 767–781. [Google Scholar] [CrossRef]
- Kim, H.J.; Ahn, S.J.; Woo, S.J.; Hong, H.K.; Suh, E.J.; Ahn, J.; Park, J.H.; Ryoo, N.K.; Lee, J.E.; Kim, K.W.; et al. Proteomics-based identification and validation of novel plasma biomarkers phospholipid transfer protein and mannan-binding lectin serine protease-1 in age-related macular degeneration. Sci. Rep. 2016, 6, 32548. [Google Scholar] [CrossRef] [PubMed]
- Claudepierre, T.; Paques, M.; Simonutti, M.; Buard, I.; Sahel, J.; Maue, R.A.; Picaud, S.; Pfrieger, F.W. Lack of Niemann-Pick type C1 induces age-related degeneration in the mouse retina. Mol. Cell. Neurosci. 2010, 43, 164–176. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; He, Z.; Shu, X. Therapeutic potential of translocator protein ligands for age-related macular degeneration. Neural. Regen. Res. 2022, 17, 793–794. [Google Scholar] [PubMed]
- Wolf, A.; Herb, M.; Schramm, M.; Langmann, T. The TSPO-NOX1 axis controls phagocyte-triggered pathological angiogenesis in the eye. Nat. Commun. 2020, 11, 2709. [Google Scholar] [CrossRef] [PubMed]
- Heo, G.Y.; Bederman, I.; Mast, N.; Liao, W.L.; Turko, I.V.; Pikuleva, I.A. Conversion of 7-ketocholesterol to oxysterol metabolites by recombinant CYP27A1 and retinal pigment epithelial cells. J. Lipid Res. 2011, 52, 1117–1127. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, I.R.; Clark, M.E.; Lee, J.W.; Curcio, C.A. 7-ketocholesterol accumulates in ocular tissues as a consequence of aging and is present in high levels in drusen. Exp. Eye. Res. 2014, 128, 151–155. [Google Scholar] [CrossRef]
- Tenbrock, L.; Wolf, J.; Boneva, S.; Schlecht, A.; Agostini, H.; Wieghofer, P.; Schlunck, G.; Lange, C. Subretinal fibrosis in neovascular age-related macular degeneration: Current concepts, therapeutic avenues, and future perspectives. Cell. Tissue Res. 2021. [Google Scholar] [CrossRef]
- Yunta, M.; Lazo, P.A. Tetraspanin proteins as organisers of membrane microdomains and signalling complexes. Cell. Signal. 2003, 15, 559–564. [Google Scholar] [CrossRef]
- Wang, A.L.; Lukas, T.J.; Yuan, M.; Du, N.; Tso, M.O.; Neufeld, A.H. Autophagy and exosomes in the aged retinal pigment epithelium: Possible relevance to drusen formation and age-related macular degeneration. PLoS ONE 2009, 4, e4160. [Google Scholar] [CrossRef]
- Golestaneh, N.; Chu, Y.; Xiao, Y.Y.; Stoleru, G.L.; Theos, A.C. Dysfunctional autophagy in RPE, a contributing factor in age-related macular degeneration. Cell. Death Dis. 2017, 8, e2537. [Google Scholar] [CrossRef]
- Ibrahim, A.S.; Hussein, K.; Wang, F.; Wan, M.; Saad, N.; Essa, M.; Kim, I.; Shakoor, A.; Owen, L.A.; DeAngelis, M.M.; et al. Bone Morphogenetic Protein (BMP)4 But Not BMP2 Disrupts the Barrier Integrity of Retinal Pigment Epithelia and Induces Their Migration: A Potential Role in Neovascular Age-Related Macular Degeneration. J. Clin. Med. 2020, 9, 2293. [Google Scholar] [CrossRef] [PubMed]
- El Husseini, D.; Boulanger, M.C.; Fournier, D.; Mahmut, A.; Bossé, Y.; Pibarot, P.; Mathieu, P. High expression of the Pi-transporter SLC20A1/Pit1 in calcific aortic valve disease promotes mineralization through regulation of Akt-1. PLoS ONE 2013, 8, e53393. [Google Scholar] [CrossRef] [PubMed]
- Masuda, T.; Wahlin, K.; Wan, J.; Hu, J.; Maruotti, J.; Yang, X.; Iacovelli, J.; Wolkow, N.; Kist, R.; Dunaief, J.L.; et al. Transcription factor SOX9 plays a key role in the regulation of visual cycle gene expression in the retinal pigment epithelium. J. Biol. Chem. 2014, 289, 12908–12921. [Google Scholar] [CrossRef] [PubMed]
- Huk, D.J.; Austin, B.F.; Horne, T.E.; Hinton, R.B.; Ray, W.C.; Heistad, D.D.; Lincoln, J. Valve Endothelial Cell-Derived Tgfβ1 Signaling Promotes Nuclear Localization of Sox9 in Interstitial Cells Associated With Attenuated Calcification. Arterioscler. Thromb. Vasc. Biol. 2016, 36, 328–338. [Google Scholar] [CrossRef] [PubMed]
- Müller, F.; Rohrer, H.; Vogel-Höpker, A. Bone morphogenetic proteins specify the retinal pigment epithelium in the chick embryo. Development 2007, 134, 3483–3493. [Google Scholar] [CrossRef]
- Kang, Y.H.; Jin, J.S.; Yi, D.W.; Son, S.M. Bone morphogenetic protein-7 inhibits vascular calcification induced by high vitamin D in mice. Tohoku J. Exp. Med. 2010, 221, 299–307. [Google Scholar] [CrossRef]
- Voelkl, J.; Tuffaha, R.; Luong, T.T.D.; Zickler, D.; Masyout, J.; Feger, M.; Verheyen, N.; Blaschke, F.; Kuro, O.M.; Tomaschitz, A.; et al. Zinc Inhibits Phosphate-Induced Vascular Calcification through TNFAIP3-Mediated Suppression of NF-κB. J. Am. Soc. Nephrol. 2018, 29, 1636–1648. [Google Scholar] [CrossRef]
- Sánchez-Ferrer, A.; Rodríguez-López, J.N.; García-Cánovas, F.; García-Carmona, F. Tyrosinase: A comprehensive review of its mechanism. Biochim. Biophys. Acta. 1995, 1247, 1–11. [Google Scholar] [CrossRef]
- Ito, S. The IFPCS presidential lecture: A chemist’s view of melanogenesis. Pigment Cell. Res. 2003, 16, 230–236. [Google Scholar] [CrossRef]
- Helip-Wooley, A.; Westbroek, W.; Dorward, H.M.; Koshoffer, A.; Huizing, M.; Boissy, R.E.; Gahl, W.A. Improper trafficking of melanocyte-specific proteins in Hermansky-Pudlak syndrome type-5. J. Investig. Dermatol. 2007, 127, 1471–1478. [Google Scholar] [CrossRef]
- Li, K.; Yang, L.; Zhang, C.; Niu, Y.; Li, W.; Liu, J.J. HPS6 interacts with dynactin p150Glued to mediate retrograde trafficking and maturation of lysosomes. J. Cell. Sci. 2014, 127, 4574–4588. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Chang, B.; Naggert, J.K.; Nishina, P.M. Lysosomal Trafficking Regulator (LYST). Adv. Exp. Med. Biol. 2016, 854, 745–750. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Emri, E.; Cappa, O.; Kelly, C.; Kortvely, E.; SanGiovanni, J.P.; McKay, B.S.; Bergen, A.A.; Simpson, D.A.; Lengyel, I. Zinc Supplementation Induced Transcriptional Changes in Primary Human Retinal Pigment Epithelium: A Single-Cell RNA Sequencing Study to Understand Age-Related Macular Degeneration. Cells 2023, 12, 773. https://doi.org/10.3390/cells12050773
Emri E, Cappa O, Kelly C, Kortvely E, SanGiovanni JP, McKay BS, Bergen AA, Simpson DA, Lengyel I. Zinc Supplementation Induced Transcriptional Changes in Primary Human Retinal Pigment Epithelium: A Single-Cell RNA Sequencing Study to Understand Age-Related Macular Degeneration. Cells. 2023; 12(5):773. https://doi.org/10.3390/cells12050773
Chicago/Turabian StyleEmri, Eszter, Oisin Cappa, Caoimhe Kelly, Elod Kortvely, John Paul SanGiovanni, Brian S. McKay, Arthur A. Bergen, David A. Simpson, and Imre Lengyel. 2023. "Zinc Supplementation Induced Transcriptional Changes in Primary Human Retinal Pigment Epithelium: A Single-Cell RNA Sequencing Study to Understand Age-Related Macular Degeneration" Cells 12, no. 5: 773. https://doi.org/10.3390/cells12050773
APA StyleEmri, E., Cappa, O., Kelly, C., Kortvely, E., SanGiovanni, J. P., McKay, B. S., Bergen, A. A., Simpson, D. A., & Lengyel, I. (2023). Zinc Supplementation Induced Transcriptional Changes in Primary Human Retinal Pigment Epithelium: A Single-Cell RNA Sequencing Study to Understand Age-Related Macular Degeneration. Cells, 12(5), 773. https://doi.org/10.3390/cells12050773







