Proteomic Analysis of Human iPSC-Derived Neural Stem Cells and Motor Neurons Identifies Proteasome Structural Alterations
Abstract
:1. Introduction
2. Materials and Methods
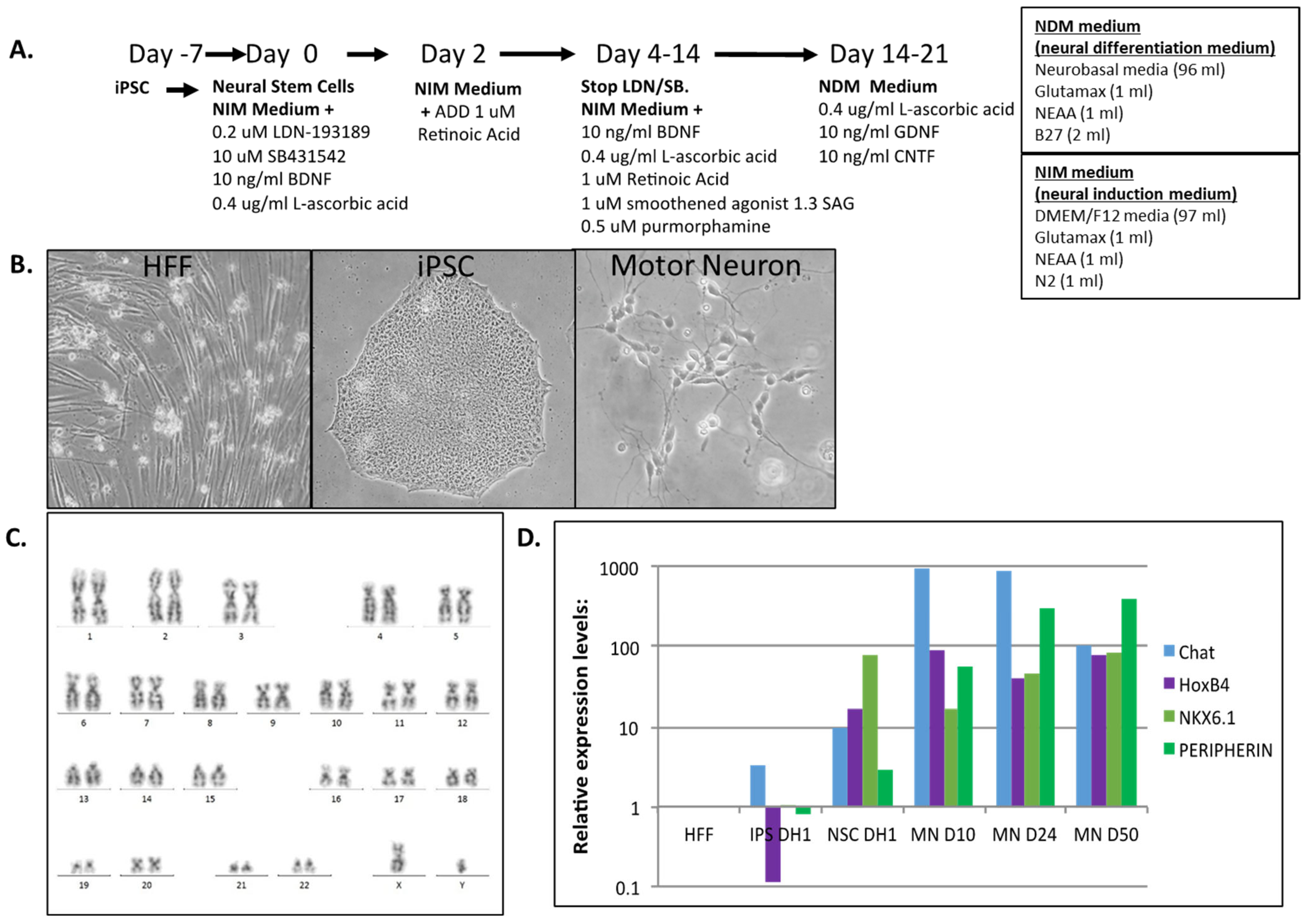
2.1. Human iPSC Differentiation to Motor Neurons
2.2. Proteomics
2.2.1. Proteomics Sample Preparation and Measurement
2.2.2. Quantitative MS Analysis
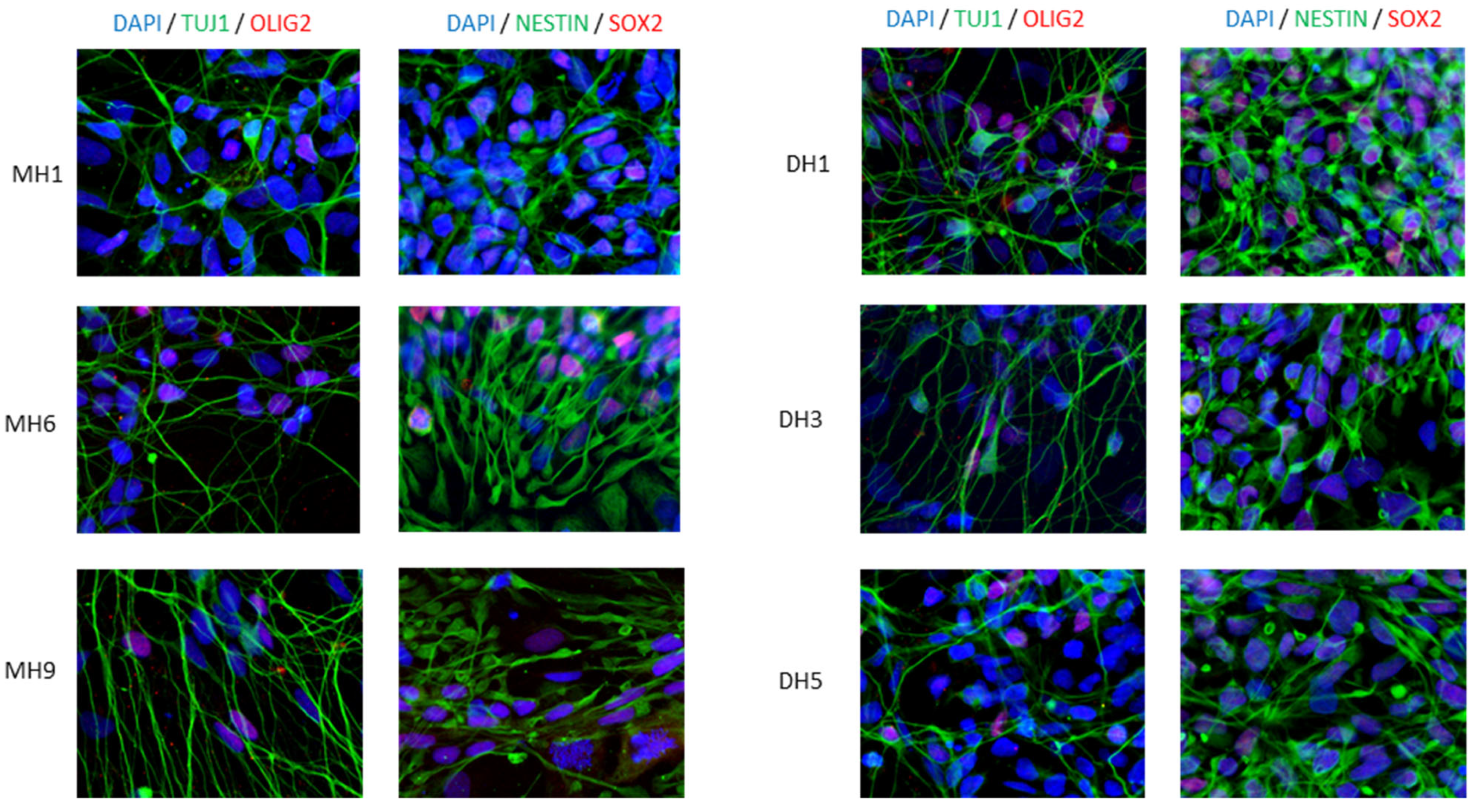
2.3. Immunofluorescence
2.4. Real Time PCR
2.5. Karyotype Analysis
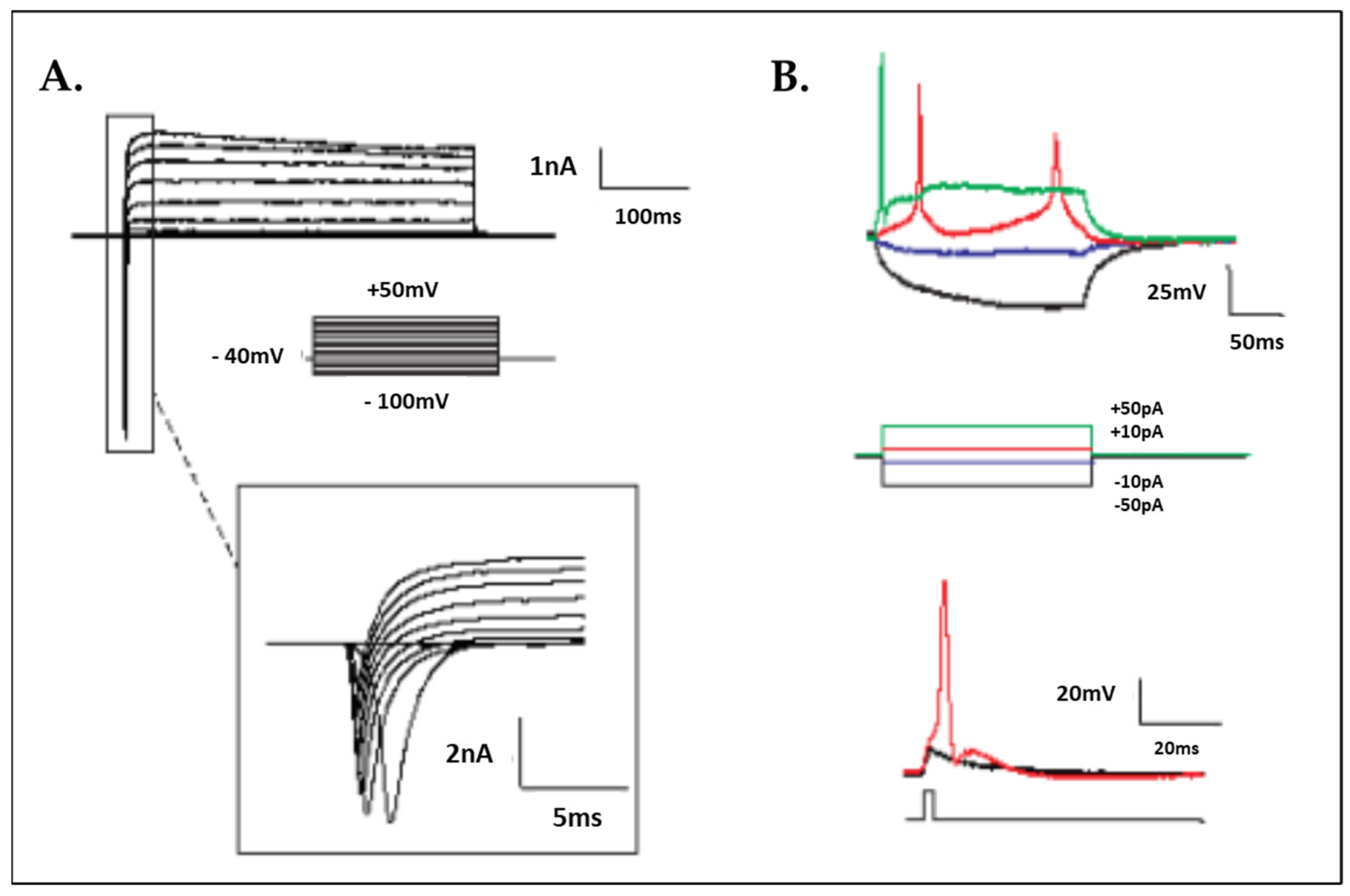
2.6. Electrophysiology
2.7. Western Blot
3. Results
3.1. Human iPSC Differentiation to Motor Neurons
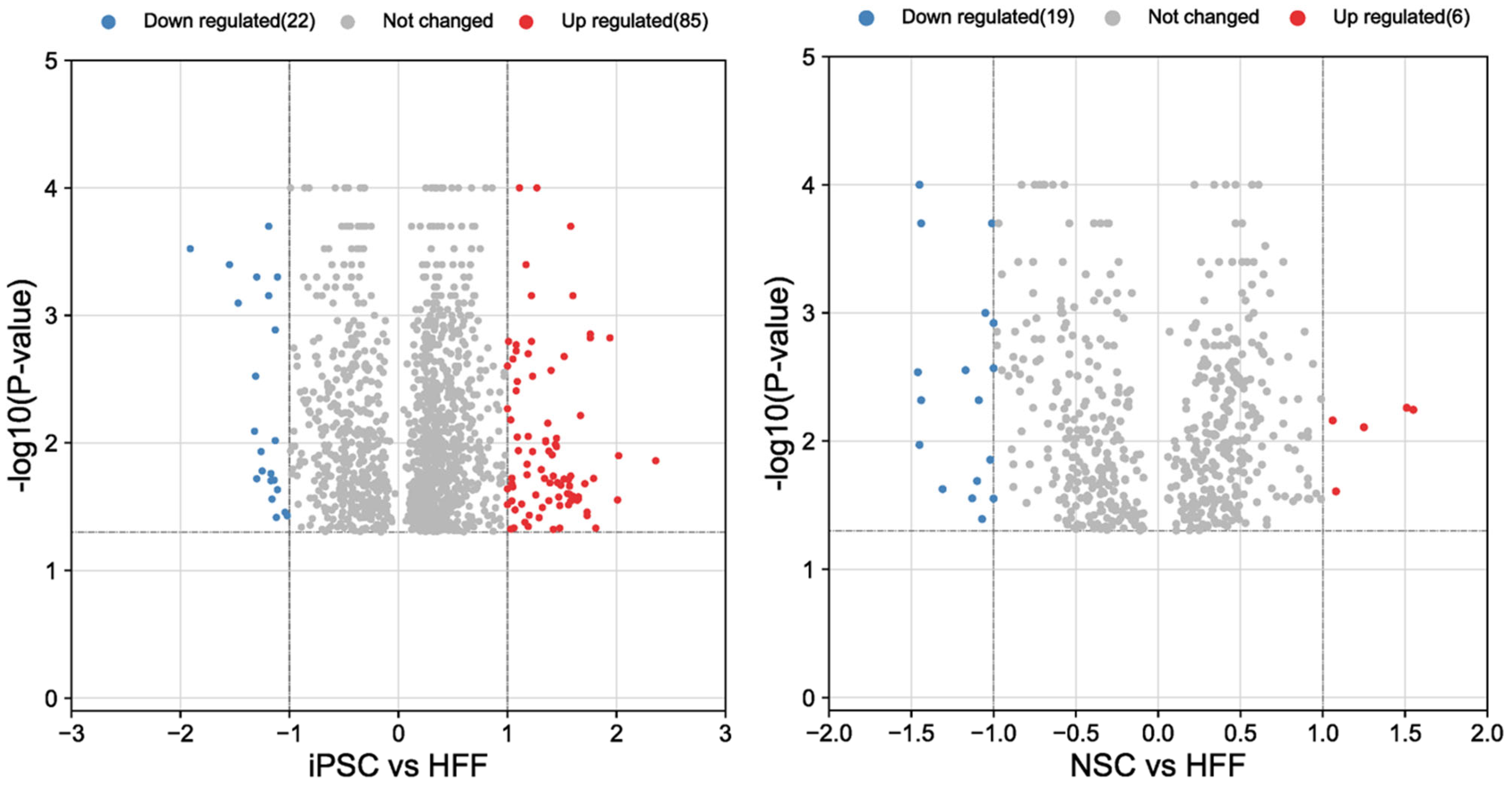
3.2. Results of Proteomic Analysis of HFFs, iPSCs and NSCs
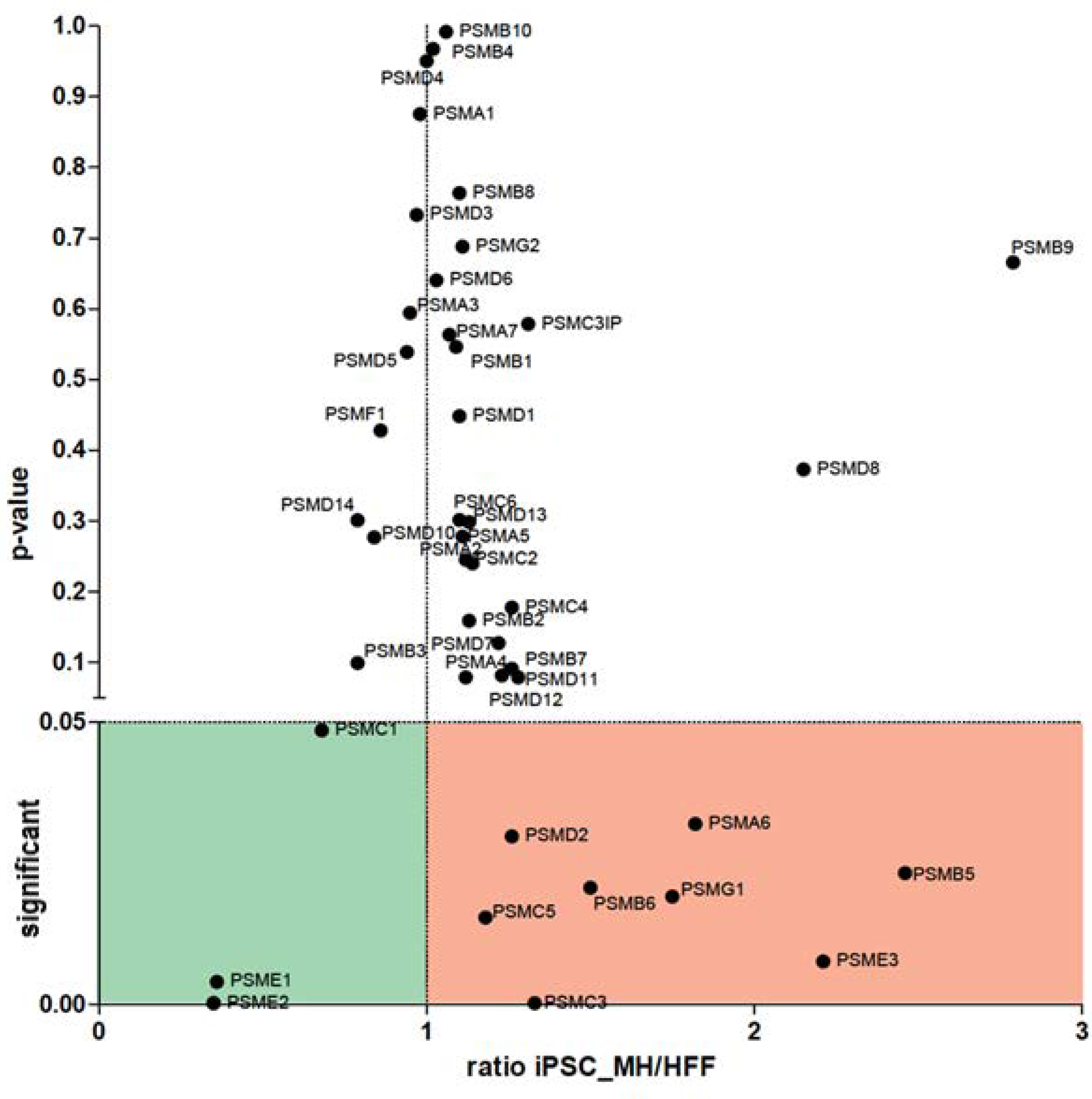
3.2.1. Analysis of HFFs vs. iPSCs
3.2.2. Analysis of HFFs Compared to iPSCs and NSCs
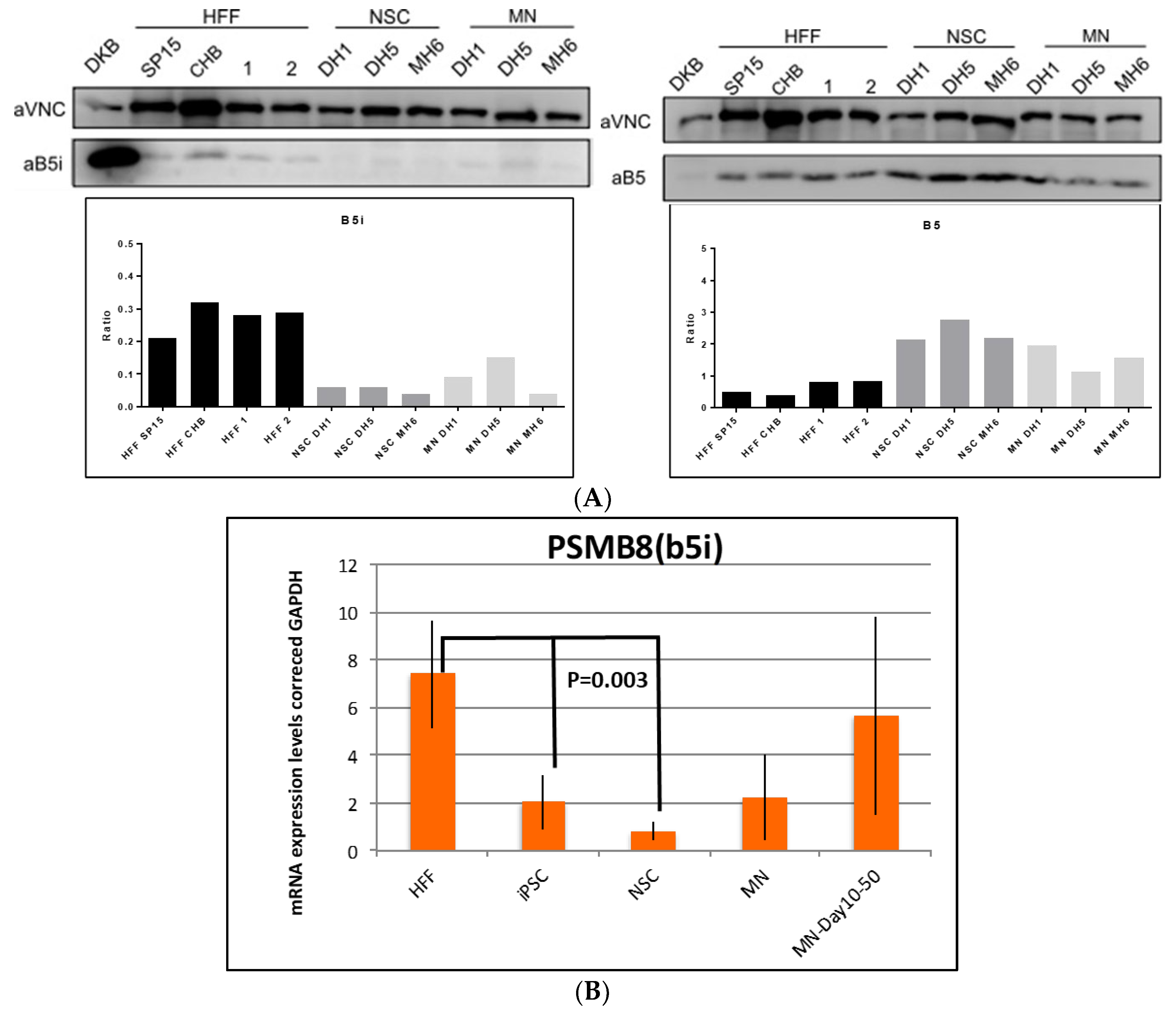
3.3. Validation of Constitutive and Immunoproteasome Findings
4. Discussion
4.1. Proteomics and Western Blot Data Validation in iPSCs and MNs
4.2. The Role of the UPS and MHC
4.3. MHC-I Expression in Disease
4.4. MHC Peptides Technique
4.5. Misfolded Proteins in Neuronal Disease
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Okada, Y.; Aoi, T.; Okada, A.; Takahashi, K.; Okita, K.; Nakagawa, M.; Koyanagi, M.; Tanabe, K.; Ohnuki, M.; et al. Variation in the safety of induced pluripotent stem cell lines. Nat. Biotechnol. 2009, 27, 743–745. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Palomo, A.B.; Requena-Osete, J.; Delgado-Morales, R.; Moreno-Manzano, V.; Grau-Bove, C.; Tejera, A.M.; Otero, M.J.; Barrot, C.; Santos-Barriopedro, I.; Vaquero, A.; et al. A Synthetic mRNA Cell Reprogramming Method Using CYCLIN D1 Promotes DNA repair, generating improved genetically stable human induced pluripotent stem cells. Stem Cells 2021, 39, 866–881. [Google Scholar] [CrossRef] [PubMed]
- Elmer, B.M.; McAllister, A.K. Major histocompatibility complex class I proteins in brain development and plasticity. Trends Neurosci. 2012, 35, 660–670. [Google Scholar] [CrossRef] [PubMed]
- Shatz, C.J. MHC Class I: An unexpected role in neuronal plasticity. Neuron 2009, 64, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Murata, S.; Sasaki, K.; Kishimoto, T.; Niwa, S.-I.; Hayashi, H.; Takahama, Y.; Tanaka, K. Regulation of CD8+ T cell development by thymus-specific proteasomes. Science 2007, 316, 1349–1353. [Google Scholar] [CrossRef] [PubMed]
- Murata, S.; Yashiroda, H.; Tanaka, K. Molecular mechanisms of proteasome assembly. Nat. Rev. Mol. Cell Biol. 2009, 10, 104–115. [Google Scholar] [CrossRef]
- Paul, S. Dysfunction of the ubiquitin–proteasome system in multiple disease conditions: Therapeutic approaches. BioEssays 2008, 30, 1172–1184. [Google Scholar] [CrossRef]
- Yerbury, J.J.; Ooi, L.; Dillin, A.; Saunders, D.N.; Hatters, D.M.; Beart, P.M.; Cashman, N.R.; Wilson, M.R.; Ecroyd, H. Walking the tightrope: Proteostasis and neurodegenerative disease. J. Neurochem. 2016, 137, 489–505. [Google Scholar] [CrossRef]
- Williams, K.L.; Topp, S.; Yang, S.; Smith, B.; Fifita, J.A.; Warraich, S.T.; Zhang, K.Y.; Farrawell, N.; Vance, C.; Hu, X.; et al. CCNF mutations in amyotrophic lateral sclerosis and frontotemporal dementia. Nat. Commun. 2016, 7, 11253. [Google Scholar] [CrossRef]
- Deng, H.X.; Chen, W.; Hong, S.T.; Boycott, K.M.; Gorrie, G.H.; Siddique, N.; Yang, Y.; Fecto, F.; Shi, Y.; Zhai, H.; et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature 2011, 477, 211–215. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.O.; Mandrioli, J.; Benatar, M.; Abramzon, Y.; Van Deerlin, V.M.; Trojanowski, J.Q.; Gibbs, J.R.; Brunetti, M.; Gronka, S.; Wuu, J.; et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron 2010, 68, 857–864. [Google Scholar] [CrossRef] [PubMed]
- Fecto, F.; Yan, J.; Vemula, S.P.; Liu, E.; Yang, Y.; Chen, W.; Zheng, J.G.; Shi, Y.; Siddique, N.; Arrat, H.; et al. SQSTM1 mutations in familial and sporadic amyotrophic lateral sclerosis. Arch. Neurol. 2011, 68, 1440–1446. [Google Scholar] [CrossRef] [PubMed]
- Dlamini, N.; Josifova, D.J.; Paine, S.M.; Wraige, E.; Pitt, M.; Murphy, A.J.; King, A.; Buk, S.; Smith, F.; Abbs, S.; et al. Clinical and neuropathological features of X-linked spinal muscular atrophy (SMAX2) associated with a novel mutation in the UBA1 gene. Neuromuscul. Disord. 2013, 23, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Ramser, J.; Ahearn, M.E.; Lenski, C.; Yariz, K.O.; Hellebrand, H.; von Rhein, M.; Clark, R.D.; Schmutzler, R.K.; Lichtner, P.; Hoffman, E.P.; et al. rare missense and synonymous variants in UBE1 are associated with x-linked infantile spinal muscular atrophy. Am. J. Hum. Genet. 2008, 82, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Brockington, A.; Ning, K.; Heath, P.R.; Wood, E.; Kirby, J.; Fusi, N.; Lawrence, N.; Wharton, S.B.; Ince, P.G.; Shaw, P.J. Unravelling the enigma of selective vulnerability in neurodegeneration: Motor neurons resistant to degeneration in ALS show distinct gene expression characteristics and decreased susceptibility to excitotoxicity. Acta Neuropathol. 2012, 125, 95–109. [Google Scholar] [CrossRef] [PubMed]
- Boegel, S.; Löwer, M.; Bukur, T.; Sorn, P.; Castle, J.C.; Sahin, U. HLA and proteasome expression body map. BMC Med. Genom. 2018, 11, 36. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Lepper, M.F.; Ohmayer, U.; von Toerne, C.; Maison, N.; Ziegler, A.G.; Hauck, S.M. Proteomic Landscape of Patient-Derived CD4+ T Cells in Recent-Onset Type 1 Diabetes. J. Proteome Res. 2018, 17, 618–634. [Google Scholar] [CrossRef]
- Aasen, T.; Raya, A.; Barrero, M.J.; Garreta, E.; Consiglio, A.; Gonzalez, F.; Vassena, R.; Bilić, J.; Pekarik, V.; Tiscornia, G.; et al. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat. Biotechnol. 2008, 26, 1276–1284. [Google Scholar] [CrossRef]
- Bax, M.; McKenna, J.; Do-Ha, D.; Stevens, C.H.; Higginbottom, S.; Balez, R.; Cabral-Da-Silva, M.e.C.; Farrawell, N.E.; Engel, M.; Poronnik, P.; et al. The ubiquitin proteasome system is a key regulator of pluripotent stem cell survival and motor neuron differentiation. Cells 2019, 8, 581. [Google Scholar] [CrossRef]
- Bendotti, C.; Marino, M.; Cheroni, C.; Fontana, E.; Crippa, V.; Poletti, A.; De Biasi, S. Dysfunction of constitutive and inducible ubiquitin-proteasome system in amyotrophic lateral sclerosis: Implication for protein aggregation and immune response. Prog. Neurobiol. 2012, 97, 101–126. [Google Scholar] [CrossRef]
- Nardo, G.; Iennaco, R.; Fusi, N.; Heath, P.R.; Marino, M.; Trolese, M.C.; Ferraiuolo, L.; Lawrence, N.; Shaw, P.J.; Bendotti, C. Transcriptomic indices of fast and slow disease progression in two mouse models of amyotrophic lateral sclerosis. Brain 2013, 136 Pt 11, 3305–3332. [Google Scholar] [CrossRef]
- Thibault, P.; Perreault, C. Immunopeptidomics: Reading the Immune Signal That Defines Self from Nonself. Mol. Cell. Proteom. 2022, 21, 100234. [Google Scholar] [CrossRef]
- Nardo, G.; Trolese, M.C.; Bendotti, C. Major Histocompatibility Complex I Expression by Motor Neurons and Its Implication in Amyotrophic Lateral Sclerosis. Front. Neurol. 2016, 7, 89. [Google Scholar] [CrossRef] [PubMed]
| All Human Cells | Cell List | Cell Type | Passage | Assay |
|---|---|---|---|---|
| Fibroblasts | HFF1 | Human forskin fibroblast | p2 | Proteomic & Western Blot |
| (4 samples) | HFF2 | Human forskin fibroblast | p3 | Proteomic & Western Blot |
| SP15 | Human skin fibroblast | p4 | Proteomic & Western Blot | |
| CHB | Human skin fibroblast | p4 | Proteomic & Western Blot | |
| iPSC | DH1 | synthetic mRNA made iPSC | p9 | Proteomic & Western Blot |
| (8 samples) | DH3 | synthetic mRNA made iPSC | p9 | Proteomic & Western Blot |
| DH5 | synthetic mRNA made iPSC | p11 | Proteomic & Western Blot | |
| MH1 | synthetic mRNA made iPSC | p11 | Proteomic & Western Blot | |
| MH6 | synthetic mRNA made iPSC | p12 | Proteomic & Western Blot | |
| MH9 | synthetic mRNA made iPSC | p12 | Proteomic & Western Blot | |
| iPSC1 | Retroviral made iPSC | p10 | Western Blot | |
| iPSC2 | Retroviral made iPSC | p10 | Western Blot | |
| NSC | DH1 | derived neural stem cells | p4 | Proteomic & Western Blot |
| (3 samples) | DH3 | derived neural stem cells | p4 | Proteomic & Western Blot |
| DH5 | derived neural stem cells | p4 | Proteomic & Western Blot | |
| Motor Neurons | DH1 | Derived Motor Neurons | p3 | Western Blot |
| (3 samples) | DH3 | Derived Motor Neurons | p3 | Western Blot |
| DH5 | Derived Motor Neurons | p3 | Western Blot |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Álvarez, I.; Tirado-Herranz, A.; Alvarez-Palomo, B.; Osete, J.R.; Edel, M.J. Proteomic Analysis of Human iPSC-Derived Neural Stem Cells and Motor Neurons Identifies Proteasome Structural Alterations. Cells 2023, 12, 2800. https://doi.org/10.3390/cells12242800
Álvarez I, Tirado-Herranz A, Alvarez-Palomo B, Osete JR, Edel MJ. Proteomic Analysis of Human iPSC-Derived Neural Stem Cells and Motor Neurons Identifies Proteasome Structural Alterations. Cells. 2023; 12(24):2800. https://doi.org/10.3390/cells12242800
Chicago/Turabian StyleÁlvarez, Iñaki, Adrián Tirado-Herranz, Belén Alvarez-Palomo, Jordi Requena Osete, and Michael J. Edel. 2023. "Proteomic Analysis of Human iPSC-Derived Neural Stem Cells and Motor Neurons Identifies Proteasome Structural Alterations" Cells 12, no. 24: 2800. https://doi.org/10.3390/cells12242800
APA StyleÁlvarez, I., Tirado-Herranz, A., Alvarez-Palomo, B., Osete, J. R., & Edel, M. J. (2023). Proteomic Analysis of Human iPSC-Derived Neural Stem Cells and Motor Neurons Identifies Proteasome Structural Alterations. Cells, 12(24), 2800. https://doi.org/10.3390/cells12242800






