Measuring Single-Cell Calcium Dynamics Using a Myofilament-Localized Optical Biosensor in hiPSC-CMs Derived from DCM Patients
Abstract
1. Introduction
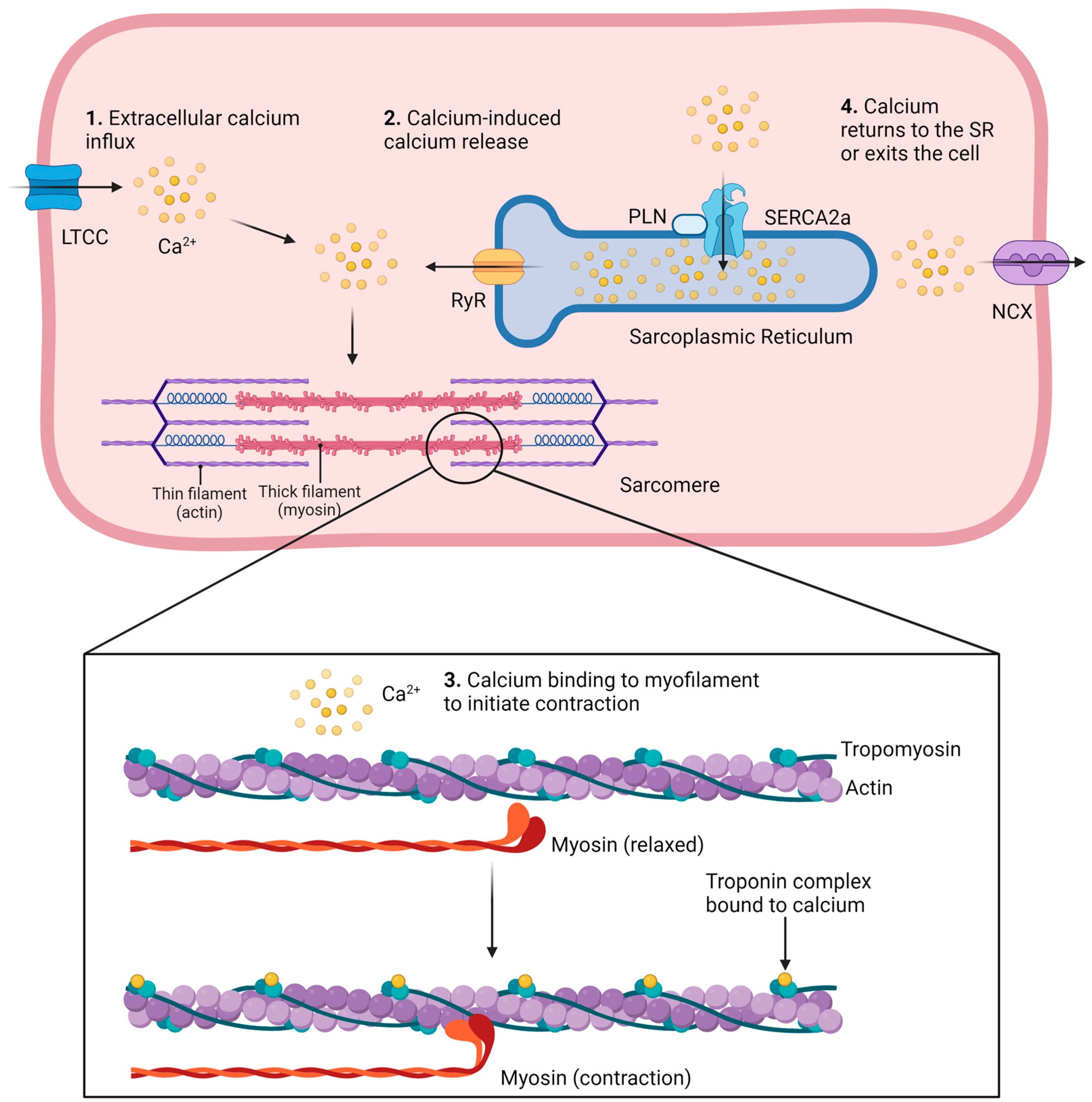
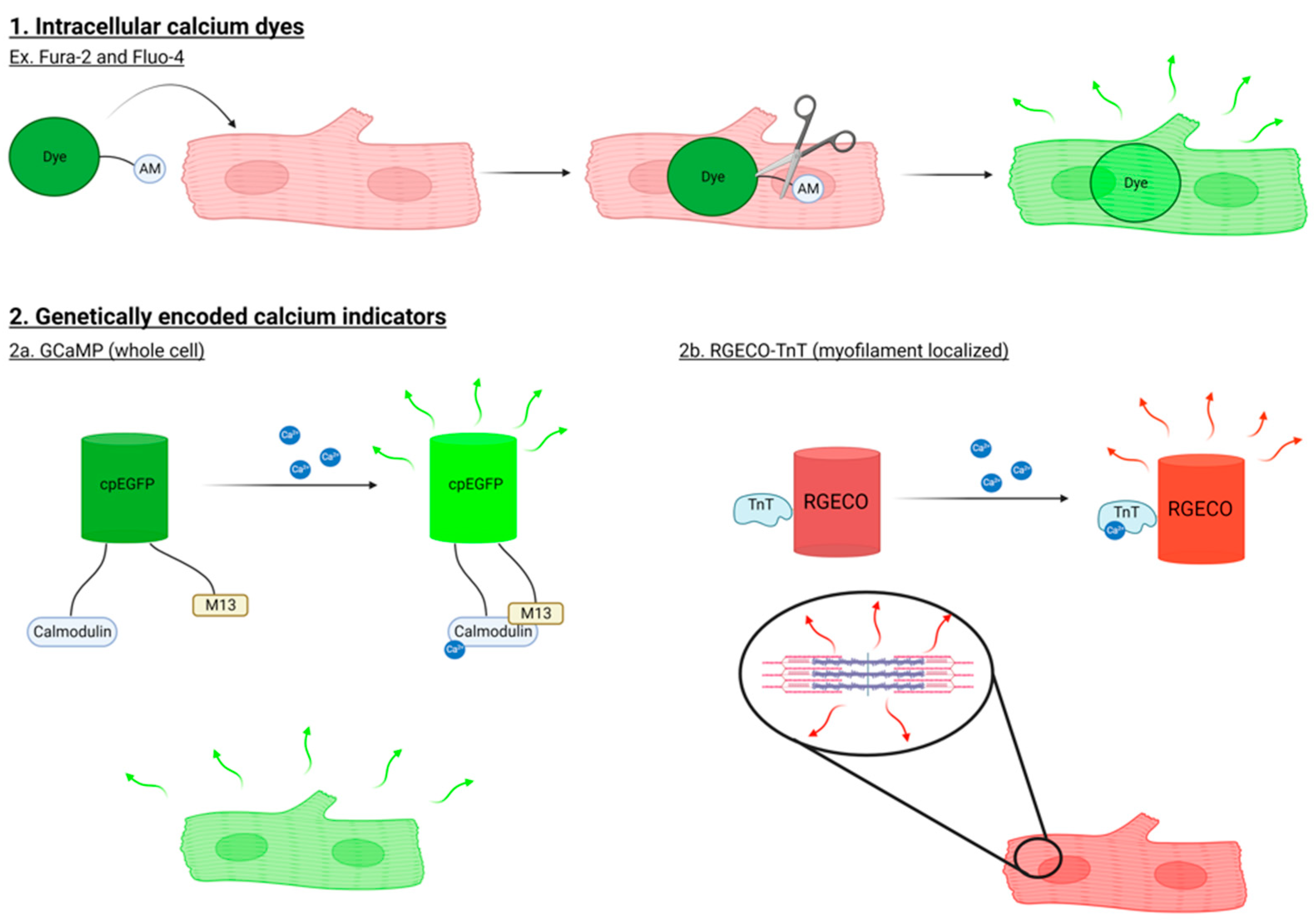
1.1. Methods to Study Ca2+ Signaling in Cardiomyocytes
1.2. Analyzing Calcium Transients Derived from hiPSC-CM Monolayers
2. Materials and Methods
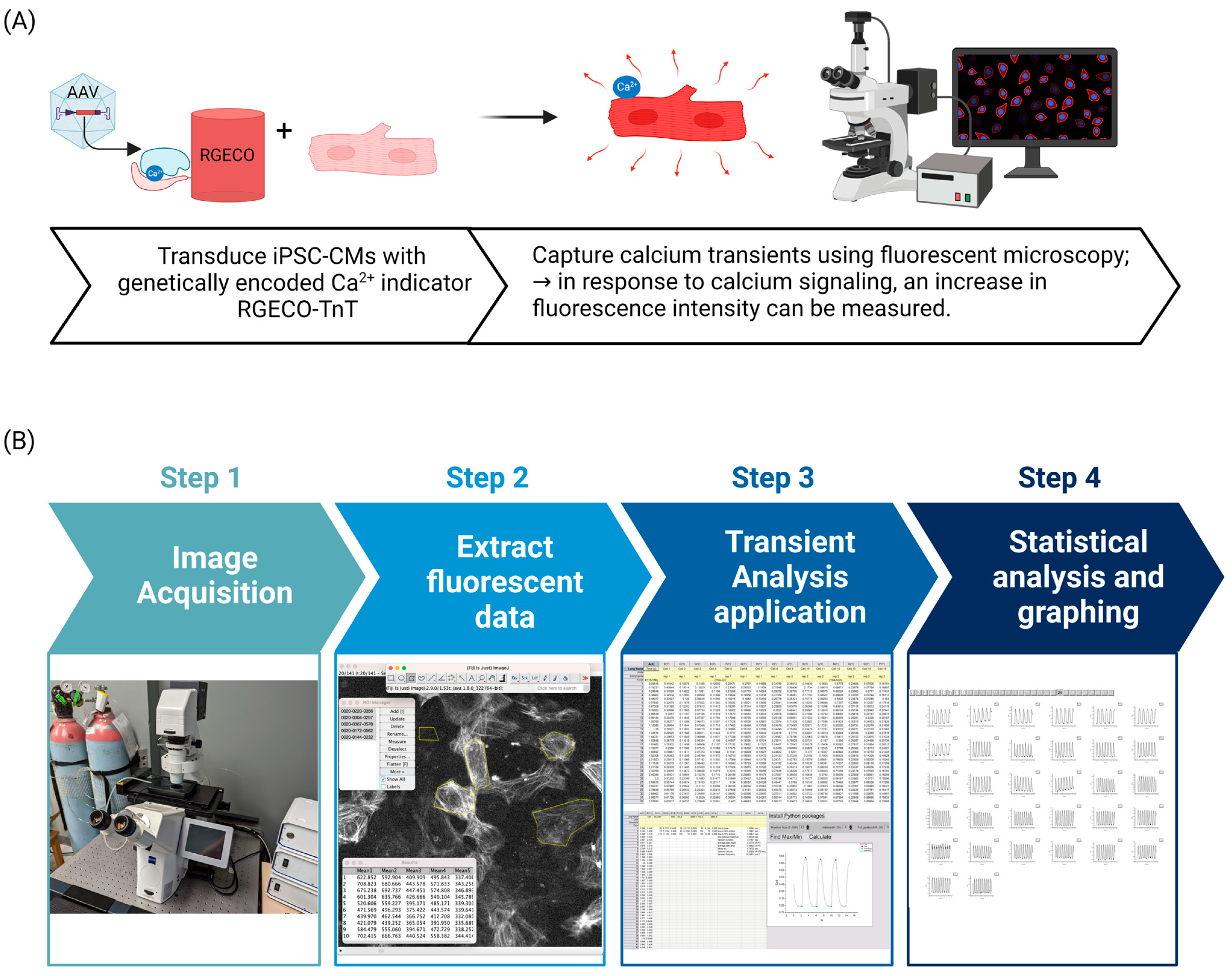
2.1. Transduction of hiPSC-CMs with AAV2/6-RGECO-TnT
- Refresh media in 96-well plate with 70 μL of warmed RPMI + B27.
- Prepare a low retention 1.5 mL tube with enough media to add 10 μL per well containing cells to be transduced (i.e., 100 μL for 9 wells, account for pipette error).
- Thaw an aliquot of AAV2/6-RGECO-TnT on ice.
- Determine the volume of virus required to infect the cells with a MOI of 5000 for all wells and transfer to the prepared 1.5 mL tube containing media using a filter tip. Mix by inverting the Eppendorf gently 3–4 times and return the tube to the ice.
- Immediately add 10 μL of virus dilution per well.
- Incubate the dish with the virus for 5 days, replacing the media with 100 μL RPMI + B27 3 days after transduction.
- On the day of the experiment, replace media with RPMI1640 (no phenol red) supplemented with B27 (with insulin).
2.2. Live Cell Ca2+ Imaging of hiPSC-CM Monolayers
- Ensure a Zeiss Filter Set 14 is installed (510–560 nm excitation, 590 nm emission, and 580 dichroic mirror).
- Enable high-speed imaging according to the manufacturer’s recommendation. Some examples to improve speed on the Axio Observer include using the GPO 0 trigger cable, on Zen Blue enable “Trigger Out” for live, and set shutter open delay to 0.00 ms. Binning may be used to improve fluorescent signal; for example, binning can be set to 3 × 3.
- In the software, under “Imaging Setup” select the appropriate filter set and set the objective lens to 20X/0.8 (ZEISS 20x PLAN APOCHROMAT, NA 0.8).
- Under time series, set to record for 15 s and enable “Camera streaming if possible” to improve speed of acquisition.
- Using “Imaging Setup” on the computer software, or the touch screen panel on the microscope, ensure the correct objective and filter set are selected. Use TL Illumination to see the cells under brightfield and RL Illumination to see the cells under fluorescent lighting. Do not have TL Illumination on during fluorescent recording.
- Allow hiPSC-CMs to stabilize in the chambered plate holder for a minimum of 15 min before acquiring measurements.
- To minimize the amount of time cells are exposed to fluorescence, first find cells under TL Illumination. Use the stage controller joystick to move the plate and focus on the cells. Once cells are found, using Zen Blue software under the Acquisition tab, select “Live” to see a live stream of the cells. Ensure the cells are in focus before selecting “Start Experiment”.
- Image stacks are recorded as .czi files that can be opened in ImageJ with the Bio-Formats plug-in.
2.3. Image Analysis
2.3.1. Extracting Fluorescence Intensities of hiPSC-CM Monolayers in Image J
- Open files from file explorer or through ImageJ (File > Open…).
- Once all images have been opened, under the “Image tab”, select “Adjust > Brightness/Contrast”. Then select “Auto” followed by “Set” and select “Propagate to all other open images” to adjust the brightness and contrast of all images simultaneously.
- Using the “Polygon” or “Freehand” selections tools, trace individual cells and press the “T” key on the keyboard following every cell to add the selected cell as a new region of interest, or ROI, to the ROI manager. Remember to select the background with no cells as the final ROI, as this is required for normalization purposes.
- In the ROI manager, select “More” >> “Multi Measure”. Ensure “Measure all slices” and “One row per slice” are selected. This will generate a table containing the fluorescent values for all ROIs selected in the image. The information found in this table can be transferred to OriginPro 2022 for further analysis.
- To perform calculations, first select the green lock in the top corner of A (X) and select “Recalculation Mode” >> “None” and select “OK” in the dialog box.
- Under column A, transform frame number to time by typing A * (time of recording/number of frames) in “F (x) = X (i.e., A * (15/156)).
- Normalize cell fluorescence intensities to background intensity by writing in F (x) = (cell column or “this” – background column)/background column (i.e., (B-F)/F or (this – F)/F).
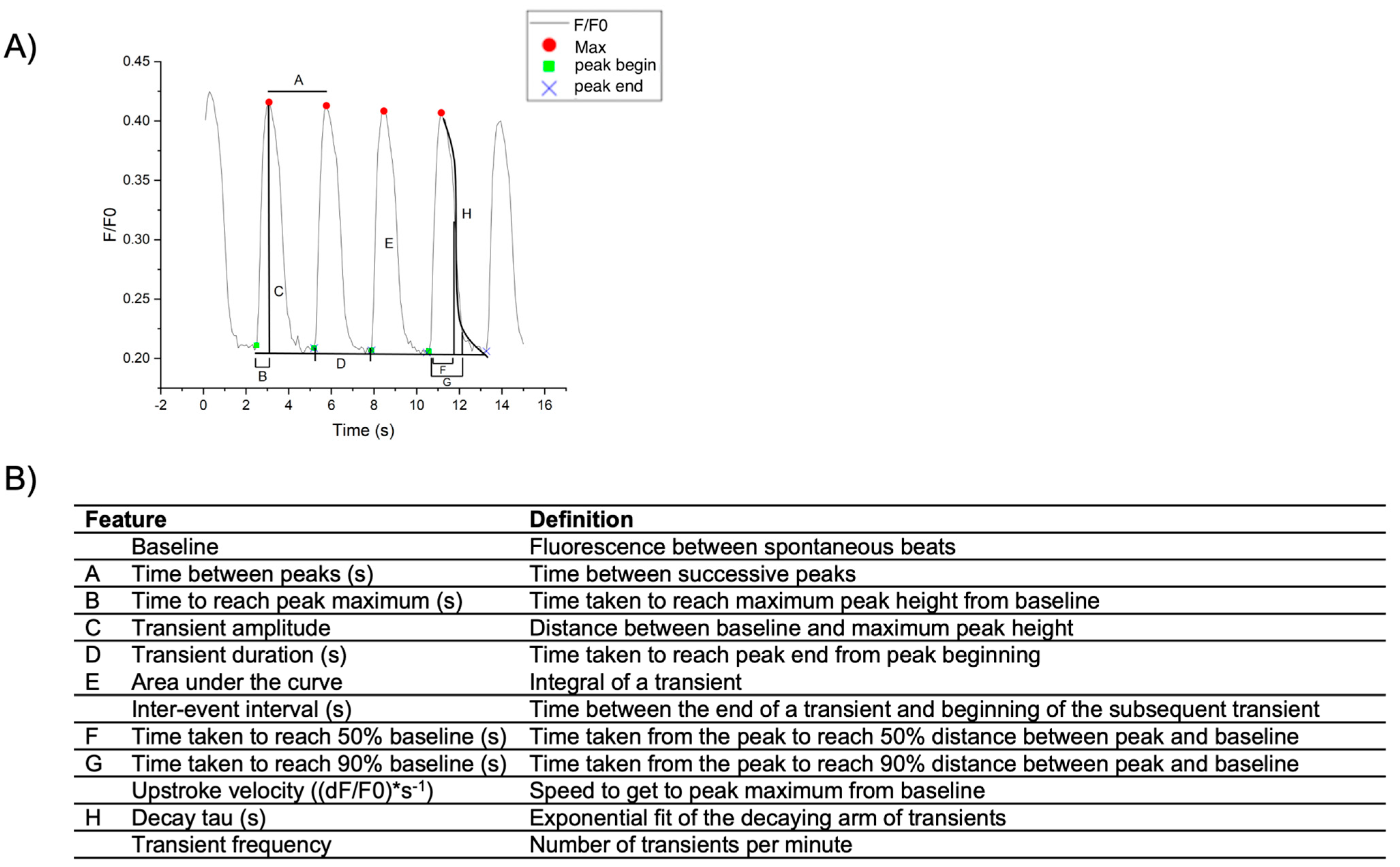
2.3.2. Calcium Transient Analysis of hiPSC-CM Monolayers Using OriginPro
2.3.3. Statistical Analysis
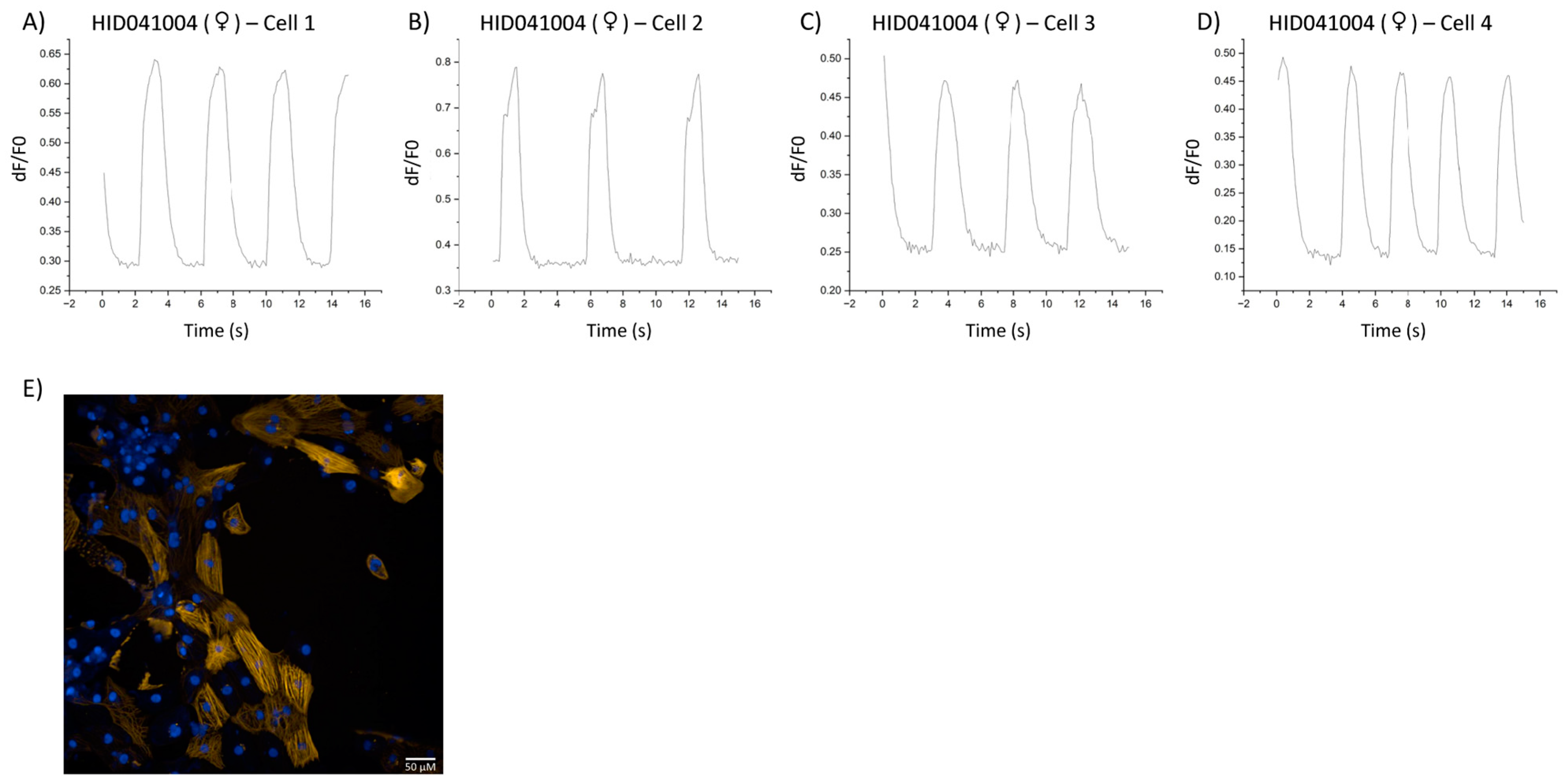
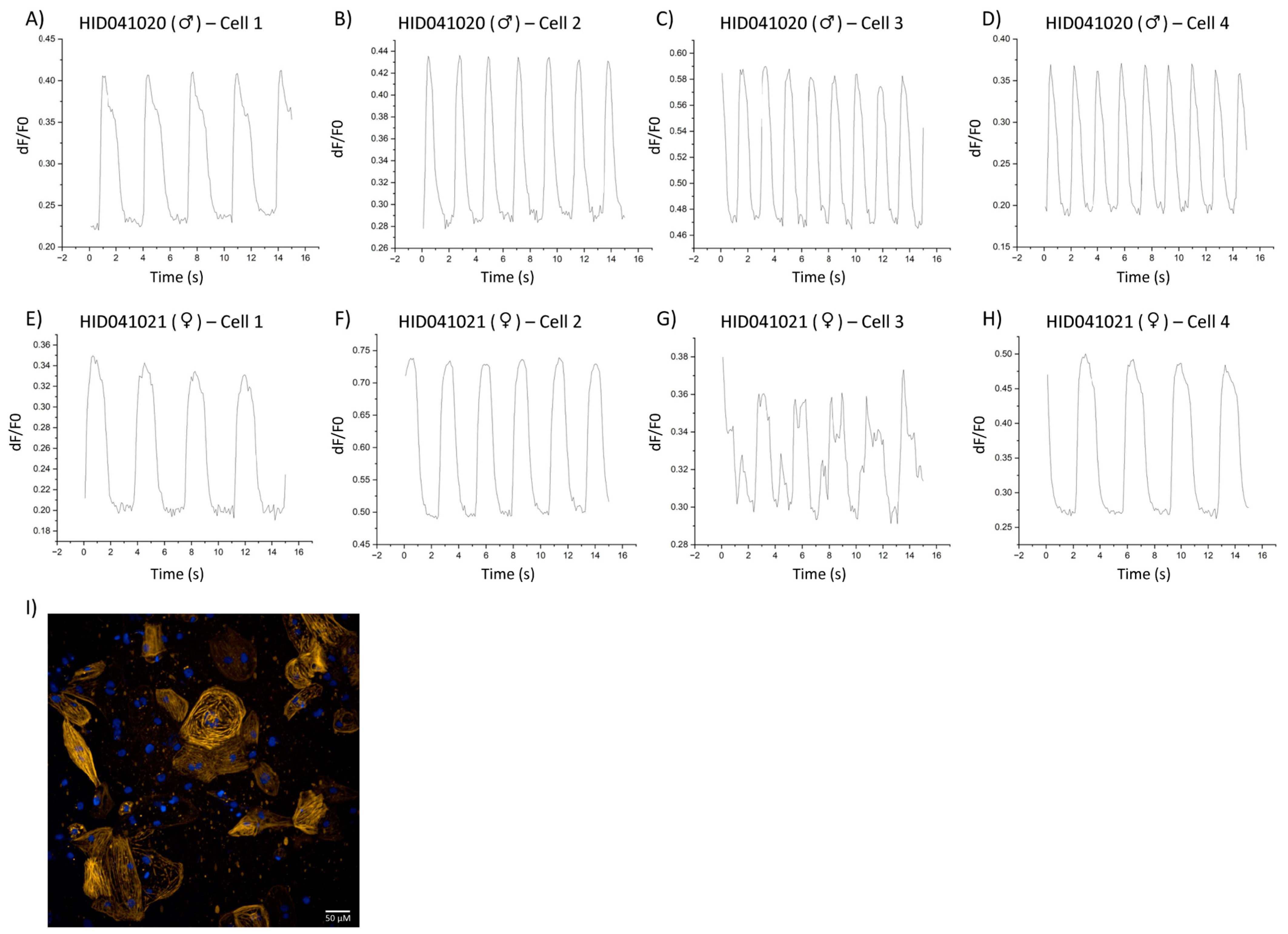
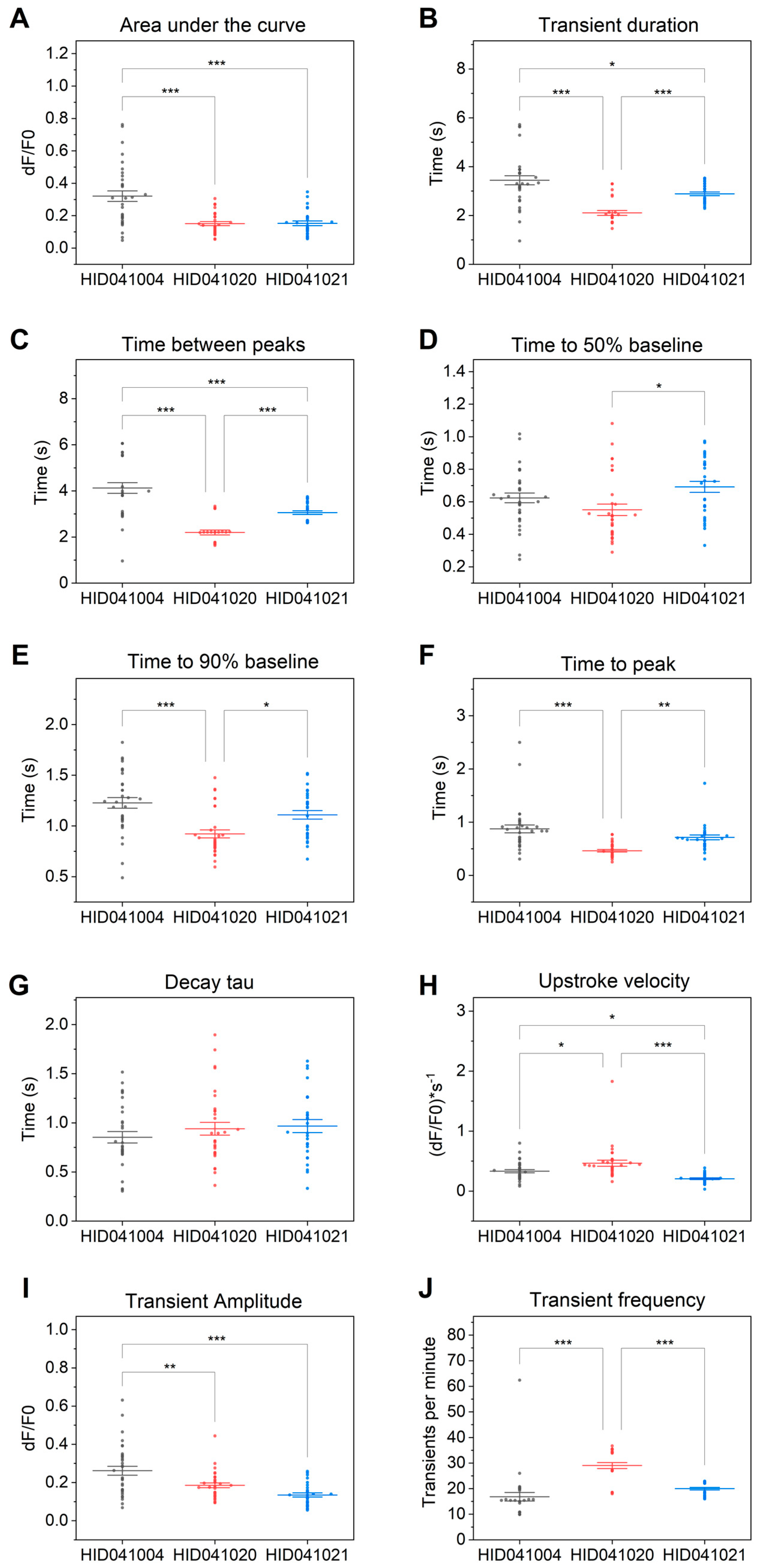
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wyles, S.P.; Li, X.; Hrstka, S.C.; Reyes, S.; Oommen, S.; Beraldi, R.; Edwards, J.; Terzic, A.; Olson, T.M.; Nelson, T.J. Modeling structural and functional deficiencies of RBM20 familial dilated cardiomyopathy using human induced pluripotent stem cells. Hum. Mol. Genet. 2016, 25, 254–265. [Google Scholar] [CrossRef] [PubMed]
- Schultheiss, H.P.; Fairweather, D.; Caforio, A.L.; Escher, F.; Hershberger, R.E.; Lipshultz, S.E.; Liu, P.P.; Matsumori, A.; Mazzanti, A.; McMurray, J.; et al. Dilated cardiomyopathy. Nat. Rev. Dis. Primers 2019, 5, 32. [Google Scholar] [CrossRef] [PubMed]
- Rosenbaum, A.N.; Agre, K.E.; Pereira, N.L. Genetics of dilated cardiomyopathy: Practical implications for heart failure management. Nat. Rev. Cardiol. 2020, 17, 286–297. [Google Scholar] [CrossRef] [PubMed]
- Hershberger, R.E.; Morales, A.; Siegfried, J.D. Clinical and genetic issues in dilated cardiomyopathy: A review for genetics professionals. Genet. Med. 2010, 12, 655–667. [Google Scholar] [CrossRef]
- Badone, B.; Ronchi, C.; Lodola, F.; Knaust, A.E.; Hansen, A.; Eschenhagen, T.; Zaza, A. Characterization of the PLN p.Arg14del Mutation in Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Int. J. Mol. Sci. 2021, 22, 13500. [Google Scholar] [CrossRef]
- Dai, Y.; Amenov, A.; Ignatyeva, N.; Koschinski, A.; Xu, H.; Soong, P.L.; Tiburcy, M.; Linke, W.A.; Zaccolo, M.; Hasenfuss, G.; et al. Troponin destabilization impairs sarcomere-cytoskeleton interactions in iPSC-derived cardiomyocytes from dilated cardiomyopathy patients. Sci. Rep. 2020, 10, 209. [Google Scholar] [CrossRef]
- Briganti, F.; Sun, H.; Wei, W.; Wu, J.; Zhu, C.; Liss, M.; Karakikes, I.; Rego, S.; Cipriano, A.; Snyder, M.; et al. iPSC Modeling of RBM20-Deficient DCM Identifies Upregulation of RBM20 as a Therapeutic Strategy. Cell Rep. 2020, 32, 108117. [Google Scholar] [CrossRef]
- Wyles, S.P.; Hrstka, S.; Reyes, S.; Terzic, A.; Olson, T.; Nelson, T. Pharmacological Modulation of Calcium Homeostasis in Familial Dilated Cardiomyopathy: An In Vitro Analysis From an RBM20 Patient-Derived iPSC Model. Clin. Transl. Sci. 2016, 9, 158–167. [Google Scholar] [CrossRef]
- Stroik, D.R.; Ceholski, D.K.; Bidwell, P.A.; Mleczko, J.; Thanel, P.F.; Kamdar, F.; Autry, J.M.; Cornea, R.L.; Thomas, D.D. Viral expression of a SERCA2a-activating PLB mutant improves calcium cycling and synchronicity in dilated cardiomyopathic hiPSC-CMs. J. Mol. Cell Cardiol. 2020, 138, 59–65. [Google Scholar] [CrossRef]
- Lenzi, F.; Caniggia, A. Nature of myocardial contraction and of action potentials; importance of the cationic gradient. Acta Med. Scand. 1953, 146, 300–312. [Google Scholar] [CrossRef]
- Reddy, Y.S.; Honig, C.R. Ca2+-binding and Ca2+-sensitizing functions of cardiac native tropomyosin, troponin, and tropomyosin. Biochim. Biophys. Acta 1972, 275, 453–463. [Google Scholar] [CrossRef]
- Meyer, M.; Schillinger, W.; Pieske, B.; Holubarsch, C.; Heilmann, C.; Posival, H.; Kuwajima, G.; Mikoshiba, K.; Just, H.; Hasenfuss, G. Alterations of sarcoplasmic reticulum proteins in failing human dilated cardiomyopathy. Circulation 1995, 92, 778–784. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, J.P.; Kamisago, M.; Asahi, M.; Li, G.H.; Ahmad, F.; Mende, U.; Kranias, E.G.; MacLennan, D.H.; Seidman, J.G.; Seidman, C.E. Dilated cardiomyopathy and heart failure caused by a mutation in phospholamban. Science 2003, 299, 1410–1413. [Google Scholar] [CrossRef] [PubMed]
- Law, M.L.; Cohen, H.; Martin, A.A.; Angulski, A.B.B.; Metzger, J.M. Dysregulation of Calcium Handling in Duchenne Muscular Dystrophy-Associated Dilated Cardiomyopathy: Mechanisms and Experimental Therapeutic Strategies. J. Clin. Med. 2020, 9, 520. [Google Scholar] [CrossRef] [PubMed]
- Zanin, S.; Lidron, E.; Rizzuto, R.; Pallafacchina, G. Methods to Measure Intracellular Ca(2+) Concentration Using Ca(2+)-Sensitive Dyes. Methods Mol. Biol. 2019, 1925, 43–58. [Google Scholar]
- Di Virgilio, F.; Jiang, L.-H.; Falzoni, S.R.S.; Sarti, A.C.; Vultaggio-Poma, V.; Chiozzi, P.; Adinolfi, E. Structure, function and techniques of investigation of the P2 × 7 receptor (P2 × 7R) in mammalian cells. Methods Enzymol. 2019, 629, 115–150. [Google Scholar]
- Nakai, J.; Ohkura, M.; Imoto, K. A high signal-to-noise Ca(2+) probe composed of a single green fluorescent protein. Nat. Biotechnol. 2001, 19, 137–141. [Google Scholar] [CrossRef]
- Tallini, Y.N.; Ohkura, M.; Choi, B.-R.; Ji, G.; Imoto, K.; Doran, R.; Lee, J.; Plan, P.; Wilson, J.; Xin, H.-B.; et al. Imaging cellular signals in the heart in vivo: Cardiac expression of the high-signal Ca2+ indicator GCaMP2. Proc. Natl. Acad. Sci. USA 2006, 103, 4753–4758. [Google Scholar] [CrossRef]
- Tian, L.; Hires, S.A.; Mao, T.; Huber, D.; Chiappe, M.E.; Chalasani, S.H.; Petreanu, L.; Akerboom, J.; McKinney, S.A.; Schreiter, E.R.; et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat. Methods 2009, 6, 875–881. [Google Scholar] [CrossRef] [PubMed]
- Akerboom, J.; Chen, T.-W.; Wardill, T.J.; Tian, L.; Marvin, J.S.; Mutlu, S.; Calderón, N.C.; Esposti, F.; Borghuis, B.G.; Sun, X.R.; et al. Optimization of a GCaMP calcium indicator for neural activity imaging. J. Neurosci. 2012, 32, 13819–13840. [Google Scholar] [CrossRef]
- Inoue, M. Genetically encoded calcium indicators to probe complex brain circuit dynamics in vivo. Neurosci. Res. 2021, 169, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Araki, S.; Wu, J.; Teramoto, T.; Chang, Y.F.; Nakano, M.; Abdelfattah, A.S.; Fujiwara, M.; Ishihara, T.; Nagai, T.; et al. An expanded palette of genetically encoded Ca(2)(+) indicators. Science 2011, 333, 1888–1891. [Google Scholar] [CrossRef] [PubMed]
- Reddish, F.N.; Miller, C.L.; Deng, X.; Dong, B.; Patel, A.A.; Ghane, M.A.; Mosca, B.; McBean, C.; Wu, S.; Solntsev, K.M.; et al. Rapid subcellular calcium responses and dynamics by calcium sensor G-CatchER+. iScience 2021, 24, 102129. [Google Scholar] [CrossRef]
- Kaestner, L.; Scholz, A.; Tian, Q.; Ruppenthal, S.; Tabellion, W.; Wiesen, K.; Katus, H.A.; Müller, O.J.; Kotlikoff, M.I.; Lipp, P.; et al. Genetically encoded Ca2+ indicators in cardiac myocytes. Circ. Res. 2014, 114, 1623–1639. [Google Scholar] [CrossRef]
- Sparrow, A.J.; Sievert, K.; Patel, S.; Chang, Y.-F.; Broyles, C.N.; Brook, F.A.; Watkins, H.; Geeves, M.A.; Redwood, C.S.; Robinson, P.; et al. Measurement of Myofilament-Localized Calcium Dynamics in Adult Cardiomyocytes and the Effect of Hypertrophic Cardiomyopathy Mutations. Circ. Res. 2019, 124, 1228–1239. [Google Scholar] [CrossRef] [PubMed]
- Greensmith, D.J. Ca analysis: An Excel based program for the analysis of intracellular calcium transients including multiple, simultaneous regression analysis. Comput. Methods Programs Biomed. 2014, 113, 241–250. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Velez Rueda, A.J.; Gonano, L.A.; Smith, A.G.; Parisi, G.; Fornasari, M.S.; Sommese, L.M. CardIAP: Calcium transients confocal image analysis tool. Front. Bioinform. 2023, 3, 1137815. [Google Scholar] [CrossRef]
- Penttinen, K.; Siirtola, H.; Àvalos-Salguero, J.; Vainio, T.; Juhola, M.; Aalto-Setälä, K. Novel Analysis Software for Detecting and Classifying Ca2+ Transient Abnormalities in Stem Cell-Derived Cardiomyocytes. PLoS ONE 2015, 10, e0135806. [Google Scholar] [CrossRef] [PubMed]
- Psaras, Y.; Margara, F.; Cicconet, M.; Sparrow, A.J.; Repetti, G.G.; Schmid, M.; Steeples, V.; Wilcox, J.A.; Bueno-Orovio, A.; Redwood, C.S.; et al. CalTrack: High-Throughput Automated Calcium Transient Analysis in Cardiomyocytes. Circ. Res. 2021, 129, 326–341. [Google Scholar] [CrossRef]
- Jung, P.; Seibertz, F.; Fakuade, F.E.; Ignatyeva, N.; Sampathkumar, S.; Ritter, M.; Li, H.; Mason, F.E.; Ebert, A.; Voigt, N. Increased cytosolic calcium buffering contributes to a cellular arrhythmogenic substrate in iPSC-cardiomyocytes from patients with dilated cardiomyopathy. Basic. Res. Cardiol. 2022, 117, 5. [Google Scholar] [CrossRef]
- Streckfuss-Bomeke, K.; Tiburcy, M.; Fomin, A.; Luo, X.; Li, W.; Fischer, C.; Özcelik, C.; Perrot, A.; Sossalla, S.; Haas, J.; et al. Severe DCM phenotype of patient harboring RBM20 mutation S635A can be modeled by patient-specific induced pluripotent stem cell-derived cardiomyocytes. J. Mol. Cell Cardiol. 2017, 113, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Sun, N.; Yazawa, M.; Liu, J.; Han, L.; Sanchez-Freire, V.; Abilez, O.J.; Navarrete, E.G.; Hu, S.; Wang, L.; Lee, A.; et al. Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci. Transl. Med. 2012, 4, 130ra47. [Google Scholar] [CrossRef]
- Liu, G.S.; Morales, A.; Vafiadaki, E.; Lam, C.K.; Cai, W.F.; Haghighi, K.; Adly, G.; Hershberger, R.E.; Kranias, E.G. A novel human R25C-phospholamban mutation is associated with super-inhibition of calcium cycling and ventricular arrhythmia. Cardiovasc. Res. 2015, 107, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Bellinger, A.M.; Reiken, S.; Carlson, C.; Mongillo, M.; Liu, X.; Rothman, L.; Matecki, S.; Lacampagne, A.; Marks, A.R. Hypernitrosylated ryanodine receptor calcium release channels are leaky in dystrophic muscle. Nat. Med. 2009, 15, 325–330. [Google Scholar] [CrossRef]
- Lian, X.; Zhang, J.; Azarin, S.M.; Zhu, K.; Hazeltine, L.B.; Bao, X.; Hsiao, C.; Kamp, T.J.; Palecek, S.P. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/beta-catenin signaling under fully defined conditions. Nat. Protoc. 2013, 8, 162–175. [Google Scholar] [CrossRef] [PubMed]
| If the number of peaks is not accurately recognized. | Lower Window Size to recognize more peaks and raise Window Size to recognize less peaks |
| If the beginning of the transient is not accurately recognized. | Lower Tol_peakstart to shift green indicators to the left and raise Tol_peakstart to shift green indicators to the right |
| If the end of the transient is not accurately recognized. | Lower Tol_peakend to shift blue x indicators to the left and raise Tol_peakend to shift blue x indicators to the right. If the final transient is incomplete, delete the last point from the spreadsheet. |
| If one indicator is out of place while the others are accurate. | Select on the indicator, select Data at the top of the screen, and select Move Data Points. Select “Yes” in the dialog box. Select and drag the indicator to the desired position then return to the workbook. |
| For transients that are noisy (i.e., poor signal intensity during acquisition). | A smoothing filter may be applied. From the tab with the transient to be smoothed, use the top toolbar to select “Analysis” > “Signal Processing” > “Smooth”. Open a new dialog window. Set recalculate to “Manual” and select column B (Y1). Use the desired smoothing method, for example FFT Filter. For output select <input>: XY = input XY to replace the original data with the smooth filtered data. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hawey, C.; Bourque, K.; Alim, K.; Derish, I.; Rody, E.; Khan, K.; Gendron, N.; Cecere, R.; Giannetti, N.; Hébert, T.E. Measuring Single-Cell Calcium Dynamics Using a Myofilament-Localized Optical Biosensor in hiPSC-CMs Derived from DCM Patients. Cells 2023, 12, 2526. https://doi.org/10.3390/cells12212526
Hawey C, Bourque K, Alim K, Derish I, Rody E, Khan K, Gendron N, Cecere R, Giannetti N, Hébert TE. Measuring Single-Cell Calcium Dynamics Using a Myofilament-Localized Optical Biosensor in hiPSC-CMs Derived from DCM Patients. Cells. 2023; 12(21):2526. https://doi.org/10.3390/cells12212526
Chicago/Turabian StyleHawey, Cara, Kyla Bourque, Karima Alim, Ida Derish, Elise Rody, Kashif Khan, Natalie Gendron, Renzo Cecere, Nadia Giannetti, and Terence E. Hébert. 2023. "Measuring Single-Cell Calcium Dynamics Using a Myofilament-Localized Optical Biosensor in hiPSC-CMs Derived from DCM Patients" Cells 12, no. 21: 2526. https://doi.org/10.3390/cells12212526
APA StyleHawey, C., Bourque, K., Alim, K., Derish, I., Rody, E., Khan, K., Gendron, N., Cecere, R., Giannetti, N., & Hébert, T. E. (2023). Measuring Single-Cell Calcium Dynamics Using a Myofilament-Localized Optical Biosensor in hiPSC-CMs Derived from DCM Patients. Cells, 12(21), 2526. https://doi.org/10.3390/cells12212526






