A Combinatorial Code for CPEB-Mediated c-myc Repression
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmids
2.2. Northern Blotting and Western Blotting
2.3. Cell Culture and Transfection
2.4. Lentivirus Production
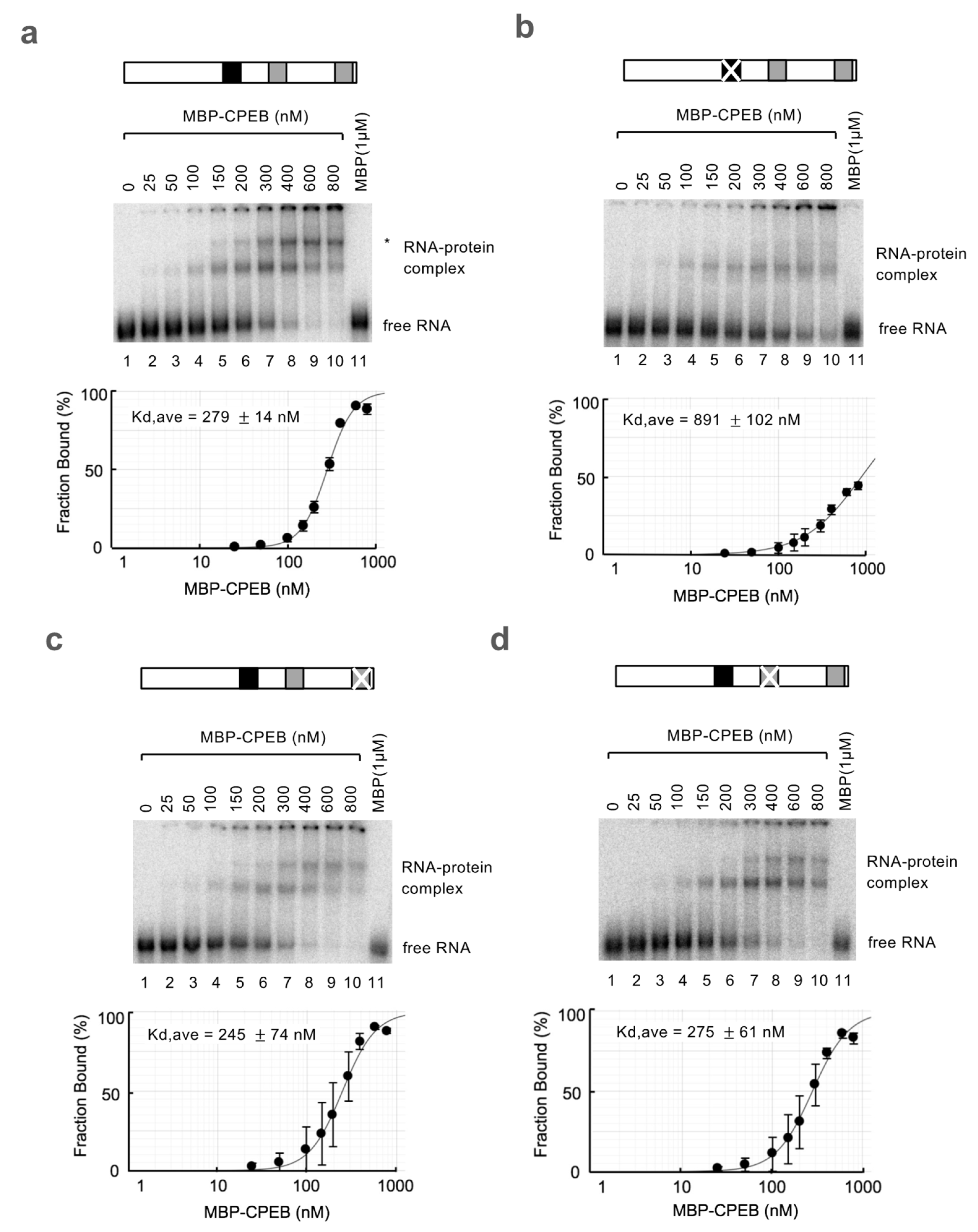
2.5. RNA Electrophoretic Mobility Shift Assay (REMSA)
3. Results
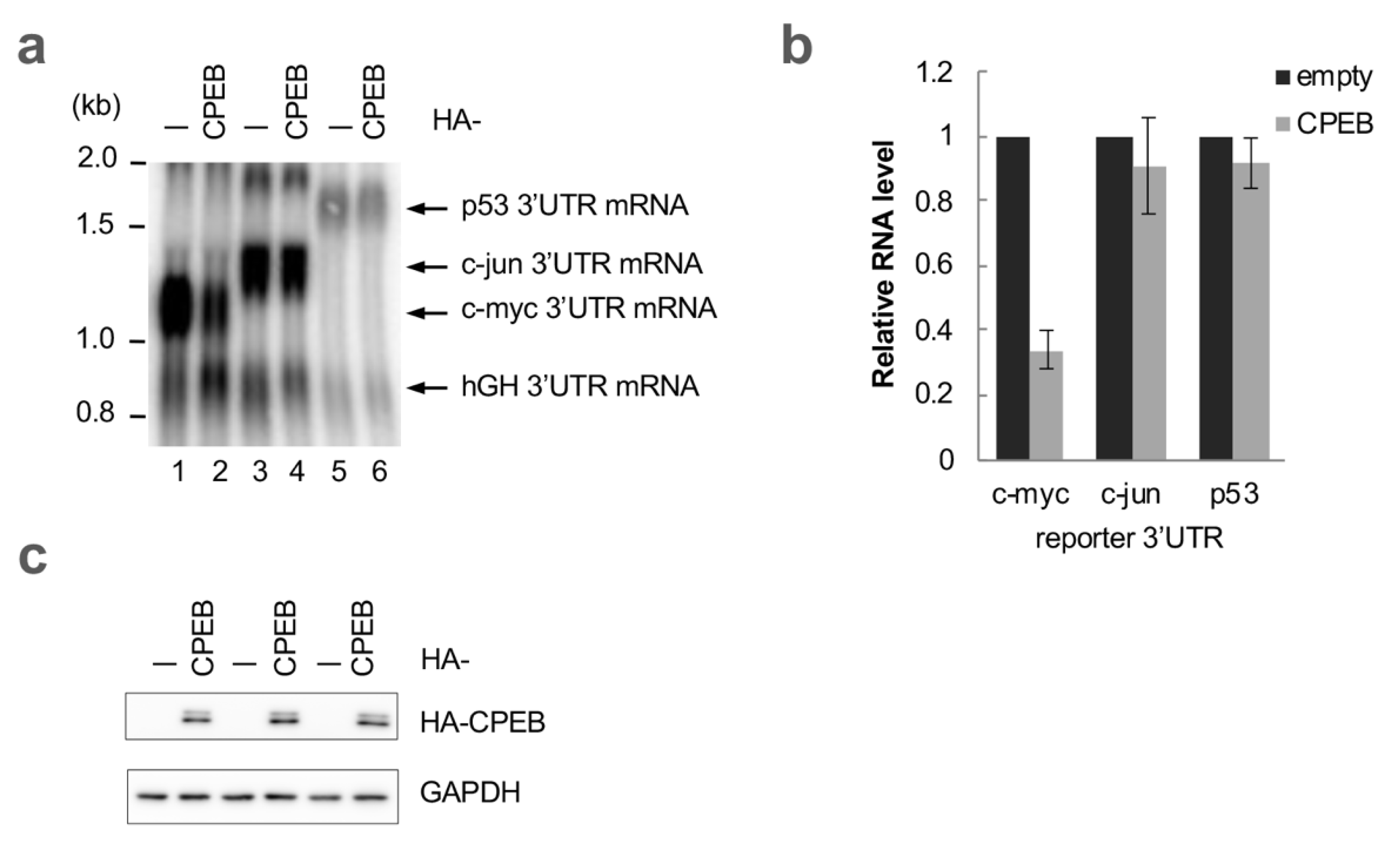
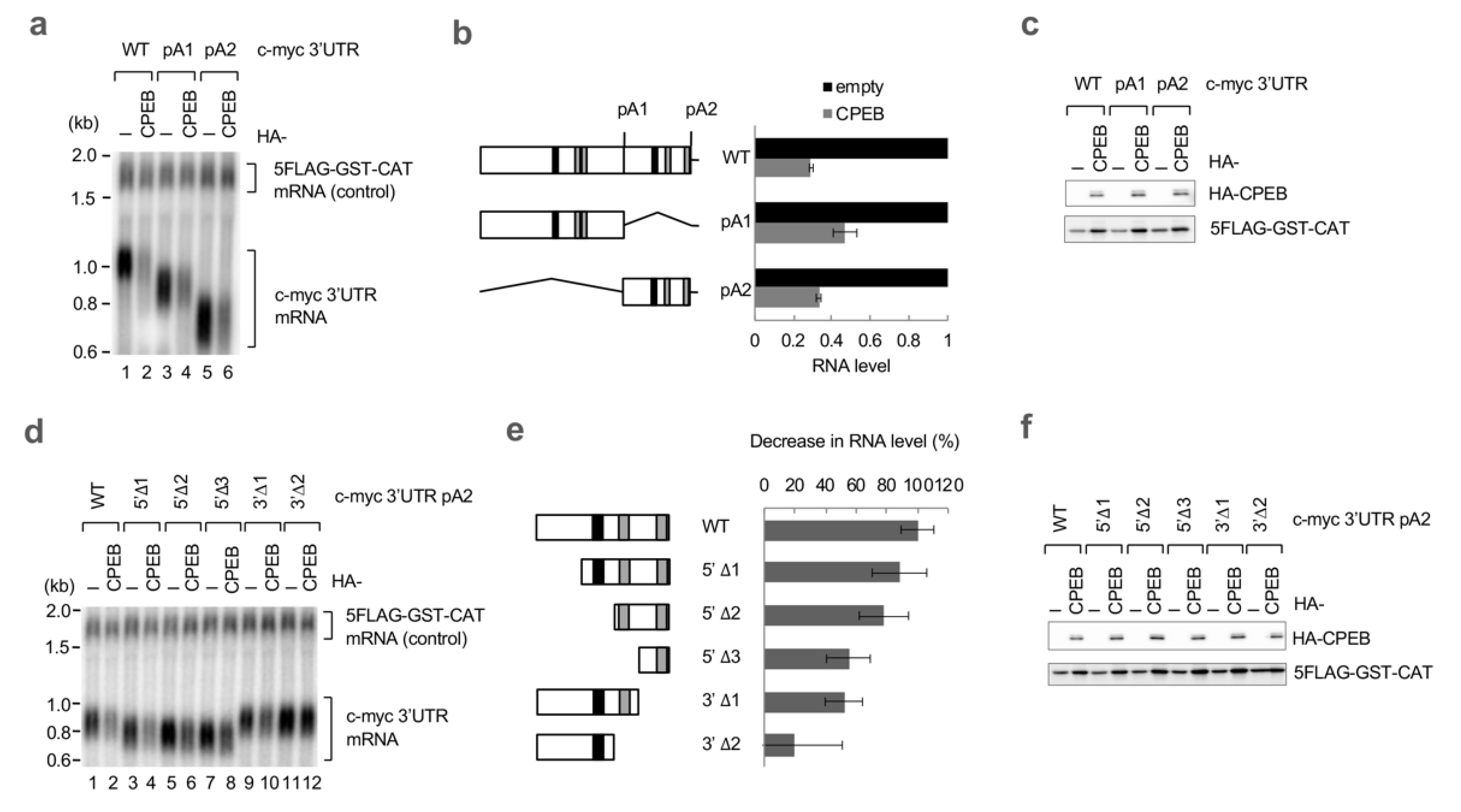
3.1. c-myc mRNA Is Specifically Degraded by CPEB
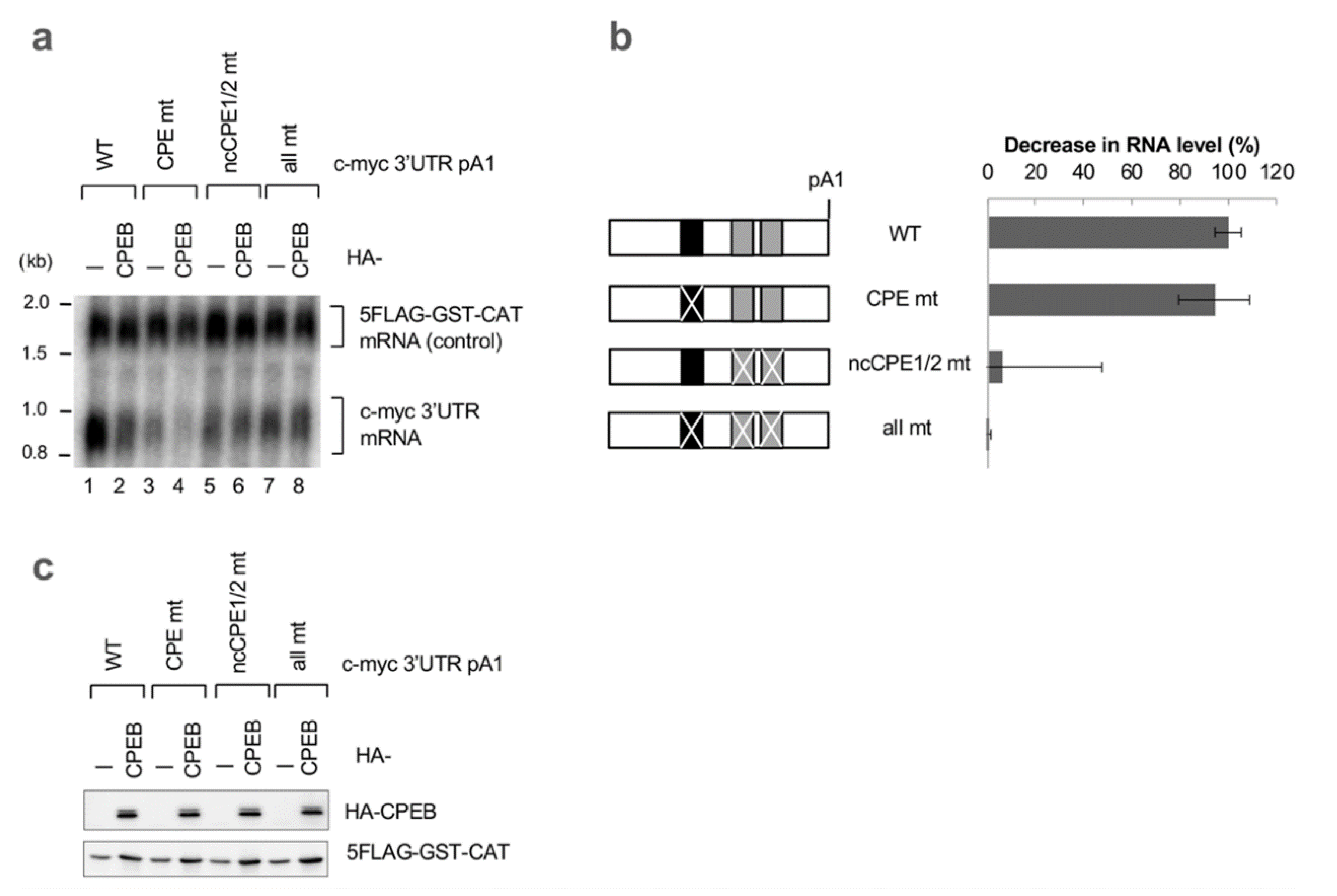
3.2. ncCPE Sequences Are Indispensable for CPEB-Mediated c-myc mRNA Decay
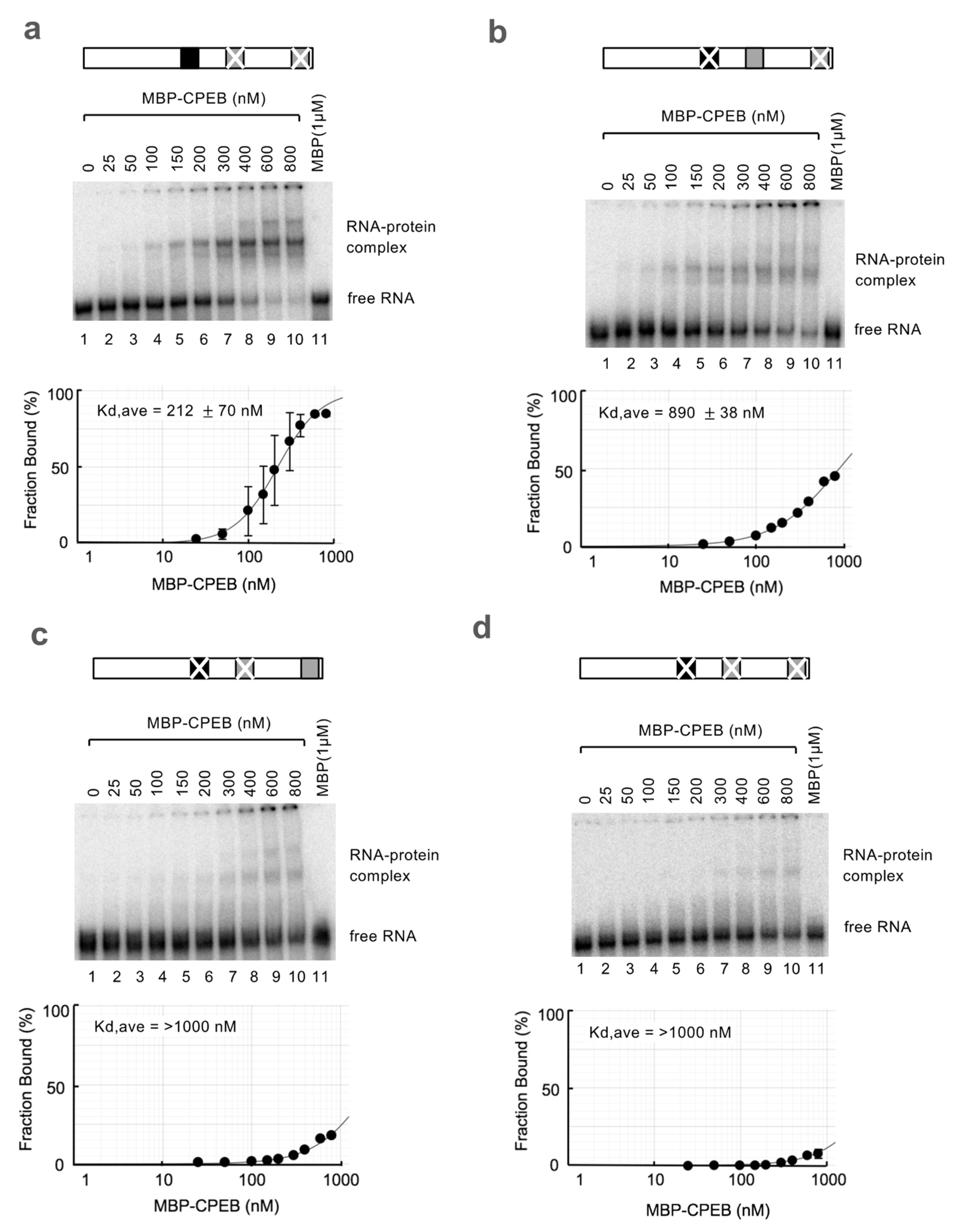
3.3. sCPE Helps a Second CPEB Bind to ncCPE Sequences
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gerstberger, S.; Hafner, M.; Tuschl, T. A census of human RNA-binding proteins. Nat. Rev. Genet. 2014, 15, 829–845. [Google Scholar] [CrossRef] [PubMed]
- Novoa, I.; Gallego, J.; Ferreira, P.G.; Mendez, R. Mitotic cell-cycle progression is regulated by CPEB1 and CPEB4-dependent translational control. Nat. Cell. Biol. 2010, 12, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Burns, D.M.; Richter, J.D. CPEB regulation of human cellular senescence, energy metabolism, and p53 mRNA translation. Genes Dev. 2008, 22, 3449–3460. [Google Scholar] [CrossRef] [PubMed]
- Burns, D.M.; D’Ambrogio, A.; Nottrott, S.; Richter, J.D. CPEB and two poly(A) polymerases control miR-122 stability and p53 mRNA translation. Nature 2011, 473, 105–108. [Google Scholar] [CrossRef] [PubMed]
- Inagaki, H.; Hosoda, N.; Tsuiji, H.; Hoshino, S.I. Direct evidence that Ataxin-2 is a translational activator mediating cytoplasmic polyadenylation. J. Biol. Chem. 2020, 295, 15810–15825. [Google Scholar] [CrossRef] [PubMed]
- Kojima, S.; Sher-Chen, E.L.; Green, C.B. Circadian control of mRNA polyadenylation dynamics regulates rhythmic protein expression. Genes Dev. 2012, 26, 2724–2736. [Google Scholar] [CrossRef] [PubMed]
- Ogami, K.; Hosoda, N.; Funakoshi, Y.; Hoshino, S. Antiproliferative protein Tob directly regulates c-myc proto-oncogene expression through cytoplasmic polyadenylation element-binding protein CPEB. Oncogene 2014, 33, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, M.; Hosoda, N.; Chifu, K.; Hoshino, S.I. TDP-43 accelerates deadenylation of target mRNAs by recruiting Caf1 deadenylase. FEBS Lett. 2019, 593, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Hosoda, N.; Funakoshi, Y.; Hirasawa, M.; Yamagishi, R.; Asano, Y.; Miyagawa, R.; Ogami, K.; Tsujimoto, M.; Hoshino, S. Anti-proliferative protein Tob negatively regulates CPEB3 target by recruiting Caf1 deadenylase. EMBO J. 2011, 30, 1311–1323. [Google Scholar] [CrossRef] [PubMed]
- Richter, J.D. CPEB: A life in translation. Trends Biochem. Sci. 2007, 32, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Miranda, G.; Méndez, R. The CPEB-family of proteins, translational control in senescence and cancer. Ageing. Res. Rev. 2012, 11, 460–472. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Richter, J.D. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol. Cell. 2006, 24, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Groisman, I.; Ivshina, M.; Marin, V.; Kennedy, N.J.; Davis, R.J.; Richter, J.D. Control of cellular senescence by CPEB. Genes. Dev. 2006, 20, 2701–2712. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zearfoss, N.R.; Alarcon, J.M.; Trifilieff, P.; Kandel, E.; Richter, J.D. A molecular circuit composed of CPEB-1 and c-Jun controls growth hormone-mediated synaptic plasticity in the mouse hippocampus. J. Neurosci. 2008, 28, 8502–8509. [Google Scholar] [CrossRef] [PubMed]
- Afroz, T.; Skrisovska, L.; Belloc, E.; Guillén-Boixet, J.; Méndez, R.; Allain, F.H. A fly trap mechanism provides sequence-specific RNA recognition by CPEB proteins. Genes. Dev. 2014, 28, 1498–1514. [Google Scholar] [CrossRef] [PubMed]
- Weill, L.; Belloc, E.; Castellazzi, C.L.; Méndez, R. Musashi 1 regulates the timing and extent of meiotic mRNA translational activation by promoting the use of specific CPEs. Nat. Struct. Mol. Biol. 2017, 24, 672–681. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ogami, K.; Ogawa, K.; Sanpei, S.; Ichikawa, F.; Udagawa, T.; Hoshino, S.-i. A Combinatorial Code for CPEB-Mediated c-myc Repression. Cells 2023, 12, 2410. https://doi.org/10.3390/cells12192410
Ogami K, Ogawa K, Sanpei S, Ichikawa F, Udagawa T, Hoshino S-i. A Combinatorial Code for CPEB-Mediated c-myc Repression. Cells. 2023; 12(19):2410. https://doi.org/10.3390/cells12192410
Chicago/Turabian StyleOgami, Koichi, Keima Ogawa, Shoko Sanpei, Fumito Ichikawa, Tsuyoshi Udagawa, and Shin-ichi Hoshino. 2023. "A Combinatorial Code for CPEB-Mediated c-myc Repression" Cells 12, no. 19: 2410. https://doi.org/10.3390/cells12192410
APA StyleOgami, K., Ogawa, K., Sanpei, S., Ichikawa, F., Udagawa, T., & Hoshino, S.-i. (2023). A Combinatorial Code for CPEB-Mediated c-myc Repression. Cells, 12(19), 2410. https://doi.org/10.3390/cells12192410






