Mechanical Properties and Nanomotion of BT-20 and ZR-75 Breast Cancer Cells Studied by Atomic Force Microscopy and Optical Nanomotion Detection Method
Abstract
1. Introduction
2. Materials and Methods
3. Results
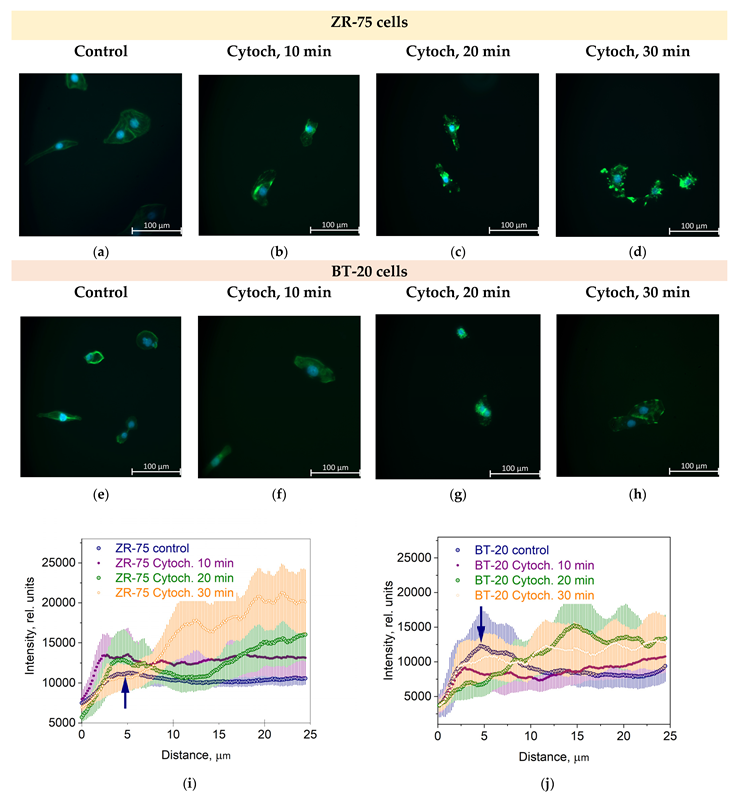
3.1. Morphology and the Actin Cytoskeleton Organization of ZR-75 and BT-20 Cells
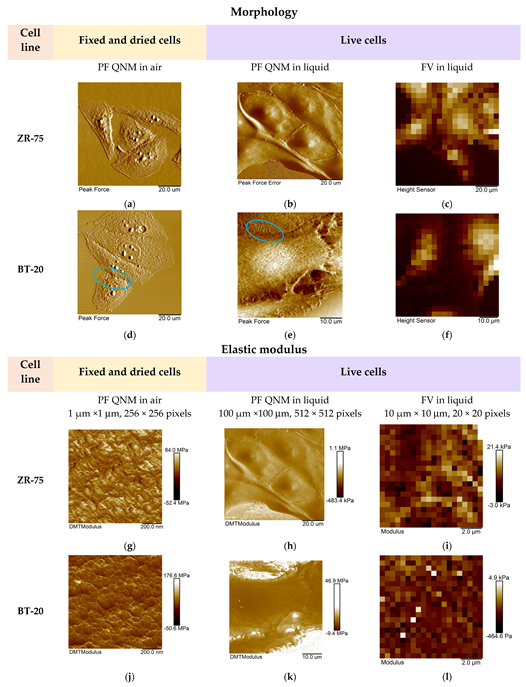
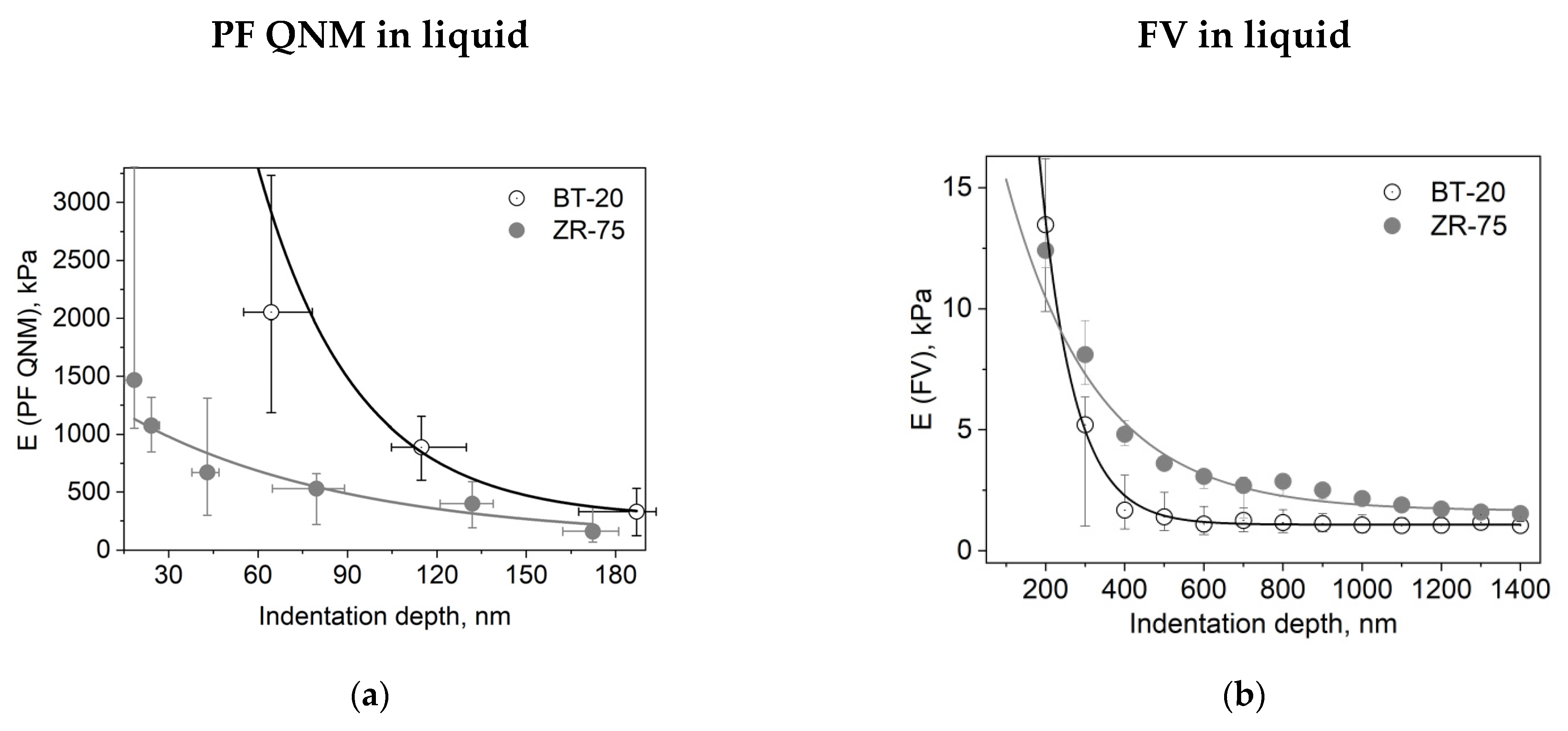
3.2. Elastic Properties of ZR-75 and BT-20 Cells Measured by Force Volume and Peak Force QNM Modes of AFM
3.3. Viscoelastic Properties of BT-20 and ZR-75 Breast Cancer Cells Measured Using FV AFM Mode
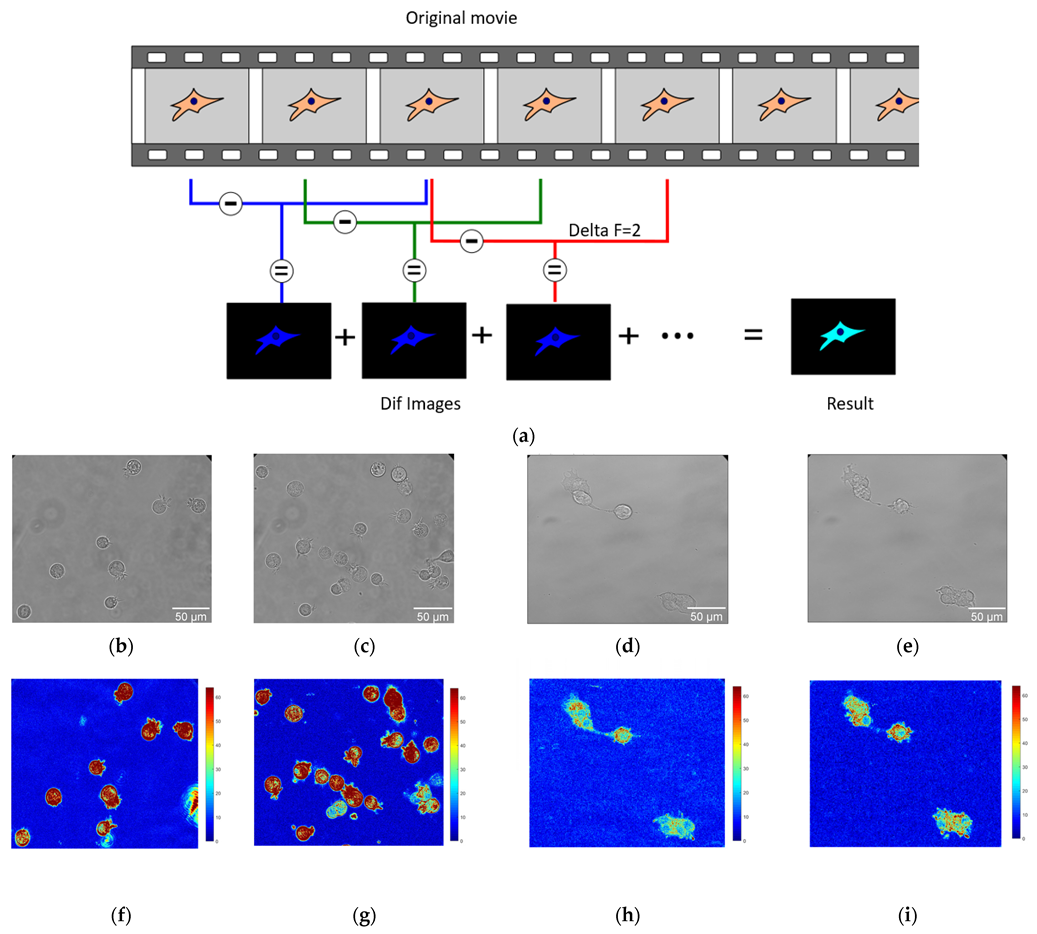
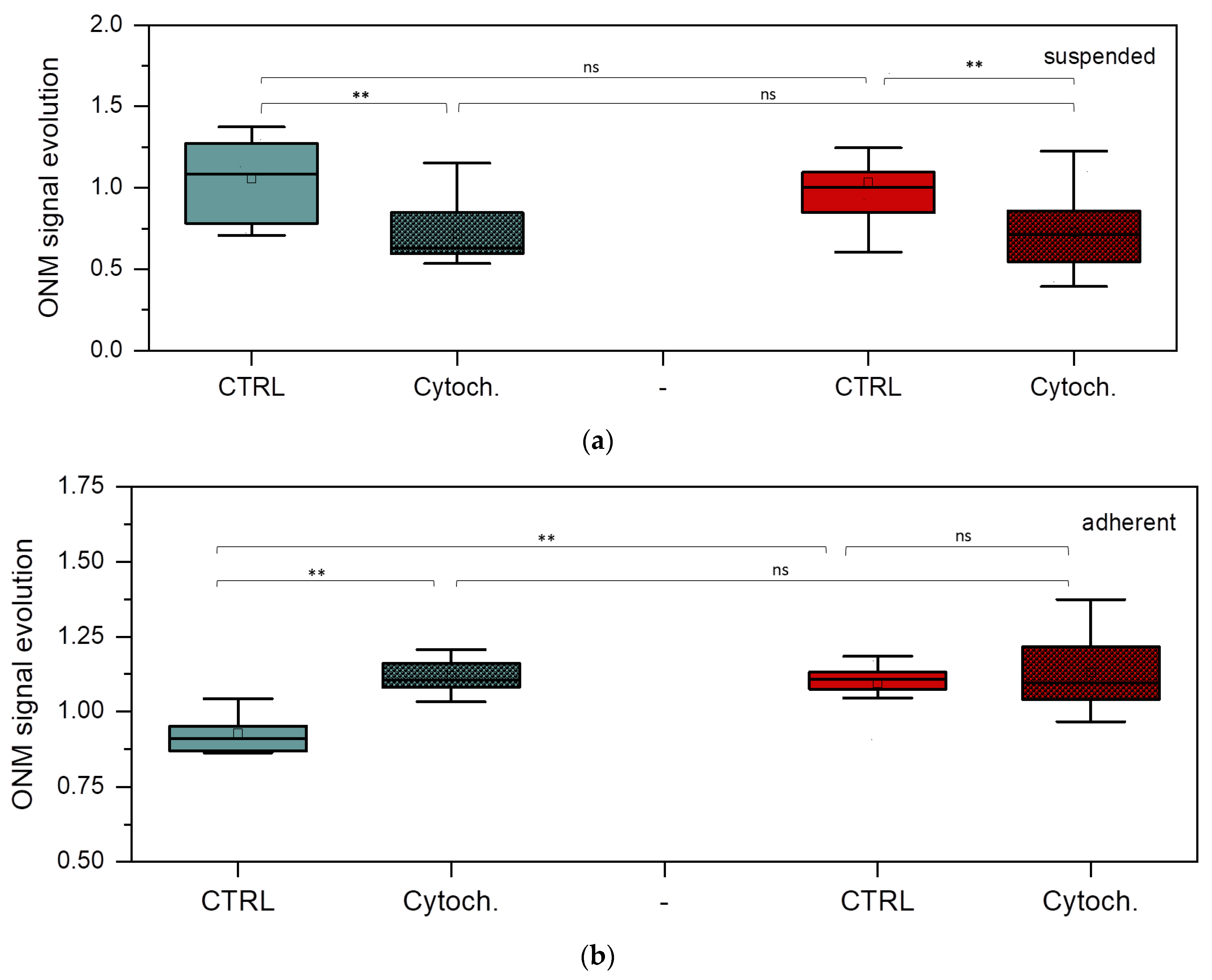
3.4. Nanomotion Parameters for BT-20 and ZR-75 Breast Cancer Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Reference | Cell | Modulus, kPa | Mode |
|---|---|---|---|
| Li et al. (2008) [63] | MCF-7 | 0.31–0.81 | FS |
| MCF-10A | 0.61–161 | ||
| Schirbaum et al. (2017) [44] | MDA-MB-231 | 4.00–6.00 | FCFM |
| MCF-7 | 2.50–3.50 | ||
| MCF-10A | 10.00–30.00 | ||
| Lekka et al. (2012) [14] | T47D | 1.20 ± 0.28 | FV |
| MCF7 | 1.24 ± 0.46 | ||
| 184A | 2.26 ± 0.56 | ||
| Sajeesh et al. (2016) [38] | MDA-MB-231 | 1.00 ± 0.10 | FV |
| MCF-7 | 3.43 ± 0.38 | ||
| Park (2018) [39] | MCF-7 | 1.35 ± 0.12 | FV |
| MCF-7/ADR | 1.67 ± 0.14 | ||
| Kwon et al. (2020) [40] | MCF-7 | 9.24 ± 1.39 | FV |
| MCF-10A | 13.69 ± 1.90 | ||
| T47D | 8.39 ± 1.24 | ||
| MDA-MB-231 | 9.57 ± 1.38 | ||
| Li et al. (2021) [42] | SKBR3 | 0.63 ± 0.31 | FV |
| MCF-7 | ~0.65 | ||
| BT474 | ~0.42 | ||
| MDA-MB-231 | 0.27 ± 0.09 | ||
| Amiri, Hastert, Dietz (2020) [41] | BT-20 | 10.00 | PFT |
| Xu et al. (2014) [43] | MCF-7 | 1.50–3.90 | QI |
| MCF-10A | 2.10–8.40 |
References
- Dai, X.; Cheng, H.; Bai, Z.; Li, J. Breast cancer cell line classification and its relevance with breast tumor subtyping. J. Cancer 2017, 8, 3131–3141. [Google Scholar] [CrossRef] [PubMed]
- Testa, U.; Castelli, G.; Pelosi, E. Breast cancer: A molecularity heterogeneous disease needing subtype-specific treatments. Med. Sci. 2020, 8, 18. [Google Scholar] [CrossRef] [PubMed]
- Quan, F.-S.; Kyung, S.K. Medical applications of the intrinsic mechanical properties of single cells. Acta Biochim. Biophys. Sin. 2016, 48, 865–871. [Google Scholar] [CrossRef] [PubMed]
- Lekka, M. Applicability of applicability of atomic force microscopy to determine cancer-related changes in cells. Phil. Trans. R. Soc. A 2022, 380, 20210346. [Google Scholar] [CrossRef] [PubMed]
- Olson, M.F.; Sahai, E. The actin cytoskeleton in cancer cell motility. Clin. Exp. Metastasis 2009, 26, 273–287. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Qin, F.; Yin, J.; Li, D.; Huang, Y.; Hu, L.; He, L.; Lv, C.; Li, X.; Li, S.; et al. Role and mechanism of actin-related protein 2/3 complex signaling in cancer invasion and metastasis: A review. Medicine 2023, 102, e33158. [Google Scholar] [CrossRef]
- Curry, N.; Ghézali, G.; Kaminski Schierle, G.S.; Rouach, N.; Kaminski, C.F. Correlative STED and atomic force microscopy on live astrocytes reveals plasticity of cytoskeletal structure and membrane physical properties during polarized migration. Front. Cell Neurosci. 2017, 11, 104. [Google Scholar] [CrossRef]
- Su, X.; Zhang, L.; Kang, H.; Zhang, B.; Bao, G.; Wang, J. Mechanical, nanomorphological and biological reconstruction of early-stage apoptosis in HeLa cells induced by cytochalasin B. Oncol. Rep. 2019, 41, 928–938. [Google Scholar] [CrossRef]
- Zieliński, T.; Pabijan, J.; Zapotoczny, B.; Zemła, J.; Wesołowska, J.; Pera, J.; Lekka, M. Changes in nanomechanical properties of single neuroblastoma cells as a model for oxygen and glucose deprivation (OGD). Sci. Rep. 2022, 12, 16276. [Google Scholar] [CrossRef]
- Pollard, T.D.; Cooper, J.A. Actin, a central player in cell shape and movement. Science 2009, 326, 1208–1212. [Google Scholar] [CrossRef]
- Kuznetsova, T.G.; Starodubtseva, M.N.; Yegorenkov, N.I.; Chizhik, S.A.; Zhdanov, R.I. Atomic force microscopy probing of cell elasticity. Micron 2007, 38, 824–833. [Google Scholar] [CrossRef] [PubMed]
- Samani, A.; Zubovits, J.; Plewes, D. Elastic moduli of normal and pathological human breast tissues: An inversion-technique-based investigation of 169 samples. Phys. Med. Biol. 2007, 52, 1565–1576. [Google Scholar] [CrossRef] [PubMed]
- Olubowale, O.H.; Biswas, S.; Azom, G.; Prather, B.L.; Owoso, S.D.; Rinee, K.C.; Marroquin, K.; Gates, K.A.; Chambers, M.B.; Xu, A.; et al. “May the Force Be with You!” force–volume mapping with atomic force microscopy. ACS Omega 2021, 6, 25860–25875. [Google Scholar] [CrossRef] [PubMed]
- Lekka, M.; Gil, D.; Pogoda, K.; Dulińska-Litewka, J.; Jach, R.; Gostek, J.; Klymenko, O.; Prauzner-Bechcicki, S.; Stachura, Z.; Wiltowska-Zuber, J.; et al. Cancer cell detection in tissue sections using AFM. Arch. Biochem. Biophys. 2012, 518, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Mohammadalipour, A.; Burdick, M.M.; Tees, D.F.J. Deformability of breast cancer cells in correlation with surface markers and cell rolling. FASEB J. 2018, 32, 1806–1817. [Google Scholar] [CrossRef]
- Hu, J.; Zhou, Y.; Obayemi, J.D.; Du, J.; Soboyejo, W.O. An investigation of the viscoelastic properties and the actin cytoskeletal structure of triple negative breast cancer cells. J. Mech. Behav. Biomed. Mater. 2018, 86, 1–13. [Google Scholar] [CrossRef]
- Wu, P.H.; Aroush, D.R.; Asnacios, A.; Chen, W.C.; Dokukin, M.E.; Doss, B.L.; Durand-Smet, P.; Ekpenyong, A.; Guck, J.; Guz, N.V.; et al. A comparison of methods to assess cell mechanical properties. Nat. Methods 2018, 15, 491–498. [Google Scholar] [CrossRef]
- Amiri, A.; Hastert, F.; Stühna, L.; Dietz, C. Structural analysis of healthy and cancerous epithelial-type breast cells by nanomechanical spectroscopy allows us to obtain peculiarities of the skeleton and junctions. Nanoscale Adv. 2019, 1, 4853–4862. [Google Scholar] [CrossRef]
- Kashani, A.S.; Packirisamy, M. Cancer cells optimize elasticity for efficient migration. R. Soc. Open Sci. 2020, 7, 7200747200747. [Google Scholar] [CrossRef]
- Husson, J. Measuring Cell mechanical properties using microindentation. Methods Mol. Biol. 2023, 2600, 3–23. [Google Scholar]
- Yang, Y.; Xiao, X.; Peng, Y.; Yang, C.; Wu, S.; Liu, Y.; Yue, T.; Pu, H.; Liu, N.; Jiang, H. The comparison between force volume and peakforce quantitative nanomechanical mode of atomic force microscope in detecting cell’s mechanical properties. Microsc. Res. Tech. 2019, 82, 1843–1851. [Google Scholar] [CrossRef] [PubMed]
- Starodubtseva, M.N. Mechanical properties of the cell surface layer measured by contact atomic force microscopy. In Contact Problems for Soft. Biological and Bioinspired Materials; Borodich, F.M., Jin, X., Eds.; Springer: Cham, Switzerland, 2022; Volume 15, pp. 51–72. [Google Scholar]
- Yang, C.T.; Méjard, R.; Griesser, H.J.; Bagnaninchi, P.O.; Thierry, B. cellular micromotion monitored by long-range surface plasmon resonance with optical fluctuation analysis. Anal. Chem. 2015, 87, 1456–1461. [Google Scholar] [CrossRef] [PubMed]
- Opp, D.; Wafula, B.; Lim, J.; Huang, E.; Lo, J.C.; Lo, C.M. Use of electric cell-substrate impedance sensing to assess in vitro cytotoxicity. Biosens. Bioelectron. 2009, 24, 2625–2629. [Google Scholar] [CrossRef] [PubMed]
- Lovelady, D.C.; Richmond, T.C.; Maggi, A.N.; Lo, C.M.; Rabson, D.A. Distinguishing cancerous from noncancerous cells through analysis of electrical noise. Phys. Rev. E. Stat. Nonlin. Soft. Matter. Phys. 2007, 76, 041908. [Google Scholar] [CrossRef] [PubMed]
- Tarantola, M.; Marel, A.K.; Sunnick, E.; Adam, H.; Wegener, J.; Janshoff, A. Dynamics of human cancer cell lines monitored by electrical and acoustic fluctuation analysis. Integr. Biol. 2010, 2, 139–150. [Google Scholar] [CrossRef]
- Longo, G.; Alonso-Sarduy, L.; Rio, L.M.; Bizzini, A.; Trampuz, A.; Notz, J.; Dietler, G.; Kasas, S. Rapid detection of bacterial resistance to antibiotics using AFM cantilevers as nanomechanical sensors. Nat. Nanotechnol. 2013, 8, 522–526. [Google Scholar] [CrossRef]
- Kohler, A.C.; Venturelli, L.; Longo, G.; Dietler, G.; Kasas, S. Nanomotion detection based on atomic force microscopy cantilevers. Cell Surf. 2019, 5, 100021. [Google Scholar] [CrossRef]
- Venturelli, L.; Harrold, Z.R.; Murray, A.E.; Villalba, M.I.; Lundin, E.M.; Dietler, G.; Kasas, S.; Foschia, R. Nanomechanical bio-sensing for fast and reliable detection of viability and susceptibility of microorganisms. Sens. Actuators B Chem. 2021, 348, 130650. [Google Scholar] [CrossRef]
- Stupar, P.; Podolski-Renić, A.; Villalba, M.I.; Dragoj, M.; Jovanović Stojanov, S.; Pešić, M.; Kasas, S. Nano-Motion Analysis for Rapid and Label Free Assessing of Cancer Cell Sensitivity to Chemotherapeutics. Medicina 2021, 57, 446. [Google Scholar] [CrossRef]
- Villalba, M.I.; Rossetti, E.; Bonvallat, A.; Yvanoff, C.; Radonicic, V.; Willaert, R.G.; Kasas, S. Simple optical nanomotion method for single-bacterium viability and antibiotic response testing. Proc. Natl. Acad. Sci. USA 2023, 120, e2221284120. [Google Scholar] [CrossRef]
- Willaert, R.G.; Vanden Boer, P.; Malovichko, A.; Alioscha-Perez, M.; Radotić, K.; Bartolić, D.; Kalauzi, A.; Villalba, M.I.; Sanglard, D.; Dietler, G.; et al. Single yeast cell nanomotions correlate with cellular activity. Sci. Adv. 2020, 6, eaba3139. [Google Scholar] [CrossRef] [PubMed]
- Radonicic, V.; Yvanoff, C.; Villalba, M.I.; Devreese, B.; Kasas, S.; Willaert, R.G. Single-Cell Optical Nanomotion of Candida albicans in Microwells for Rapid Antifungal Susceptibility Testing. Fermentation 2023, 9, 365. [Google Scholar] [CrossRef]
- Parmar, P.; Villalba, M.I.; Horii Huber, A.S.; Kalauzi, A.; Bartolić, D.; Radotić, K.; Willaert, R.G.; MacFabe, D.F.; Kasas, S. Mitochondrial nanomotion measured by optical microscopy. Front. Microbiol. 2023, 14, 1133773. [Google Scholar] [CrossRef] [PubMed]
- Ting, T.C.T. The contact stresses between a rigid indenter and a viscoelastic half-space. J. Appl. Mech. 1966, 33, 845–854. [Google Scholar] [CrossRef]
- Efremov, Y.M.; Wang, W.; Hardy, S.D.; Geahlen, R.L.; Raman, A. Measuring nanoscale viscoelastic parameters of cells directly from AFM force-displacement curves. Sci. Rep. 2017, 7, 1541. [Google Scholar]
- Efremov, Y.M.; Kotova, S.L.; Akovantseva, A.A.; Timashev, P.S. Nanomechanical properties of enucleated cells: Contribution of the nucleus to the passive cell mechanics. J. Nanobiotechnology 2020, 18, 134. [Google Scholar] [CrossRef]
- Sajeesh, P.; Raj, A.; Doble, M.; Sen, A.K. Characterization and sorting of cells based on stiffness contrast in a microfluidic channel. RSC Adv. 2016, 6, 74704–74714. [Google Scholar] [CrossRef]
- Park, S. Mechanical alteration associated with chemotherapeutic resistance of breast cancer cells. J. Cancer Prev. 2018, 23, 87–92. [Google Scholar] [CrossRef]
- Kwon, S.; Yang, W.; Moon, D.; Kim, K.S. Comparison of cancer cell elasticity by cell type. J. Cancer 2020, 11, 5403–5412. [Google Scholar] [CrossRef]
- Amiri, A.; Hastert, F.D.; Dietz, C. Carcinomas with occult metastasis potential: Diagnosis/prognosis accuracy improvement by means of force spectroscopy. Adv. Biosyst. 2020, 4, e2000042. [Google Scholar] [CrossRef]
- Li, M.; Xi, N.; Wang, Y.C.; Liu, L.Q. Atomic force microscopy for revealing micro/nanoscale mechanics in tumor metastasis: From single cells to microenvironmental cues. Acta. Pharmacol. Sin. 2021, 42, 323–339. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Wang, Y.; Jiang, N.; Yang, H.; Lin, J.; Xie, S. Elasticity measurement of breast cancer cells by atomic force microscopy. In Proceedings of the Twelfth International Conference on Photonics and Imaging in Biology and Medicine, Wuhan, China, 14–17 June 2014. [Google Scholar] [CrossRef]
- Schirbaum, N.; Reinlaender, J.; Schaffer, T.H. Viscoelastic properties of normal and cancerous human breasts cells are affected differently by contact to adjacent cells. Acta Biomater. 2017, 55, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Zambito, M.; Viti, F.; Bosio, A.G.; Ceccherini, I.; Florio, T.; Vassalli, M. The Impact of Experimental Conditions on Cell Mechanics as Measured with Nanoindentation. Nanomaterials 2023, 13, 1190. [Google Scholar] [CrossRef] [PubMed]
- Starodubtseva, M.N.; Nadyrov, E.A.; Shkliarava, N.M.; Tsukanava, A.U.; Matveyenkau, M.V.; Nedoseikina, M.S. Heterogeneity of nanomechanical properties of the human umbilical vein endothelial cell surface. Microvasc. Res. 2021, 136, 104168. [Google Scholar] [CrossRef]
- Gil-Redondo, J.C.; Weber, A.; Zbiral, B.; Vivanco, M.D.; Toca-Herrera, J.L. Substrate stiffness modulates the viscoelastic properties of MCF-7 cells. J. Mech. Behav. Biomed. Mater. 2022, 125, 104979. [Google Scholar] [CrossRef]
- Onwudiwe, K.; Hu, J.; Obayemi, J.; Uzonwanne, V.; Ani, C.; Nwazojie, C.; Onyekanne, C.; Ezenwafor, T.; Odusanya, O.; Soboyejo, W. Actin cytoskeletal structure and the statistical variations of the mechanical properties of non-tumorigenic breast and triple-negative breast cancer cells. J. Mech. Behav. Biomed. Mater. 2021, 119, 104505. [Google Scholar] [CrossRef]
- Smolyakov, G.; Thiebot, B.; Campillo, C.; Labdi, S.; Severac, C.; Pelta, J.; Dague, É. Elasticity, adhesion, and tether extrusion on breast cancer cells provide a signature of their invasive potential. ACS Appl. Mater. Interfaces 2016, 8, 27426–27431. [Google Scholar] [CrossRef]
- Li, M.; Dang, D.; Liu, L.; Xi, N.; Wang, Y. Atomic Force Microscopy in Characterizing Cell Mechanics for Biomedical Applications: A Review. IEEE Trans Nanobioscience 2017, 16, 523–540. [Google Scholar] [CrossRef]
- Liu, Y.; Sokolov, I.; Dokukin, M.E.; Xiong, Y.; Peng, P. If AFM can be used to measure absolute values of the Young’s modulus of nanocomposite materials down to the nanoscale? Nanoscale 2020, 12, 12432–12443. [Google Scholar] [CrossRef]
- Dokukin, M.E.; Sokolov, I. On the measurements of rigidity modulus of soft materials in nanoindentation experiments at small depth. Macromolecules 2012, 45, 4277–4288. [Google Scholar] [CrossRef]
- Guz, N.; Dokukin, M.; Kalaparthi, V.; Sokolov, I. If cell mechanics can be described by elastic modulus: Study of different models and probes used in indentation experiments. Biophys. J. 2014, 107, 564–575. [Google Scholar] [CrossRef]
- Kontomaris, S.V.; Stylianou, A.; Chliveros, G.; Malamou, A. AFM indentation on highly heterogeneous materials using different indenter geometries. Appl. Mech. 2023, 4, 460–475. [Google Scholar] [CrossRef]
- Mathur, A.B.; Collinsworth, A.M.; Reichert, W.M.; Kraus, W.E.; Truskey, G.A. Endothelial, cardiac muscle and skeletal muscle exhibit different viscous and elastic properties as determined by atomic force microscopy. J. Biomech. 2001, 34, 1545–1553. [Google Scholar] [CrossRef] [PubMed]
- Brill-Karniely, Y. Mechanical Measurements of Cells Using AFM: 3D or 2D Physics? Front. Bioeng. Biotechnol. 2020, 8, 605153. [Google Scholar] [CrossRef] [PubMed]
- Hohmann, T.; Dehghani, F. The Cytoskeleton—A Complex Interacting Meshwork. Cells, 2019; 8, 362. [Google Scholar]
- Kasas, S.; Wang, X.; Hirling, H.; Marsault, R.; Huni, B.; Yersin, A.; Regazzi, R.; Grenningloh, G.; Riederer, B.; Forrò, L.; et al. Superficial and deep changes of cellular mechanical properties following cytoskeleton disassembly. Cell Motil. Cytoskelet. 2005, 62, 124–132. [Google Scholar] [CrossRef]
- Kubiak, A.; Chighizola, M.; Schulte, C.; Bryniarska, N.; Wesołowska, J.; Pudełek, M.; Lasota, M.; Ryszawy, D.; Basta-Kaim, A.; Laidler, P.; et al. Stiffening of DU145 prostate cancer cells driven by actin filaments—Microtubule crosstalk conferring resistance to microtubule-targeting drugs. Nanoscale 2021, 13, 6212–6226. [Google Scholar] [CrossRef]
- Fritzsche, M.; Fernandes, R.A.; Chang, V.T.; Colin-York, H.; Clausen, M.P.; Felce, J.H.; Galiani, S.; Erlenkämper, C.; Santos, A.M.; Heddleston, J.M.; et al. Cytoskeletal actin dynamics shape a ramifying actin network underpinning immunological synapse formation. Sci. Adv. 2017, 3, e1603032. [Google Scholar] [CrossRef]
- Graminha, A.E.; Honorato, J.; Correa, R.S.; Cominetti, M.R.; Menezes, A.C.S.; Batista, A.A. A novel ruthenium(ii) gallic acid complex disrupts the actin cytoskeleton and inhibits migration, invasion and adhesion of triple negative breast tumor cells. Dalton Trans. 2021, 50, 323–335. [Google Scholar] [CrossRef]
- Arumugam, A.; Subramani, R.; Lakshmanaswamy, R. Involvement of actin cytoskeletal modifications in the inhibition of triple-negative breast cancer growth and metastasis by nimbolide. Mol. Ther. Oncolytics 2021, 20, 596–606. [Google Scholar] [CrossRef]
- Li, Q.S.; Lee, G.Y.; Ong, C.N.; Lim, C.T. AFM indentation study of breast cancer cells. Biochem. Biophys. Res. Commun. 2008, 374, 609–613. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Starodubtseva, M.N.; Shkliarava, N.M.; Chelnokova, I.A.; Villalba, M.I.; Krylov, A.Y.; Nadyrov, E.A.; Kasas, S. Mechanical Properties and Nanomotion of BT-20 and ZR-75 Breast Cancer Cells Studied by Atomic Force Microscopy and Optical Nanomotion Detection Method. Cells 2023, 12, 2362. https://doi.org/10.3390/cells12192362
Starodubtseva MN, Shkliarava NM, Chelnokova IA, Villalba MI, Krylov AY, Nadyrov EA, Kasas S. Mechanical Properties and Nanomotion of BT-20 and ZR-75 Breast Cancer Cells Studied by Atomic Force Microscopy and Optical Nanomotion Detection Method. Cells. 2023; 12(19):2362. https://doi.org/10.3390/cells12192362
Chicago/Turabian StyleStarodubtseva, Maria N., Nastassia M. Shkliarava, Irina A. Chelnokova, María I. Villalba, Andrei Yu. Krylov, Eldar A. Nadyrov, and Sandor Kasas. 2023. "Mechanical Properties and Nanomotion of BT-20 and ZR-75 Breast Cancer Cells Studied by Atomic Force Microscopy and Optical Nanomotion Detection Method" Cells 12, no. 19: 2362. https://doi.org/10.3390/cells12192362
APA StyleStarodubtseva, M. N., Shkliarava, N. M., Chelnokova, I. A., Villalba, M. I., Krylov, A. Y., Nadyrov, E. A., & Kasas, S. (2023). Mechanical Properties and Nanomotion of BT-20 and ZR-75 Breast Cancer Cells Studied by Atomic Force Microscopy and Optical Nanomotion Detection Method. Cells, 12(19), 2362. https://doi.org/10.3390/cells12192362






