FNBP1 Facilitates Cervical Cancer Cell Survival by the Constitutive Activation of FAK/PI3K/AKT/mTOR Signaling
Abstract
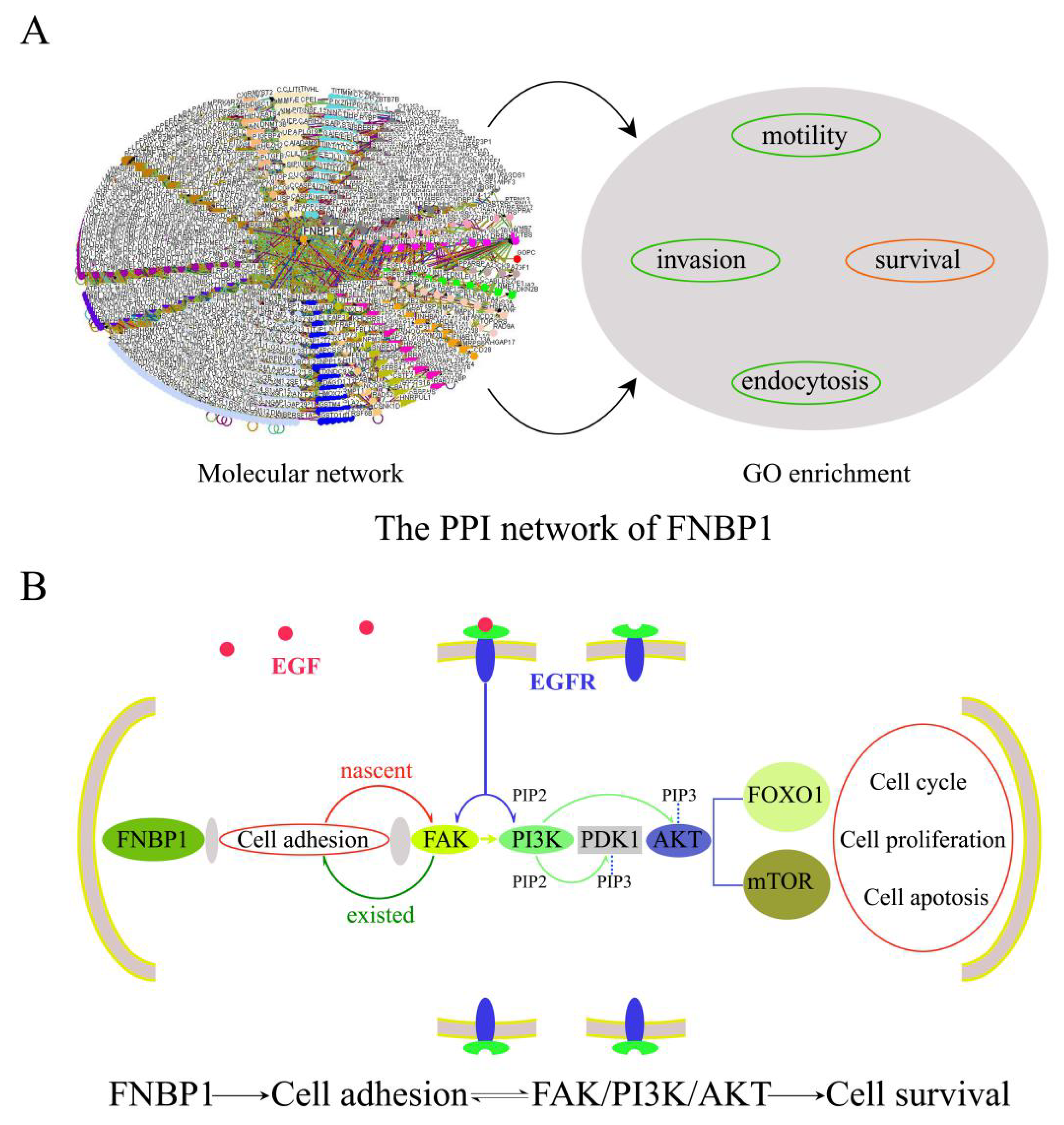
1. Introduction
2. Materials and Methods
2.1. Antibodies and Reagents
2.2. Cell Culture and Viability Assay
2.3. Construct of shRNA Plasmids and Transfection
2.4. Quantitative RT-PCR (qPCR)
2.5. Western Blot
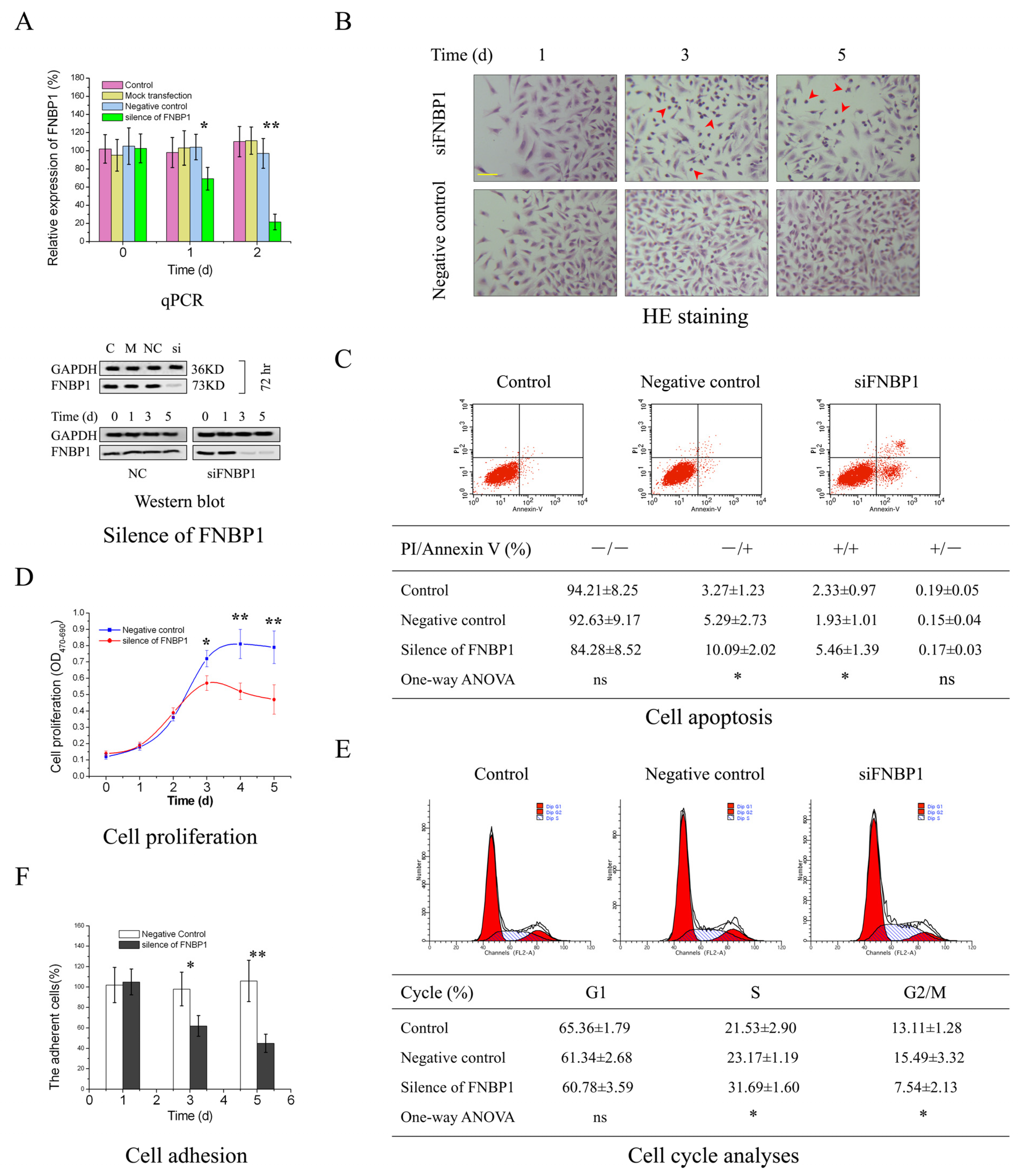
2.6. Hematoxylin–Eosin (HE) Staining
2.7. Cell Proliferation
2.8. Cell Cycle Analysis with Flow Cytometry
2.9. Cell Apoptosis Analysis
2.10. Cell Adhesion Assay
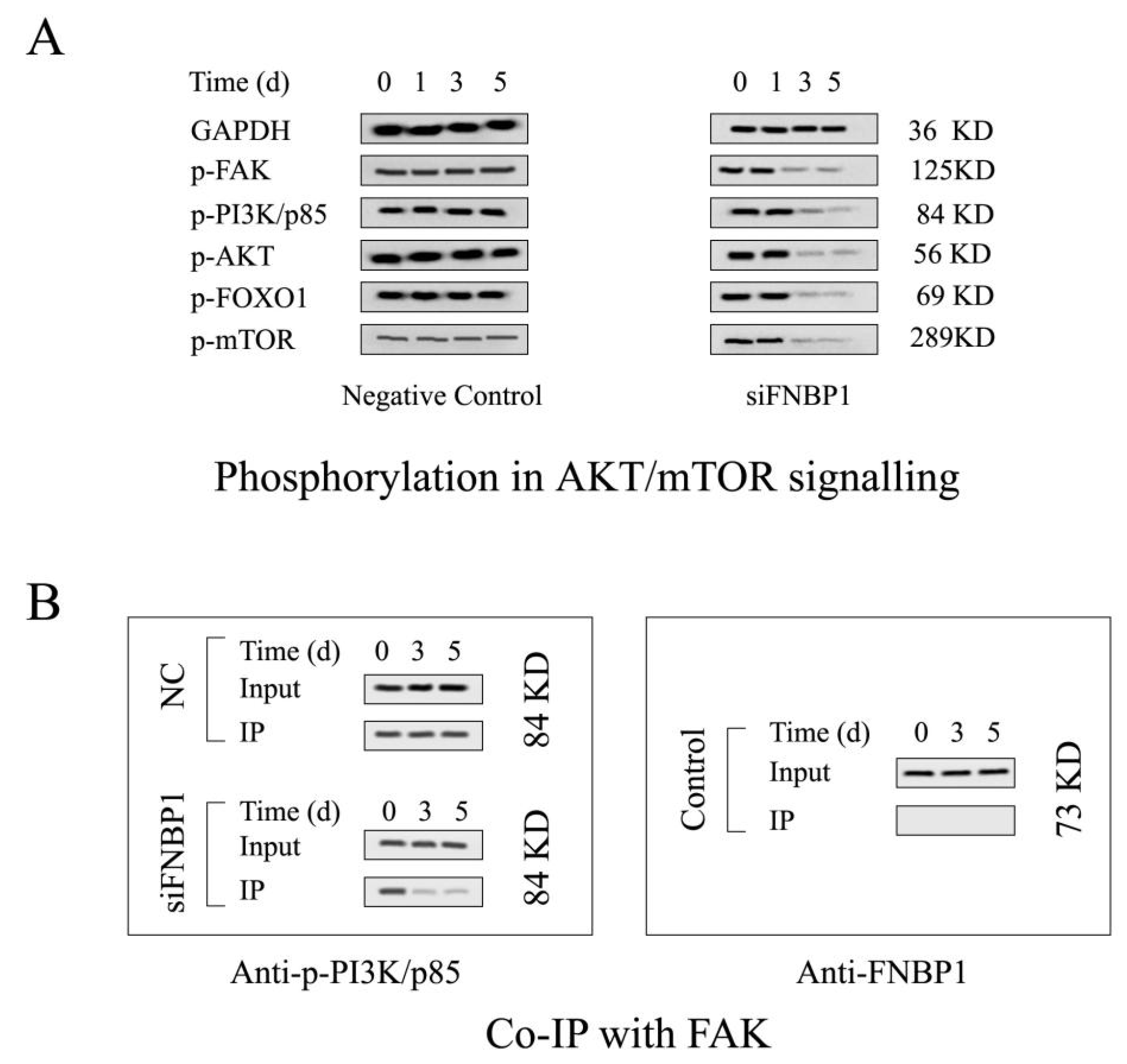
2.11. Co-Immunoprecipitation (Co-IP)
2.12. Bioinformatic and Statistical Analyses
3. Results
3.1. The Biological Effects of FNBP1 Knockdown on Cervical Cancer Cells
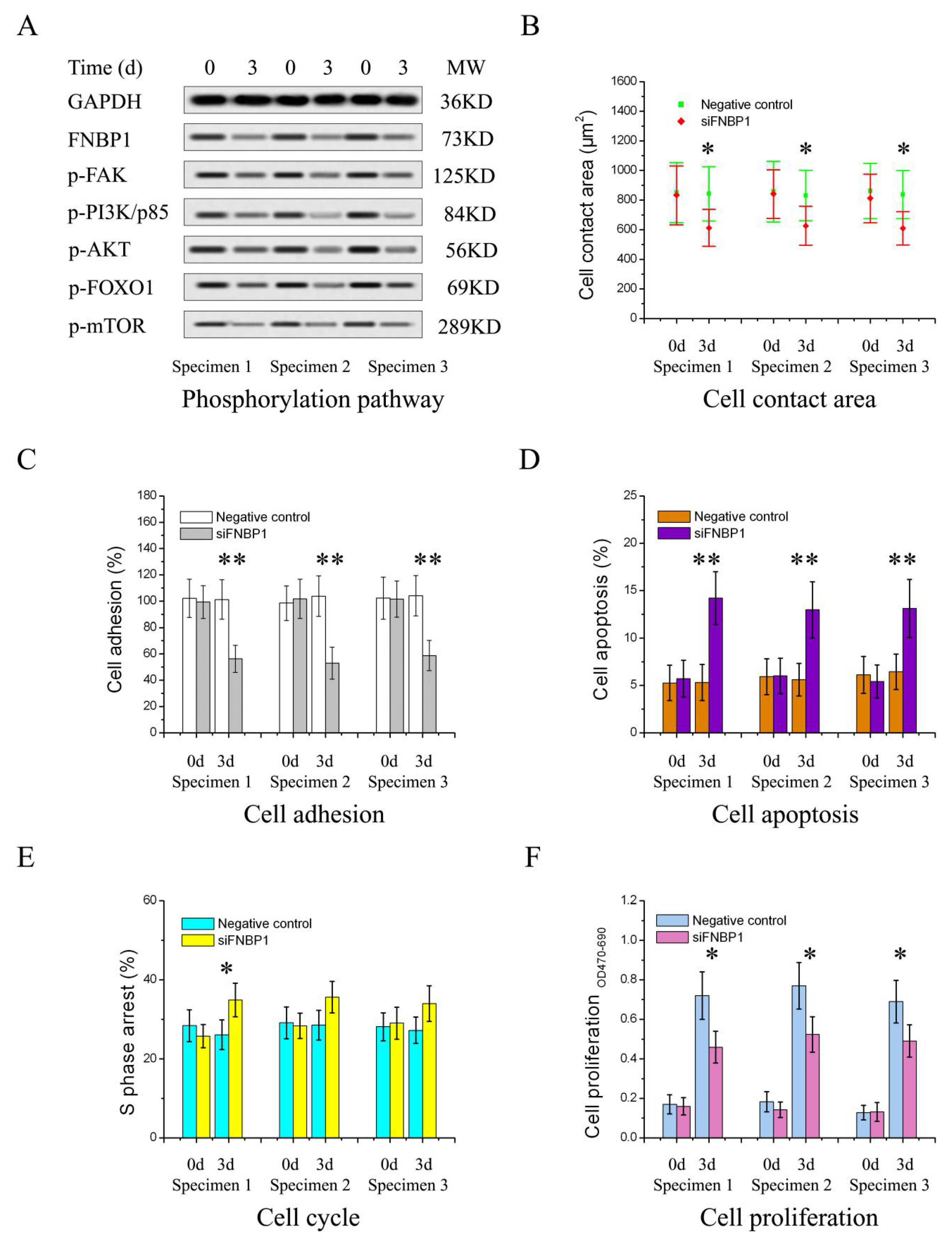
3.2. FNBP1 Konckdown Mediated Biological Effects Relieved by AKT Reactivation
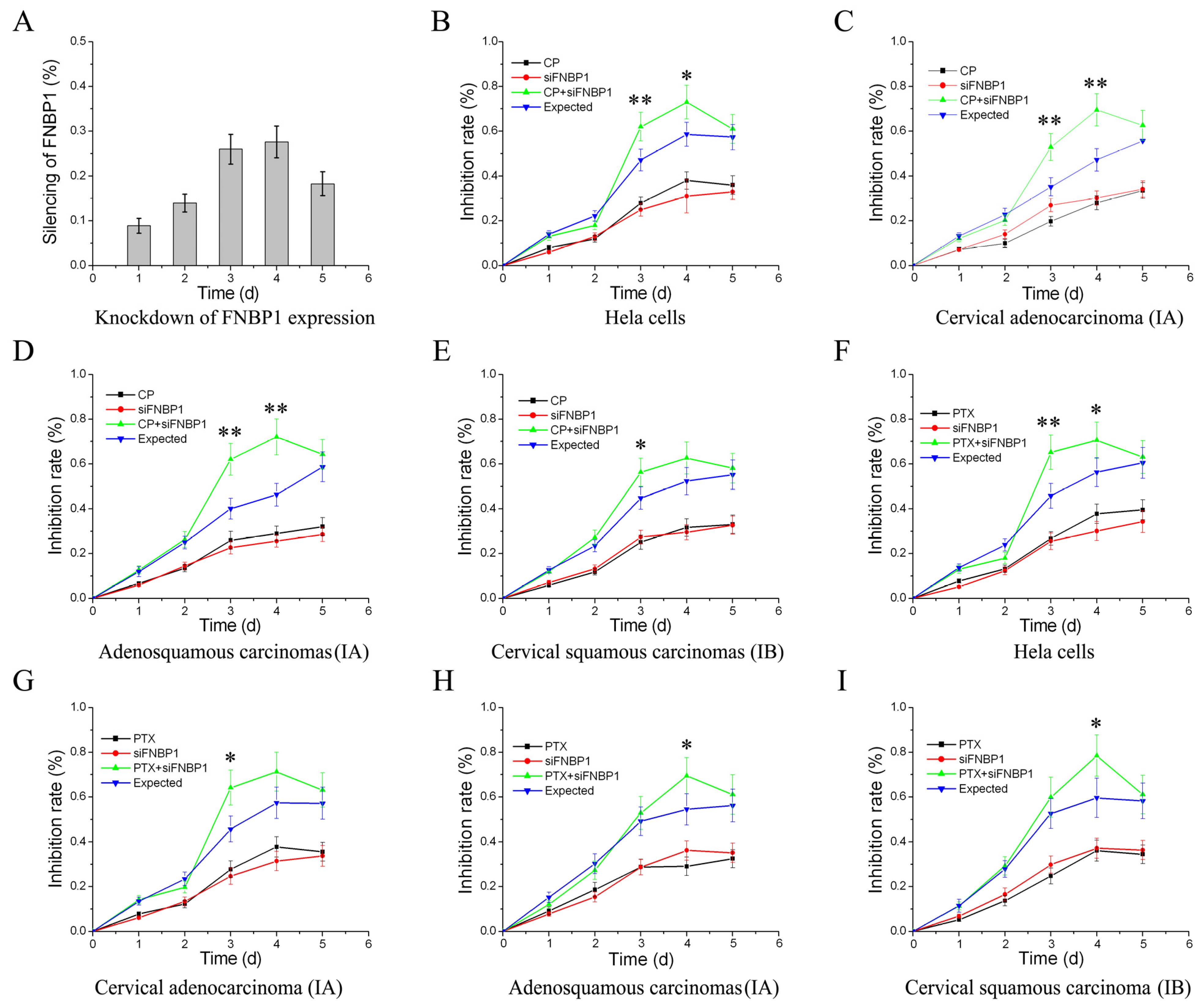
3.3. Reactivation of FAK/PI3K/AKT/mTOR Signaling by Addition of Exogenous EGF
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gupta, S.; Kumar, P.; Das, B.C. HPV: Molecular pathways and targets. Curr. Probl. Cancer. 2018, 42, 161–174. [Google Scholar] [CrossRef] [PubMed]
- Bossler, F.; Hoppe-Seyler, K.; Hoppe-Seyler, F. PI3K/AKT/mTOR Signaling Regulates the Virus/Host Cell Crosstalk in HPV-Positive Cervical Cancer Cells. Int. J. Mol. Sci. 2019, 20, 2188. [Google Scholar] [CrossRef] [PubMed]
- Buskwofie, A.; David-West, G.; Clare, C.A. A Review of Cervical Cancer: Incidence and Disparities. J. Natl. Med. Assoc. 2020, 112, 229–232. [Google Scholar] [CrossRef]
- Burmeister, C.A.; Khan, S.F.; Schäfer, G.; Mbatani, N.; Adams, T.; Moodley, J.; Prince, S. Cervical cancer therapies: Current challenges and future perspectives. Tumour Virus Res. 2022, 13, 200238. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Lee, J.Y.; Lee, Y.Y.; Shim, S.H.; Suh, D.H.; Kim, J.W. Major clinical research advances in gynecologic cancer in 2021. J. Gynecol. Oncol. 2022, 33, e43. [Google Scholar] [CrossRef]
- Wilailak, S.; Kengsakul, M.; Kehoe, S. Worldwide initiatives to eliminate cervical cancer. Int. J. Gynaecol. Obstet. 2021, 155 (Suppl. S1), 102–106. [Google Scholar] [CrossRef]
- Wirtz, C.; Mohamed, Y.; Engel, D.; Sidibe, A.; Holloway, M.; Bloem, P.; Kumar, S.; Brotherton, J.; Reis, V.; Morgan, C. Integrating HPV vaccination programs with enhanced cervical cancer screening and treatment, a systematic review. Vaccine 2022, 40 (Suppl. S1), A116–A123. [Google Scholar] [CrossRef]
- Zhao, S.; Huang, L.; Basu, P.; Domingo, E.J.; Supakarapongkul, W.; Ling, W.Y.; Ocviyanti, D.; Rezhake, R.; Qiao, Y.; Tay, E.H.; et al. Cervical cancer burden, status of implementation and challenges of cervical cancer screening in Association of Southeast Asian Nations (ASEAN) countries. Cancer Lett. 2022, 525, 22–32. [Google Scholar] [CrossRef]
- Bruni, L.; Serrano, B.; Roura, E.; Alemany, L.; Cowan, M.; Herrero, R.; Poljak, M.; Murillo, R.; Broutet, N.; Riley, L.M.; et al. Cervical cancer screening programmes and age-specific coverage estimates for 202 countries and territories worldwide: A review and synthetic analysis. Lancet Glob. Health 2022, 10, e1115–e1127. [Google Scholar] [CrossRef]
- Colombo, N.; Dubot, C.; Lorusso, D.; Caceres, M.V.; Hasegawa, K.; Shapira-Frommer, R.; Tewari, K.S.; Salman, P.; Usta, E.H.; Yañez, E.; et al. KEYNOTE-826 Investigators, Pembrolizumab for Persistent, Recurrent, or Metastatic Cervical Cancer. N. Engl. J. Med. 2021, 385, 1856–1867. [Google Scholar] [CrossRef]
- Marquina, G.; Manzano, A.; Casado, A. Targeted Agents in Cervical Cancer: Beyond Bevacizumab. Curr. Oncol. Rep. 2018, 20, 40. [Google Scholar] [CrossRef] [PubMed]
- Maene, C.; Salihi, R.R.; Van Nieuwenhuysen, E.; Han, S.N.; Concin, N.; Vergote, I. Combination of weekly paclitaxel-carboplatin plus standard bevacizumab as neoadjuvant treatment in stage IB-IIB cervical cancer. Int. J. Gynecol. Cancer 2021, 31, 824–828. [Google Scholar] [CrossRef]
- Hak, L.C.W.; Khan, S.; Di Meglio, I.; Law, A.-L.; Häsler, S.L.A.; Quintaneiro, L.M.; Ferreira, A.P.A.; Krause, M.; McMahon, H.T.; Boucrot, E. FBP17 and CIP4 recruit SHIP2 and lamellipodin to prime the plasma membrane for fast endophilin-mediated endocytosis. Nat. Cell Biol. 2018, 20, 1023–1031. [Google Scholar] [CrossRef]
- Tsuboi, S.; Takada, H.; Hara, T.; Mochizuki, N.; Funyu, T.; Saitoh, H.; Terayama, Y.; Yamaya, K.; Ohyama, C.; Nonoyama, S.; et al. FBP17 Mediates a Common Molecular Step in the Formation of Podosomes and Phagocytic Cups in Macrophages. J. Biol. Chem. 2009, 284, 8548–8556. [Google Scholar] [CrossRef]
- Tsujita, K.; Suetsugu, S.; Sasaki, N.; Furutani, M.; Oikawa, T.; Takenawa, T. Coordination between the actin cytoskeleton and membrane deformation by a novel membrane tubulation domain of PCH proteins is involved in endocytosis. J. Cell Biol. 2006, 172, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Tanaka-Takiguchi, Y.; Itoh, T.; Tsujita, K.; Yamada, S.; Yanagisawa, M.; Fujiwara, K.; Yamamoto, A.; Ichikawa, M.; Takiguchi, K. Physicochemical analysis from real-time imaging of liposome tubulation reveals the characteristics of individual F-BAR domain proteins. Langmuir 2013, 29, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Tsujita, K.; Takenawa, T.; Itoh, T. Feedback regulation between plasma membrane tension and membrane-bending proteins organizes cell polarity during leading edge formation. Nat. Cell Biol. 2015, 17, 749–758. [Google Scholar] [CrossRef] [PubMed]
- Shimada, A.; Niwa, H.; Tsujita, K.; Suetsugu, S.; Nitta, K.; Hanawa-Suetsugu, K.; Akasaka, R.; Nishino, Y.; Toyama, M.; Chen, L.; et al. Curved EFC/F-BAR-domain dimers are joined end to end into a filament for membrane invagination in endocytosis. Cell 2007, 129, 761–772. [Google Scholar] [CrossRef]
- Suman, P.; Mishra, S.; Chander, H. High formin binding protein 17 (FBP17) expression indicates poor differentiation and invasiveness of ductal carcinomas. Sci. Rep. 2020, 10, 11543. [Google Scholar] [CrossRef]
- Yamamoto, H.; Sutoh, M.; Hatakeyama, S.; Hashimoto, Y.; Yoneyama, T.; Koie, T.; Saitoh, H.; Yamaya, K.; Funyu, T.; Nakamura, T.; et al. Requirement for FBP17 in invadopodia formation by invasive bladder tumor cells. J. Urol. 2011, 185, 1930–1938. [Google Scholar] [CrossRef]
- BYoon, B.K.; Hwang, N.; Chun, K.-H.; Lee, Y.; Duarte, T.P.M.; Kim, J.-W.; Kim, T.-H.; Cheong, J.-H.; Fang, S.; Kim, J.-W. Sp1-Induced FNBP1 Drives Rigorous 3D Cell Motility in EMT-Type Gastric Cancer Cells. Int. J. Mol. Sci. 2021, 22, 6784. [Google Scholar] [CrossRef]
- Cao, R.; Shao, J.; Zhang, W.; Lin, Y.; Huang, Z.; Li, Z. Silencing long intergenic non-protein coding RNA 00987 inhibits proliferation, migration, and invasion of osteosarcoma cells by sponging miR-376a-5p to regulate FNBP1 expression. Discov. Oncol. 2021, 12, 18. [Google Scholar] [CrossRef]
- Suman, P.; Mishra, S.; Chander, H. High expression of FBP17 in invasive breast cancer cells promotes invadopodia formation. Med. Oncol. 2018, 35, 71. [Google Scholar] [CrossRef]
- Suman, P.; Mehta, V.; Craig, A.W.B.; Chander, H. Wild-type p53 suppresses formin-binding protein-17 (FBP17) to reduce invasion. Carcinogenesis 2022, 43, 494–503. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Tian, Z.; Song, X.; Zhang, J. Membrane tension sensing molecule-FNBP1 is a prognostic biomarker related to immune infiltration in BRCA, LUAD and STAD. BMC Immunol. 2022, 23, 1. [Google Scholar] [CrossRef] [PubMed]
- Khezri, M.R.; Jafari, R.; Yousefi, K.; Zolbanin, N.M. The PI3K/AKT signaling pathway in cancer: Molecular mechanisms and possible therapeutic interventions. Exp. Mol. Pathol. 2022, 127, 104787. [Google Scholar] [CrossRef] [PubMed]
- Roudsari, N.M.; Lashgari, N.-A.; Momtaz, S.; Abaft, S.; Jamali, F.; Safaiepour, P.; Narimisa, K.; Jackson, G.; Bishayee, A.; Rezaei, N.; et al. Inhibitors of the PI3K/Akt/mTOR Pathway in Prostate Cancer Chemoprevention and Intervention. Pharmaceutics 2021, 13, 1195. [Google Scholar] [CrossRef]
- Rascio, F.; Spadaccino, F.; Rocchetti, M.T.; Castellano, G.; Stallone, G.; Netti, G.S.; Ranieri, E. The Pathogenic Role of PI3K/AKT Pathway in Cancer Onset and Drug Resistance: An Updated Review. Cancers 2021, 13, 3949. [Google Scholar] [CrossRef]
- Liu, Y.-Z.; Wang, T.-T.; Zhang, Y.-Z. A modified method for the culture of naturally HPV-infected high-grade cervical intraepithelial neoplasia keratinocytes from human neoplastic cervical biopsies. Oncol. Lett. 2016, 11, 1457–1462. [Google Scholar] [CrossRef]
- Ikeda, K.; Tate, G.; Suzuki, T.; Mitsuya, T. Coordinate expression of cytokeratin 8 and cytokeratin 17 immunohistochemical staining in cervical intraepithelial neoplasia and cervical squamous cell carcinoma: An immunohistochemical analysis and review of the literature. Gynecol. Oncol. 2008, 108, 598–602. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, J.; Zhou, Y.-F.; Ren, X.-Y.; Lin, M.-M.; Zhang, Q.-Q.; Wang, Y.-H.; Li, X. Rich1 negatively regulates the epithelial cell cycle, proliferation and adhesion by CDC42/RAC1-PAK1-Erk1/2 pathway. Cell. Signal. 2015, 27, 1703–1712. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Cheng, C.; He, C.-L.; Zhou, Y.-J.; Cao, Y. The expression of Bcl-XL, Bcl-XS and p27Kip1 in topotecan-induced apoptosis in hepatoblastoma HepG2 cell line. Cancer Investig. 2008, 26, 456–463. [Google Scholar] [CrossRef] [PubMed]
- Adamo, M.P.; Monzón, M.L.; Cuffini, C.; Pedranti, M.; Zapata, M. Detection of rubella-virus-induced apoptosis in Vero cell cultures with hematoxylin and eosin staining. Rev. Argent. Microbiol. 2002, 34, 177–185. [Google Scholar] [PubMed]
- Takano, K.; Toyooka, K.; Suetsugu, S. EFC/F-BAR proteins and the N-WASP-WIP complex induce membrane curvature-dependent actin polymerization. EMBO J. 2008, 27, 2817–2828. [Google Scholar] [CrossRef]
- Katoh, M.; Katoh, M. Identification and characterization of human FNBP1L gene in silico. Int. J. Mol. Med. 2004, 13, 157–162. [Google Scholar] [CrossRef]
- Frost, A.; Perera, R.; Roux, A.; Spasov, K.; Destaing, O.; Egelman, E.H.; De Camilli, P.; Unger, V.M. Structural basis of membrane invagination by F-BAR domains. Cell 2008, 132, 807–817. [Google Scholar] [CrossRef]
- Ma, Y.Y.; Wei, S.J.; Li, Y.C.; Lung, J.C.; Chang, T.C.; Whang-Peng, J.; Liu, J.M.; Yang, D.M.; Yang, W.K.; Shen, C.Y. PIK3CA as an oncogene in cervical cancer. Oncogene 2000, 19, 2739–2744. [Google Scholar] [CrossRef]
- Manning, B.D.; Toker, A. AKT/PKB Signaling: Navigating the Network. Cell 2017, 169, 381–405. [Google Scholar] [CrossRef]
- Song, M.; Bode, A.M.; Dong, Z.; Lee, M.H. AKT as a Therapeutic Target for Cancer. Cancer Res. 2019, 79, 1019–1031. [Google Scholar] [CrossRef]
- Formisano, L.; Napolitano, F.; Rosa, R.; D’Amato, V.; Servetto, A.; Marciano, R.; De Placido, P.; Bianco, C.; Bianco, R. Mechanisms of resistance to mTOR inhibitors. Crit. Rev. Oncol. Hematol. 2020, 147, 102886. [Google Scholar] [CrossRef]
- Lee, E.Y.; Muller, W.J. Oncogenes and tumor suppressor genes. Cold Spring Harb. Perspect. Biol. 2010, 2, a003236. [Google Scholar] [CrossRef]
- Cesur-Ergün, B.; Demir-Dora, D. Gene therapy in cancer. J. Gene Med. 2023, e3550. [Google Scholar] [CrossRef] [PubMed]
- Song, F.; Dai, Q.; Grimm, M.O.; Steinbach, D. The Antithetic Roles of IQGAP2 and IQGAP3 in Cancers. Cancers 2023, 15, 1115. [Google Scholar] [CrossRef] [PubMed]
- Hahn, W.C.; Bader, J.S.; Braun, T.P.; Califano, A.; Clemons, P.A.; Druker, B.J.; Ewald, A.J.; Fu, H.; Jagu, S.; Kemp, C.J.; et al. An expanded universe of cancer targets. Cell 2021, 184, 1142–1155. [Google Scholar] [CrossRef] [PubMed]
- Petroni, G.; Buqué, A.; Coussens, L.M.; Galluzzi, L. Targeting oncogene and non-oncogene addiction to inflame the tumour microenvironment. Nat. Rev. Drug Discov. 2022, 21, 440–462. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Li, X.; Zhou, Y.; Lin, M.; Zhang, Q.; Wang, Y. FNBP1 Facilitates Cervical Cancer Cell Survival by the Constitutive Activation of FAK/PI3K/AKT/mTOR Signaling. Cells 2023, 12, 1964. https://doi.org/10.3390/cells12151964
Zhang J, Li X, Zhou Y, Lin M, Zhang Q, Wang Y. FNBP1 Facilitates Cervical Cancer Cell Survival by the Constitutive Activation of FAK/PI3K/AKT/mTOR Signaling. Cells. 2023; 12(15):1964. https://doi.org/10.3390/cells12151964
Chicago/Turabian StyleZhang, Jun, Xin Li, Yunfei Zhou, Mingming Lin, Qianying Zhang, and Yunhong Wang. 2023. "FNBP1 Facilitates Cervical Cancer Cell Survival by the Constitutive Activation of FAK/PI3K/AKT/mTOR Signaling" Cells 12, no. 15: 1964. https://doi.org/10.3390/cells12151964
APA StyleZhang, J., Li, X., Zhou, Y., Lin, M., Zhang, Q., & Wang, Y. (2023). FNBP1 Facilitates Cervical Cancer Cell Survival by the Constitutive Activation of FAK/PI3K/AKT/mTOR Signaling. Cells, 12(15), 1964. https://doi.org/10.3390/cells12151964






