Epigenetic Reprogramming in Mice and Humans: From Fertilization to Primordial Germ Cell Development
Abstract
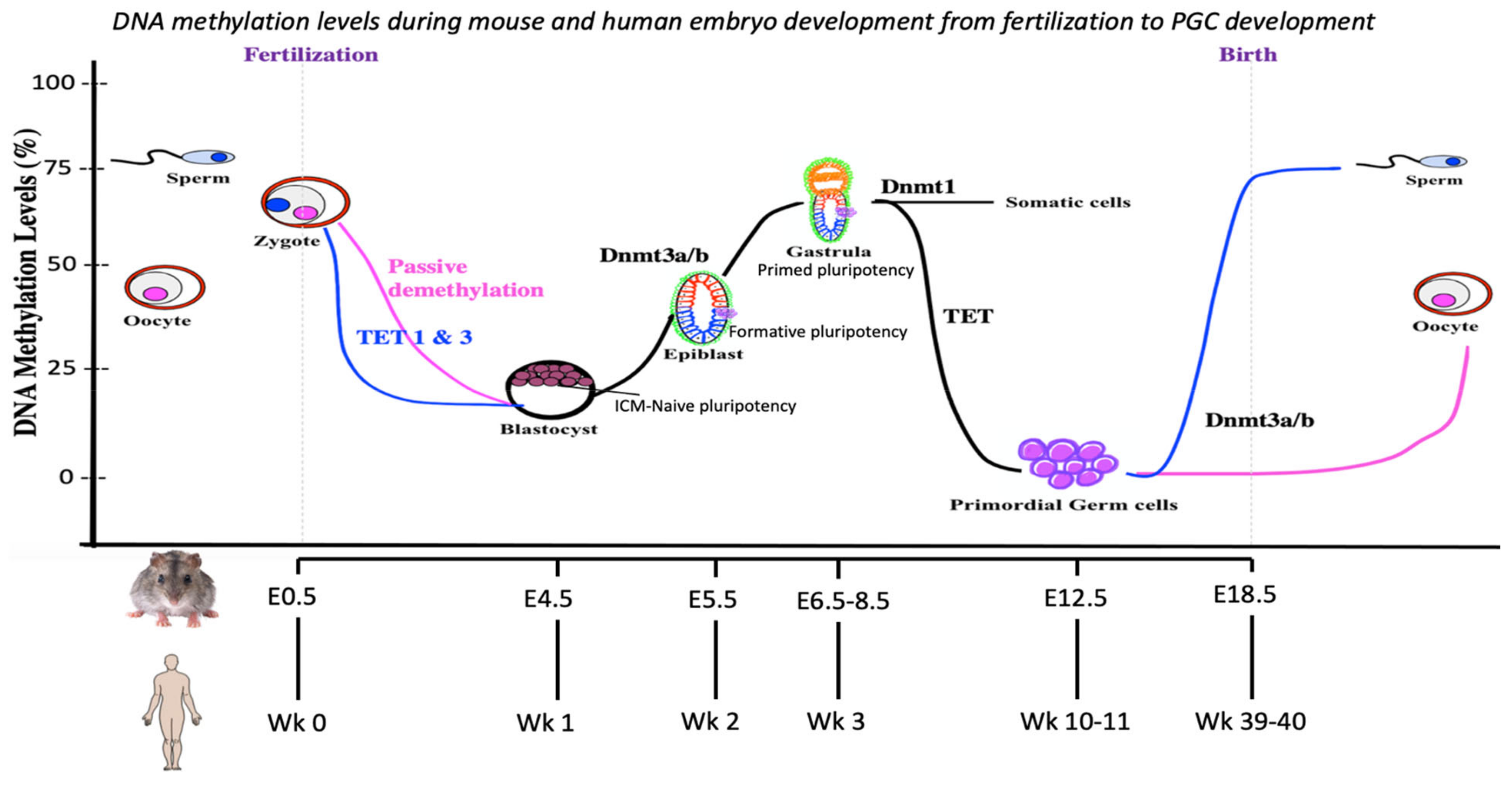
1. Epigenetic Reprogramming: Overview
2. DNA Demethylation: First Wave after Fertilization
3. Tet Mutants and Knockouts
4. De Novo DNA Methylation after the Blastocyst ICM Nadir
5. Maintenance of Methylation by Dnmt1
6. Tet Enzymes Antagonistically Regulate DNA Methylation Mediated by Dnmt Enzymes
7. PGC Migration during Embryonic Development
8. Epigenetic Reprogramming and Random X-Chromosome Inactivation (XCI)
9. Understanding Genomic Imprinting by ICR and Imprinted DMRs in Epigenetics
10. Exploring the Potential of ICRs for Studying Windows of Early Exposures to Toxicants and their Effects on Genomic Imprinting
11. Endocrine-Disrupting Chemicals and Their Potential Effects on Epigenetic Reprogramming and Transgenerational Inheritance
12. DNA Demethylation in PGCs after Allocation from Formative Pluripotent Epiblasts: Second Wave
13. Consequences of Altered Epigenetic Reprogramming in PGCs for Germ Cell Development, Reproductive Health, and Offspring Health
14. Non-CpG Methylation (CpA, CpT and CpC) and Its Unique Role in Epigenetic Reprogramming
15. Histone Modifications during Epigenetic Reprogramming
16. Placental Epigenetics: How the Imprinting and X-Inactivation Differs from Embryo
17. Concluding Remarks
18. Future Directions
| Aspect | Mice | Humans |
|---|---|---|
| DNA Methylation | DNA methylation undergoes erasure during PGC development [163]. | DNA methylation is partially retained in mature gametes [10] |
| Histone-to-Protamine Transition | Histones are replaced by protamines during sperm maturation [164] | Histones are replaced by protamines during sperm maturation [164] |
| DNA Methylation Dynamics in Spermatogenesis | Re-establishment of DNA methylation during spermatogenesis [165] | Mechanisms of DNA methylation re-establishment not well explored |
| DNA Methylation Patterns in Oocytes | Unique bimodal pattern, predominantly in gene bodies (~40% methylation in oocytes) [1] | Higher average DNA methylation, predominantly in gene bodies (~54% methylation in oocytes) [166] |
| DNMT3L Expression | Essential for de-novo methylation in mouse oocytes [167] | Not expressed in human oocytes [168] |
| Retained Histones in Mature Sperm | Few nucleosomes are retained in mature sperm [169] | More nucleosomes are retained in mature sperm [170]. |
| DNA Methylation | Paternal protamines replaced by maternal histones, erasure of almost all paternal DNA methylation. Maternal DNA methylation largely preserved. | Global reprogramming of DNA methylation in the pre-implantation embryo, with substantial retention of maternal methylation. Less passive demethylation, possibly due to a more active role of DNMT1 [139]. |
| Zygotic Genome Activation (ZGA) | Major wave of ZGA at the 2-cell stage [171]. | Major wave of ZGA at the 8-cell stage [172]. |
| Chromatin Remodeling | Relaxed chromatin state in zygotes gradually resolved to a more canonical state by the blastocyst stage [173]. | Widespread open chromatin in pre-ZGA embryos, rapidly remodeled upon ZGA. Temporal regulation of chromatin accessibility dependent on transcriptional activation [174]. |
| Transcriptome | Differences in the transcriptome compared to humans. Similar transcription factors, but divergent regulation and networks [175]. | Similar transcription factors, but temporal regulation and networks can differ [175]. |
| De novo Methylation | Two phases of de novo methylation: first in the paternal genome in the zygote, second between the 4- and 8-cell stage coinciding with ZGA. Transient methylation of repeat elements [176]. | De novo methylation observed during pre-implantation development. Two phases: early-to-mid pronuclear stage in the paternal genome, and between the 4- and 8-cell stage coinciding with ZGA. Methylation of repeat elements, transient in subsequent developmental stages [177]. |
| Function of Repressive Chromatin | Repressive chromatin marks such as H3K27me3 play a role in reinforcing lineage specification in both mice and humans [178]. | The targeted gain of H3K27me3 is observed in the post-implantation embryo in mice, but the specific mechanisms and extent of H3K27me3’s function in humans are largely unexplored [178]. |
| Role of H3K9me2 | H3K9me2 is associated with methylated DNA in the post-implantation embryo in both mice and humans. However, its functional role is specialized and not required for the genome-wide gain of DNA methylation [179]. | H3K9me2’s specific functions in the post-implantation embryo in humans are not well understood, and its role may be different from mice. |
| Active Chromatin Marks | Active histone modifications like H3K4me3 and H3K27ac likely play a role in transcriptional regulation during lineage specification in both mice and humans [178]. | The specific requirements and effects of H3K4me3 and H3K27ac in establishing and reinforcing the transcriptional program during lineage specification may vary between mice and humans [178]. |
| Imprinted Gene Clusters | Conserved in methylation status, allelic expression, and synteny [17] | Conserved in methylation status, allelic expression, and synteny, with several exceptions [17] |
| Number of Imprinted Genes | ~151 [180] | 50–90 [181] |
| Identification Methods | Sequencing approaches over SNPs, genomic imbalances [181] | Sequencing approaches over SNPs, genomic imbalances [181] |
| Regulatory Complexity | Imprinted gene expression and methylation may be more widespread and variable [180] | Imprinted gene expression and methylation may be more widespread and variable than mice [182] |
| Maintenance of Imprints | Requires ZFP57 and other genetic factors during later stages [183] | Maintenance of imprints during human reprogramming is not well understood [178] |
| Placental Imprinting | Limited number of placental-specific imprinted gDMRs [184] | More than 1500 placental-specific imprinted gDMRs, mostly not conserved between species [185] |
| Imprinting mechanisms | DNA methylation, H3K27me3 (extra-embryonic lineages) [184] | DNA methylation, H3K27me3 (unknown if present) |
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kobayashi, H.; Sakurai, T.; Imai, M.; Takahashi, N.; Fukuda, A.; Yayoi, O.; Sato, S.; Nakabayashi, K.; Hata, K.; Sotomaru, Y.; et al. Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genet. 2012, 8, e1002440. [Google Scholar] [CrossRef]
- Smallwood, S.A.; Tomizawa, S.I.; Krueger, F.; Ruf, N.; Carli, N.; Segonds-Pichon, A.; Sato, S.; Hata, K.; Andrews, S.R.; Kelsey, G. Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat. Genet. 2011, 43, 811–814. [Google Scholar] [CrossRef]
- Veselovska, L.; Smallwood, S.A.; Saadeh, H.; Stewart, K.R.; Krueger, F.; Maupetit-Méhouas, S.; Arnaud, P.; Tomizawa, S.I.; Andrews, S.; Kelsey, G. Deep sequencing and de novo assembly of the mouse oocyte transcriptome define the contribution of transcription to the DNA methylation landscape. Genome Biol. 2015, 16, 209. [Google Scholar] [CrossRef]
- Lee, J.; Inoue, K.; Ono, R.; Ogonuki, N.; Kohda, T.; Kaneko-Ishino, T.; Ogura, A.; Ishino, F. Erasing genomic imprinting memory in mouse clone embryos produced from day 11.5 primordial germ cells. Development 2002, 129, 1807–1817. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, Q.; Tang, F.; Yan, L.; Qiao, J. Epigenetic Regulation and Risk Factors During the Development of Human Gametes and Early Embryos. Annu. Rev. Genomics Hum. Genet. 2019, 20, 21–40. [Google Scholar] [CrossRef]
- Wu, S.C.; Zhang, Y. Active DNA demethylation: Many roads lead to Rome. Nat. Rev. Mol. Cell Biol. 2010, 11, 607–620. [Google Scholar] [CrossRef]
- Fang, F.; Fan, S.; Zhang, X.; Zhang, M.Q. Predicting methylation status of CpG islands in the human brain. Bioinformatics 2006, 22, 2204–2209. [Google Scholar] [CrossRef]
- Ramsahoye, B.H.; Biniszkiewicz, D.; Lyko, F.; Clark, V.; Bird, A.P.; Jaenisch, R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl. Acad. Sci. USA 2000, 97, 5237–5242. [Google Scholar] [CrossRef]
- Long, M.D.; Smiraglia, D.J.; Campbell, M.J. The Genomic Impact of DNA CpG Methylation on Gene Expression; Relationships in Prostate Cancer. Biomolecules 2017, 7, 15. [Google Scholar] [CrossRef]
- Guo, F.; Yan, L.; Guo, H.; Li, L.; Hu, B.; Zhao, Y.; Yong, J.; Hu, Y.; Wang, X.; Wei, Y.; et al. The Transcriptome and DNA Methylome Landscapes of Human Primordial Germ Cells. Cell 2015, 161, 1437–1452. [Google Scholar] [CrossRef]
- Fukuda, K.; Makino, Y.; Kaneko, S.; Shimura, C.; Okada, Y.; Ichiyanagi, K.; Shinkai, Y. Potential role of KRAB-ZFP binding and transcriptional states on DNA methylation of retroelements in human male germ cells. elife 2022, 11, e76822. [Google Scholar] [CrossRef]
- Wang, J.; Huang, J.; Shi, G. Retrotransposons in pluripotent stem cells. Cell Regen. 2020, 9, 4. [Google Scholar] [CrossRef]
- Tanaka, S.S.; Toyooka, Y.; Akasu, R.; Katoh-Fukui, Y.; Nakahara, Y.; Suzuki, R.; Yokoyama, M.; Noce, T. The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes. Dev. 2000, 14, 841–853. [Google Scholar] [CrossRef]
- Dansereau, D.A.; Lasko, P. The development of germline stem cells in Drosophila. Methods Mol. Biol. 2008, 450, 3–26. [Google Scholar] [CrossRef]
- Boellaard, W.P.A.; Stoop, H.; Gillis, A.J.M.; Oosterhuis, J.W.; Looijenga, L.H.J. VASA mRNA (DDX4) detection is more specific than immunohistochemistry using poly- or monoclonal antibodies for germ cells in the male urogenital tract. Medicine 2017, 96, e7489. [Google Scholar] [CrossRef]
- Messerschmidt, D.M.; Knowles, B.B.; Solter, D. DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes. Dev. 2014, 28, 812–828. [Google Scholar] [CrossRef]
- Reik, W.; Dean, W.; Walter, J. Epigenetic reprogramming in mammalian development. Science 2001, 293, 1089–1093. [Google Scholar] [CrossRef]
- Santos, F.; Hendrich, B.; Reik, W.; Dean, W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev. Biol. 2002, 241, 172–182. [Google Scholar] [CrossRef]
- Gu, T.P.; Guo, F.; Yang, H.; Wu, H.P.; Xu, G.F.; Liu, W.; Xie, Z.G.; Shi, L.; He, X.; Jin, S.G.; et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 2011, 477, 606–610. [Google Scholar] [CrossRef]
- Loenarz, C.; Schofield, C.J. Physiological and biochemical aspects of hydroxylations and demethylations catalyzed by human 2-oxoglutarate oxygenases. Trends Biochem. Sci. 2011, 36, 7–18. [Google Scholar] [CrossRef]
- Ko, M.; An, J.; Bandukwala, H.S.; Chavez, L.; Aijo, T.; Pastor, W.A.; Segal, M.F.; Li, H.; Koh, K.P.; Lahdesmaki, H.; et al. Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature 2013, 497, 122–126. [Google Scholar] [CrossRef]
- Xu, Y.; Xu, C.; Kato, A.; Tempel, W.; Abreu, J.G.; Bian, C.; Hu, Y.; Hu, D.; Zhao, B.; Cerovina, T.; et al. Tet3 CXXC domain and dioxygenase activity cooperatively regulate key genes for Xenopus eye and neural development. Cell 2012, 151, 1200–1213. [Google Scholar] [CrossRef]
- Bortvin, A.; Goodheart, M.; Liao, M.; Page, D.C. Dppa3/Pgc7/stella is a maternal factor and is not required for germ cell specification in mice. BMC Dev. Biol. 2004, 4, 2. [Google Scholar] [CrossRef]
- Hackett, J.A.; Huang, Y.; Gunesdogan, U.; Gretarsson, K.A.; Kobayashi, T.; Surani, M.A. Tracing the transitions from pluripotency to germ cell fate with CRISPR screening. Nat. Commun. 2018, 9, 4292. [Google Scholar] [CrossRef]
- Nakamura, T.; Liu, Y.J.; Nakashima, H.; Umehara, H.; Inoue, K.; Matoba, S.; Tachibana, M.; Ogura, A.; Shinkai, Y.; Nakano, T. PGC7 binds histone H3K9me2 to protect against conversion of 5mC to 5hmC in early embryos. Nature 2012, 486, 415–419. [Google Scholar] [CrossRef]
- Petrussa, L.; Van de Velde, H.; De Rycke, M. Dynamic regulation of DNA methyltransferases in human oocytes and preimplantation embryos after assisted reproductive technologies. Mol. Hum. Reprod. 2014, 20, 861–874. [Google Scholar] [CrossRef]
- Greenberg, M.V.C.; Bourc’his, D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 2019, 20, 590–607. [Google Scholar] [CrossRef]
- Smith, Z.D.; Chan, M.M.; Mikkelsen, T.S.; Gu, H.; Gnirke, A.; Regev, A.; Meissner, A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 2012, 484, 339–344. [Google Scholar] [CrossRef]
- Argelaguet, R.; Clark, S.J.; Mohammed, H.; Stapel, L.C.; Krueger, C.; Kapourani, C.A.; Imaz-Rosshandler, I.; Lohoff, T.; Xiang, Y.; Hanna, C.W.; et al. Multi-omics profiling of mouse gastrulation at single-cell resolution. Nature 2019, 576, 487–491. [Google Scholar] [CrossRef]
- Yang, J.; Bashkenova, N.; Zang, R.; Huang, X.; Wang, J. The roles of TET family proteins in development and stem cells. Development 2020, 147, dev183129. [Google Scholar] [CrossRef]
- Dawlaty, M.M.; Ganz, K.; Powell, B.E.; Hu, Y.C.; Markoulaki, S.; Cheng, A.W.; Gao, Q.; Kim, J.; Choi, S.W.; Page, D.C.; et al. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell 2011, 9, 166–175. [Google Scholar] [CrossRef]
- Li, H.; Hu, Z.; Jiang, H.; Pu, J.; Selli, I.; Qiu, J.; Zhang, B.; Feng, J. TET1 Deficiency Impairs Morphogen-free Differentiation of Human Embryonic Stem Cells to Neuroectoderm. Sci. Rep. 2020, 10, 10343. [Google Scholar] [CrossRef]
- Dawlaty, M.M.; Breiling, A.; Le, T.; Raddatz, G.; Barrasa, M.I.; Cheng, A.W.; Gao, Q.; Powell, B.E.; Li, Z.; Xu, M.; et al. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev. Cell 2013, 24, 310–323. [Google Scholar] [CrossRef]
- Ma, L.; Tang, Q.; Gao, X.; Lee, J.; Lei, R.; Suzuki, M.; Zheng, D.; Ito, K.; Frenette, P.S.; Dawlaty, M.M. Tet-mediated DNA demethylation regulates specification of hematopoietic stem and progenitor cells during mammalian embryogenesis. Sci. Adv. 2022, 8, eabm3470. [Google Scholar] [CrossRef]
- Li, Z.; Fang, F.; Long, Y.; Zhao, Q.; Wang, X.; Ye, Z.; Meng, T.; Gu, X.; Xiang, W.; Xiong, C.; et al. The balance between NANOG and SOX17 mediated by TET proteins regulates specification of human primordial germ cell fate. Cell Biosci. 2022, 12, 181. [Google Scholar] [CrossRef]
- Smith, A. Formative pluripotency: The executive phase in a developmental continuum. Development 2017, 144, 365–373. [Google Scholar] [CrossRef]
- Dahlet, T.; Argueso Lleida, A.; Al Adhami, H.; Dumas, M.; Bender, A.; Ngondo, R.P.; Tanguy, M.; Vallet, J.; Auclair, G.; Bardet, A.F.; et al. Genome-wide analysis in the mouse embryo reveals the importance of DNA methylation for transcription integrity. Nat. Commun. 2020, 11, 3153. [Google Scholar] [CrossRef]
- Zhang, Z.M.; Lu, R.; Wang, P.; Yu, Y.; Chen, D.; Gao, L.; Liu, S.; Ji, D.; Rothbart, S.B.; Wang, Y.; et al. Structural basis for DNMT3A-mediated de novo DNA methylation. Nature 2018, 554, 387–391. [Google Scholar] [CrossRef]
- Otani, J.; Nankumo, T.; Arita, K.; Inamoto, S.; Ariyoshi, M.; Shirakawa, M. Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain. EMBO Rep. 2009, 10, 1235–1241. [Google Scholar] [CrossRef]
- Bostick, M.; Kim, J.K.; Esteve, P.O.; Clark, A.; Pradhan, S.; Jacobsen, S.E. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 2007, 317, 1760–1764. [Google Scholar] [CrossRef]
- Howell, C.Y.; Bestor, T.H.; Ding, F.; Latham, K.E.; Mertineit, C.; Trasler, J.M.; Chaillet, J.R. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell 2001, 104, 829–838. [Google Scholar] [CrossRef] [PubMed]
- Li, E.; Zhang, Y. DNA methylation in mammals. Cold Spring Harb. Perspect. Biol. 2014, 6, a019133. [Google Scholar] [CrossRef] [PubMed]
- Rountree, M.R.; Bachman, K.E.; Baylin, S.B. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet. 2000, 25, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Chuang, L.S.; Ian, H.I.; Koh, T.W.; Ng, H.H.; Xu, G.; Li, B.F. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science 1997, 277, 1996–2000. [Google Scholar] [CrossRef] [PubMed]
- Ishiyama, S.; Nishiyama, A.; Saeki, Y.; Moritsugu, K.; Morimoto, D.; Yamaguchi, L.; Arai, N.; Matsumura, R.; Kawakami, T.; Mishima, Y.; et al. Structure of the Dnmt1 Reader Module Complexed with a Unique Two-Mono-Ubiquitin Mark on Histone H3 Reveals the Basis for DNA Methylation Maintenance. Mol. Cell 2017, 68, 350–360.e7. [Google Scholar] [CrossRef]
- Takeshita, K.; Suetake, I.; Yamashita, E.; Suga, M.; Narita, H.; Nakagawa, A.; Tajima, S. Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1). Proc. Natl. Acad. Sci. USA 2011, 108, 9055–9059. [Google Scholar] [CrossRef]
- Song, J.; Rechkoblit, O.; Bestor, T.H.; Patel, D.J. Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation. Science 2011, 331, 1036–1040. [Google Scholar] [CrossRef]
- Yarychkivska, O.; Shahabuddin, Z.; Comfort, N.; Boulard, M.; Bestor, T.H. BAH domains and a histone-like motif in DNA methyltransferase 1 (DNMT1) regulate de novo and maintenance methylation in vivo. J. Biol. Chem. 2018, 293, 19466–19475. [Google Scholar] [CrossRef]
- Kyriakopoulos, C.; Nordstrom, K.; Kramer, P.L.; Gottfreund, J.Y.; Salhab, A.; Arand, J.; Muller, F.; von Meyenn, F.; Ficz, G.; Reik, W.; et al. A comprehensive approach for genome-wide efficiency profiling of DNA modifying enzymes. Cell Rep. Methods 2022, 2, 100187. [Google Scholar] [CrossRef]
- Kinoshita, M.; Barber, M.; Mansfield, W.; Cui, Y.; Spindlow, D.; Stirparo, G.G.; Dietmann, S.; Nichols, J.; Smith, A. Capture of Mouse and Human Stem Cells with Features of Formative Pluripotency. Cell Stem Cell 2021, 28, 453–471. [Google Scholar] [CrossRef]
- Ohinata, Y.; Ohta, H.; Shigeta, M.; Yamanaka, K.; Wakayama, T.; Saitou, M. A signaling principle for the specification of the germ cell lineage in mice. Cell 2009, 137, 571–584. [Google Scholar] [CrossRef]
- Yu, L.; Wei, Y.; Duan, J.; Schmitz, D.A.; Sakurai, M.; Wang, L.; Wang, K.; Zhao, S.; Hon, G.C.; Wu, J. Blastocyst-like structures generated from human pluripotent stem cells. Nature 2021, 591, 620–626. [Google Scholar] [CrossRef]
- Scholer, H.R.; Hatzopoulos, A.K.; Balling, R.; Suzuki, N.; Gruss, P. A family of octamer-specific proteins present during mouse embryogenesis: Evidence for germline-specific expression of an Oct factor. EMBO J. 1989, 8, 2543–2550. [Google Scholar] [CrossRef]
- Kehler, J.; Tolkunova, E.; Koschorz, B.; Pesce, M.; Gentile, L.; Boiani, M.; Lomeli, H.; Nagy, A.; McLaughlin, K.J.; Scholer, H.R.; et al. Oct4 is required for primordial germ cell survival. EMBO Rep. 2004, 5, 1078–1083. [Google Scholar] [CrossRef]
- Lawson, K.A.; Hage, W.J. Clonal analysis of the origin of primordial germ cells in the mouse. Ciba Found. Symp. 1994, 182, 68–84, discussion 84–91. [Google Scholar] [CrossRef]
- Yoshimizu, T.; Obinata, M.; Matsui, Y. Stage-specific tissue and cell interactions play key roles in mouse germ cell specification. Development 2001, 128, 481–490. [Google Scholar] [CrossRef]
- Brons, I.G.; Smithers, L.E.; Trotter, M.W.; Rugg-Gunn, P.; Sun, B.; Chuva de Sousa Lopes, S.M.; Howlett, S.K.; Clarkson, A.; Ahrlund-Richter, L.; Pedersen, R.A.; et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 2007, 448, 191–195. [Google Scholar] [CrossRef]
- Guo, G.; Yang, J.; Nichols, J.; Hall, J.S.; Eyres, I.; Mansfield, W.; Smith, A. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development 2009, 136, 1063–1069. [Google Scholar] [CrossRef]
- Hayashi, K.; Ohta, H.; Kurimoto, K.; Aramaki, S.; Saitou, M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell 2011, 146, 519–532. [Google Scholar] [CrossRef]
- Ulmert, I.; Henriques-Oliveira, L.; Pereira, C.F.; Lahl, K. Mononuclear phagocyte regulation by the transcription factor Blimp-1 in health and disease. Immunology 2020, 161, 303–313. [Google Scholar] [CrossRef]
- Ohinata, Y.; Payer, B.; O’Carroll, D.; Ancelin, K.; Ono, Y.; Sano, M.; Barton, S.C.; Obukhanych, T.; Nussenzweig, M.; Tarakhovsky, A.; et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature 2005, 436, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Chia, N.Y.; Chan, Y.S.; Feng, B.; Lu, X.; Orlov, Y.L.; Moreau, D.; Kumar, P.; Yang, L.; Jiang, J.; Lau, M.S.; et al. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature 2010, 468, 316–320. [Google Scholar] [CrossRef] [PubMed]
- Durcova-Hills, G.; Tang, F.; Doody, G.; Tooze, R.; Surani, M.A. Reprogramming primordial germ cells into pluripotent stem cells. PLoS ONE 2008, 3, e3531. [Google Scholar] [CrossRef] [PubMed]
- Leitch, H.G.; Blair, K.; Mansfield, W.; Ayetey, H.; Humphreys, P.; Nichols, J.; Surani, M.A.; Smith, A. Embryonic germ cells from mice and rats exhibit properties consistent with a generic pluripotent ground state. Development 2010, 137, 2279–2287. [Google Scholar] [CrossRef]
- Magnusdottir, E.; Dietmann, S.; Murakami, K.; Gunesdogan, U.; Tang, F.; Bao, S.; Diamanti, E.; Lao, K.; Gottgens, B.; Azim Surani, M. A tripartite transcription factor network regulates primordial germ cell specification in mice. Nat. Cell Biol. 2013, 15, 905–915. [Google Scholar] [CrossRef]
- Acampora, D.; Di Giovannantonio, L.G.; Simeone, A. Otx2 is an intrinsic determinant of the embryonic stem cell state and is required for transition to a stable epiblast stem cell condition. Development 2013, 140, 43–55. [Google Scholar] [CrossRef]
- Cantu, A.V.; Laird, D.J. A pilgrim’s progress: Seeking meaning in primordial germ cell migration. Stem Cell Res. 2017, 24, 181–187. [Google Scholar] [CrossRef]
- Grimaldi, C.; Raz, E. Germ cell migration-Evolutionary issues and current understanding. Semin. Cell Dev. Biol. 2020, 100, 152–159. [Google Scholar] [CrossRef]
- Barton, L.J.; LeBlanc, M.G.; Lehmann, R. Finding their way: Themes in germ cell migration. Curr. Opin. Cell Biol. 2016, 42, 128–137. [Google Scholar] [CrossRef]
- Sasaki, H.; Matsui, Y. Epigenetic events in mammalian germ-cell development: Reprogramming and beyond. Nat. Rev. Genet. 2008, 9, 129–140. [Google Scholar] [CrossRef]
- Hajkova, P.; Jeffries, S.J.; Lee, C.; Miller, N.; Jackson, S.P.; Surani, M.A. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science 2010, 329, 78–82. [Google Scholar] [CrossRef]
- De Napoles, M.; Nesterova, T.; Brockdorff, N. Early loss of Xist RNA expression and inactive X chromosome associated chromatin modification in developing primordial germ cells. PLoS ONE 2007, 2, e860. [Google Scholar] [CrossRef]
- Sugimoto, M.; Abe, K. X chromosome reactivation initiates in nascent primordial germ cells in mice. PLoS Genet. 2007, 3, e116. [Google Scholar] [CrossRef]
- Huynh, K.D.; Lee, J.T. Inheritance of a pre-inactivated paternal X chromosome in early mouse embryos. Nature 2003, 426, 857–862. [Google Scholar] [CrossRef]
- Mak, W.; Nesterova, T.B.; de Napoles, M.; Appanah, R.; Yamanaka, S.; Otte, A.P.; Brockdorff, N. Reactivation of the paternal X chromosome in early mouse embryos. Science 2004, 303, 666–669. [Google Scholar] [CrossRef]
- Okamoto, I.; Otte, A.P.; Allis, C.D.; Reinberg, D.; Heard, E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science 2004, 303, 644–649. [Google Scholar] [CrossRef]
- Namekawa, S.H.; Payer, B.; Huynh, K.D.; Jaenisch, R.; Lee, J.T. Two-step imprinted X inactivation: Repeat versus genic silencing in the mouse. Mol. Cell Biol. 2010, 30, 3187–3205. [Google Scholar] [CrossRef]
- Kalantry, S.; Purushothaman, S.; Bowen, R.B.; Starmer, J.; Magnuson, T. Evidence of Xist RNA-independent initiation of mouse imprinted X-chromosome inactivation. Nature 2009, 460, 647–651. [Google Scholar] [CrossRef]
- Murakami, K.; Araki, K.; Ohtsuka, S.; Wakayama, T.; Niwa, H. Choice of random rather than imprinted X inactivation in female embryonic stem cell-derived extra-embryonic cells. Development 2011, 138, 197–202. [Google Scholar] [CrossRef]
- Furlan, G.; Rougeulle, C. Function and evolution of the long noncoding RNA circuitry orchestrating X-chromosome inactivation in mammals. Wiley Interdiscip. Rev. RNA 2016, 7, 702–722. [Google Scholar] [CrossRef]
- Pandya-Jones, A.; Plath, K. The “lnc” between 3D chromatin structure and X chromosome inactivation. Semin. Cell Dev. Biol. 2016, 56, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Peeters, S.B.; Yang, C.; Brown, C.J. Have humans lost control: The elusive X-controlling element. Semin. Cell Dev. Biol. 2016, 56, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Pinter, S.F. A Tale of Two Cities: How Xist and its partners localize to and silence the bicompartmental X. Semin. Cell Dev. Biol. 2016, 56, 19–34. [Google Scholar] [CrossRef] [PubMed]
- Vallot, C.; Ouimette, J.F.; Rougeulle, C. Establishment of X chromosome inactivation and epigenomic features of the inactive X depend on cellular contexts. Bioessays 2016, 38, 869–880. [Google Scholar] [CrossRef] [PubMed]
- Jeon, Y.; Sarma, K.; Lee, J.T. New and Xisting regulatory mechanisms of X chromosome inactivation. Curr. Opin. Genet. Dev. 2012, 22, 62–71. [Google Scholar] [CrossRef]
- Zylicz, J.J.; Heard, E. Molecular Mechanisms of Facultative Heterochromatin Formation: An X-Chromosome Perspective. Annu. Rev. Biochem. 2020, 89, 255–282. [Google Scholar] [CrossRef]
- Dossin, F.; Pinheiro, I.; Zylicz, J.J.; Roensch, J.; Collombet, S.; Le Saux, A.; Chelmicki, T.; Attia, M.; Kapoor, V.; Zhan, Y.; et al. SPEN integrates transcriptional and epigenetic control of X-inactivation. Nature 2020, 578, 455–460. [Google Scholar] [CrossRef]
- McHugh, C.A.; Chen, C.K.; Chow, A.; Surka, C.F.; Tran, C.; McDonel, P.; Pandya-Jones, A.; Blanco, M.; Burghard, C.; Moradian, A.; et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 2015, 521, 232–236. [Google Scholar] [CrossRef]
- Zylicz, J.J.; Bousard, A.; Zumer, K.; Dossin, F.; Mohammad, E.; da Rocha, S.T.; Schwalb, B.; Syx, L.; Dingli, F.; Loew, D.; et al. The Implication of Early Chromatin Changes in X Chromosome Inactivation. Cell 2019, 176, 182–197.e23. [Google Scholar] [CrossRef]
- Bousard, A.; Raposo, A.C.; Zylicz, J.J.; Picard, C.; Pires, V.B.; Qi, Y.; Gil, C.; Syx, L.; Chang, H.Y.; Heard, E.; et al. The role of Xist-mediated Polycomb recruitment in the initiation of X-chromosome inactivation. EMBO Rep. 2019, 20, e48019. [Google Scholar] [CrossRef]
- Colognori, D.; Sunwoo, H.; Kriz, A.J.; Wang, C.Y.; Lee, J.T. Xist Deletional Analysis Reveals an Interdependency between Xist RNA and Polycomb Complexes for Spreading along the Inactive X. Mol. Cell 2019, 74, 101–117.e10. [Google Scholar] [CrossRef]
- Nesterova, T.B.; Wei, G.; Coker, H.; Pintacuda, G.; Bowness, J.S.; Zhang, T.; Almeida, M.; Bloechl, B.; Moindrot, B.; Carter, E.J.; et al. Systematic allelic analysis defines the interplay of key pathways in X chromosome inactivation. Nat. Commun. 2019, 10, 3129. [Google Scholar] [CrossRef]
- De Napoles, M.; Mermoud, J.E.; Wakao, R.; Tang, Y.A.; Endoh, M.; Appanah, R.; Nesterova, T.B.; Silva, J.; Otte, A.P.; Vidal, M.; et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev. Cell 2004, 7, 663–676. [Google Scholar] [CrossRef]
- Plath, K.; Fang, J.; Mlynarczyk-Evans, S.K.; Cao, R.; Worringer, K.A.; Wang, H.; de la Cruz, C.C.; Otte, A.P.; Panning, B.; Zhang, Y. Role of histone H3 lysine 27 methylation in X inactivation. Science 2003, 300, 131–135. [Google Scholar] [CrossRef]
- Silva, J.; Mak, W.; Zvetkova, I.; Appanah, R.; Nesterova, T.B.; Webster, Z.; Peters, A.H.; Jenuwein, T.; Otte, A.P.; Brockdorff, N. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev. Cell 2003, 4, 481–495. [Google Scholar] [CrossRef]
- Blackledge, N.P.; Farcas, A.M.; Kondo, T.; King, H.W.; McGouran, J.F.; Hanssen, L.L.P.; Ito, S.; Cooper, S.; Kondo, K.; Koseki, Y.; et al. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell 2014, 157, 1445–1459. [Google Scholar] [CrossRef]
- Kalantry, S.; Mills, K.C.; Yee, D.; Otte, A.P.; Panning, B.; Magnuson, T. The Polycomb group protein Eed protects the inactive X-chromosome from differentiation-induced reactivation. Nat. Cell Biol. 2006, 8, 195–202. [Google Scholar] [CrossRef]
- Tjalsma, S.J.D.; Hori, M.; Sato, Y.; Bousard, A.; Ohi, A.; Raposo, A.C.; Roensch, J.; Le Saux, A.; Nogami, J.; Maehara, K.; et al. H4K20me1 and H3K27me3 are concurrently loaded onto the inactive X chromosome but dispensable for inducing gene silencing. EMBO Rep. 2021, 22, e51989. [Google Scholar] [CrossRef]
- Proudhon, C.; Duffie, R.; Ajjan, S.; Cowley, M.; Iranzo, J.; Carbajosa, G.; Saadeh, H.; Holland, M.L.; Oakey, R.J.; Rakyan, V.K.; et al. Protection against de novo methylation is instrumental in maintaining parent-of-origin methylation inherited from the gametes. Mol. Cell 2012, 47, 909–920. [Google Scholar] [CrossRef]
- Xie, W.; Barr, C.L.; Kim, A.; Yue, F.; Lee, A.Y.; Eubanks, J.; Dempster, E.L.; Ren, B. Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell 2012, 148, 816–831. [Google Scholar] [CrossRef]
- Matouk, I.J.; Halle, D.; Gilon, M.; Hochberg, A. The non-coding RNAs of the H19-IGF2 imprinted loci: A focus on biological roles and therapeutic potential in Lung Cancer. J. Transl. Med. 2015, 13, 113. [Google Scholar] [CrossRef] [PubMed]
- Eggermann, T.; Perez de Nanclares, G.; Maher, E.R.; Temple, I.K.; Tumer, Z.; Monk, D.; Mackay, D.J.; Gronskov, K.; Riccio, A.; Linglart, A.; et al. Imprinting disorders: A group of congenital disorders with overlapping patterns of molecular changes affecting imprinted loci. Clin. Epigenetics 2015, 7, 123. [Google Scholar] [CrossRef] [PubMed]
- Pham, A.; Selenou, C.; Giabicani, E.; Fontaine, V.; Marteau, S.; Brioude, F.; David, L.; Mitanchez, D.; Sobrier, M.L.; Netchine, I. Maintenance of methylation profile in imprinting control regions in human induced pluripotent stem cells. Clin. Epigenetics 2022, 14, 190. [Google Scholar] [CrossRef] [PubMed]
- Jima, D.D.; Skaar, D.A.; Planchart, A.; Motsinger-Reif, A.; Cevik, S.E.; Park, S.S.; Cowley, M.; Wright, F.; House, J.; Liu, A.; et al. Genomic map of candidate human imprint control regions: The imprintome. Epigenetics 2022, 17, 1920–1943. [Google Scholar] [CrossRef]
- King, K.; Murphy, S.; Hoyo, C. Epigenetic regulation of Newborns’ imprinted genes related to gestational growth: Patterning by parental race/ethnicity and maternal socioeconomic status. J. Epidemiol. Community Health 2015, 69, 639–647. [Google Scholar] [CrossRef]
- Vidal, A.C.; Benjamin Neelon, S.E.; Liu, Y.; Tuli, A.M.; Fuemmeler, B.F.; Hoyo, C.; Murtha, A.P.; Huang, Z.; Schildkraut, J.; Overcash, F.; et al. Maternal stress, preterm birth, and DNA methylation at imprint regulatory sequences in humans. Genet. Epigenet 2014, 6, 37–44. [Google Scholar] [CrossRef]
- Heijmans, B.T.; Tobi, E.W.; Stein, A.D.; Putter, H.; Blauw, G.J.; Susser, E.S.; Slagboom, P.E.; Lumey, L.H. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc. Natl. Acad. Sci. USA 2008, 105, 17046–17049. [Google Scholar] [CrossRef]
- De Rooij, S.R.; Painter, R.C.; Phillips, D.I.; Osmond, C.; Michels, R.P.; Godsland, I.F.; Bossuyt, P.M.; Bleker, O.P.; Roseboom, T.J. Impaired insulin secretion after prenatal exposure to the Dutch famine. Diabetes Care 2006, 29, 1897–1901. [Google Scholar] [CrossRef]
- Thorvaldsen, J.L.; Bartolomei, M.S. SnapShot: Imprinted gene clusters. Cell 2007, 130, 958. [Google Scholar] [CrossRef]
- Yang, M.; Park, M.S.; Lee, H.S. Endocrine disrupting chemicals: Human exposure and health risks. J. Environ. Sci. Health C Environ. Carcinog. Ecotoxicol. Rev. 2006, 24, 183–224. [Google Scholar] [CrossRef]
- Al-Mousawi, J.; Boskovic, A. Transcriptional and epigenetic control of early life cell fate decisions. Curr. Opin. Oncol. 2022, 34, 148–154. [Google Scholar] [CrossRef]
- Smith, Z.D.; Meissner, A. DNA methylation: Roles in mammalian development. Nat. Rev. Genet. 2013, 14, 204–220. [Google Scholar] [CrossRef]
- Robles-Matos, N.; Artis, T.; Simmons, R.A.; Bartolomei, M.S. Environmental Exposure to Endocrine Disrupting Chemicals Influences Genomic Imprinting, Growth, and Metabolism. Genes 2021, 12, 1153. [Google Scholar] [CrossRef]
- Waterland, R.A.; Travisano, M.; Tahiliani, K.G. Diet-induced hypermethylation at agouti viable yellow is not inherited transgenerationally through the female. FASEB J. 2007, 21, 3380–3385. [Google Scholar] [CrossRef]
- Rosenfeld, C.S.; Sieli, P.T.; Warzak, D.A.; Ellersieck, M.R.; Pennington, K.A.; Roberts, R.M. Maternal exposure to bisphenol A and genistein has minimal effect on A(vy)/a offspring coat color but favors birth of agouti over nonagouti mice. Proc. Natl. Acad. Sci. USA 2013, 110, 537–542. [Google Scholar] [CrossRef]
- Salian, S.; Doshi, T.; Vanage, G. Perinatal exposure of rats to Bisphenol A affects the fertility of male offspring. Life Sci. 2009, 85, 742–752. [Google Scholar] [CrossRef]
- Hancock, G.V.; Wamaitha, S.E.; Peretz, L.; Clark, A.T. Mammalian primordial germ cell specification. Development 2021, 148, dev189217. [Google Scholar] [CrossRef]
- Leitch, H.G.; Tang, W.W.; Surani, M.A. Primordial germ-cell development and epigenetic reprogramming in mammals. Curr. Top. Dev. Biol. 2013, 104, 149–187. [Google Scholar] [CrossRef]
- Hajkova, P. Epigenetic reprogramming in the germline: Towards the ground state of the epigenome. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2011, 366, 2266–2273. [Google Scholar] [CrossRef]
- Hajkova, P.; Ancelin, K.; Waldmann, T.; Lacoste, N.; Lange, U.C.; Cesari, F.; Lee, C.; Almouzni, G.; Schneider, R.; Surani, M.A. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature 2008, 452, 877–881. [Google Scholar] [CrossRef]
- Seki, Y.; Hayashi, K.; Itoh, K.; Mizugaki, M.; Saitou, M.; Matsui, Y. Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev. Biol. 2005, 278, 440–458. [Google Scholar] [CrossRef] [PubMed]
- Sachs, M.; Onodera, C.; Blaschke, K.; Ebata, K.T.; Song, J.S.; Ramalho-Santos, M. Bivalent chromatin marks developmental regulatory genes in the mouse embryonic germline in vivo. Cell Rep. 2013, 3, 1777–1784. [Google Scholar] [CrossRef] [PubMed]
- Guibert, S.; Forne, T.; Weber, M. Global profiling of DNA methylation erasure in mouse primordial germ cells. Genome Res. 2012, 22, 633–641. [Google Scholar] [CrossRef] [PubMed]
- Kurimoto, K.; Yabuta, Y.; Hayashi, K.; Ohta, H.; Kiyonari, H.; Mitani, T.; Moritoki, Y.; Kohri, K.; Kimura, H.; Yamamoto, T.; et al. Quantitative Dynamics of Chromatin Remodeling during Germ Cell Specification from Mouse Embryonic Stem Cells. Cell Stem Cell 2015, 16, 517–532. [Google Scholar] [CrossRef] [PubMed]
- Ohno, R.; Nakayama, M.; Naruse, C.; Okashita, N.; Takano, O.; Tachibana, M.; Asano, M.; Saitou, M.; Seki, Y. A replication-dependent passive mechanism modulates DNA demethylation in mouse primordial germ cells. Development 2013, 140, 2892–2903. [Google Scholar] [CrossRef]
- Seisenberger, S.; Andrews, S.; Krueger, F.; Arand, J.; Walter, J.; Santos, F.; Popp, C.; Thienpont, B.; Dean, W.; Reik, W. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol. Cell 2012, 48, 849–862. [Google Scholar] [CrossRef]
- Arand, J.; Wossidlo, M.; Lepikhov, K.; Peat, J.R.; Reik, W.; Walter, J. Selective impairment of methylation maintenance is the major cause of DNA methylation reprogramming in the early embryo. Epigenetics Chromatin 2015, 8, 1. [Google Scholar] [CrossRef]
- Hargan-Calvopina, J.; Taylor, S.; Cook, H.; Hu, Z.; Lee, S.A.; Yen, M.R.; Chiang, Y.S.; Chen, P.Y.; Clark, A.T. Stage-Specific Demethylation in Primordial Germ Cells Safeguards against Precocious Differentiation. Dev. Cell 2016, 39, 75–86. [Google Scholar] [CrossRef]
- Hackett, J.A.; Sengupta, R.; Zylicz, J.J.; Murakami, K.; Lee, C.; Down, T.A.; Surani, M.A. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science 2013, 339, 448–452. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Hong, K.; Liu, R.; Inoue, A.; Shen, L.; Zhang, K.; Zhang, Y. Dynamics of 5-methylcytosine and 5-hydroxymethylcytosine during germ cell reprogramming. Cell Res. 2013, 23, 329–339. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Shen, L.; Liu, Y.; Sendler, D.; Zhang, Y. Role of Tet1 in erasure of genomic imprinting. Nature 2013, 504, 460–464. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Hong, K.; Liu, R.; Shen, L.; Inoue, A.; Diep, D.; Zhang, K.; Zhang, Y. Tet1 controls meiosis by regulating meiotic gene expression. Nature 2012, 492, 443–447. [Google Scholar] [CrossRef]
- Manikkam, M.; Guerrero-Bosagna, C.; Tracey, R.; Haque, M.M.; Skinner, M.K. Transgenerational actions of environmental compounds on reproductive disease and identification of epigenetic biomarkers of ancestral exposures. PLoS ONE 2012, 7, e31901. [Google Scholar] [CrossRef]
- Sales, V.M.; Ferguson-Smith, A.C.; Patti, M.E. Epigenetic Mechanisms of Transmission of Metabolic Disease across Generations. Cell Metab. 2017, 25, 559–571. [Google Scholar] [CrossRef]
- Laurent, L.; Wong, E.; Li, G.; Huynh, T.; Tsirigos, A.; Ong, C.T.; Low, H.M.; Kin Sung, K.W.; Rigoutsos, I.; Loring, J.; et al. Dynamic changes in the human methylome during differentiation. Genome Res. 2010, 20, 320–331. [Google Scholar] [CrossRef]
- Lister, R.; Pelizzola, M.; Dowen, R.H.; Hawkins, R.D.; Hon, G.; Tonti-Filippini, J.; Nery, J.R.; Lee, L.; Ye, Z.; Ngo, Q.M.; et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009, 462, 315–322. [Google Scholar] [CrossRef]
- Zvetkova, I.; Apedaile, A.; Ramsahoye, B.; Mermoud, J.E.; Crompton, L.A.; John, R.; Feil, R.; Brockdorff, N. Global hypomethylation of the genome in XX embryonic stem cells. Nat. Genet. 2005, 37, 1274–1279. [Google Scholar] [CrossRef]
- Lister, R.; Mukamel, E.A.; Nery, J.R.; Urich, M.; Puddifoot, C.A.; Johnson, N.D.; Lucero, J.; Huang, Y.; Dwork, A.J.; Schultz, M.D.; et al. Global epigenomic reconfiguration during mammalian brain development. Science 2013, 341, 1237905. [Google Scholar] [CrossRef]
- Guo, J.U.; Su, Y.; Shin, J.H.; Shin, J.; Li, H.; Xie, B.; Zhong, C.; Hu, S.; Le, T.; Fan, G.; et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat. Neurosci. 2014, 17, 215–222. [Google Scholar] [CrossRef]
- Varley, K.E.; Gertz, J.; Bowling, K.M.; Parker, S.L.; Reddy, T.E.; Pauli-Behn, F.; Cross, M.K.; Williams, B.A.; Stamatoyannopoulos, J.A.; Crawford, G.E.; et al. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 2013, 23, 555–567. [Google Scholar] [CrossRef]
- Arand, J.; Spieler, D.; Karius, T.; Branco, M.R.; Meilinger, D.; Meissner, A.; Jenuwein, T.; Xu, G.; Leonhardt, H.; Wolf, V.; et al. In vivo control of CpG and non-CpG DNA methylation by DNA methyltransferases. PLoS Genet. 2012, 8, e1002750. [Google Scholar] [CrossRef] [PubMed]
- Natsume, H.; Szczepaniak, K.; Yamada, H.; Iwashita, Y.; Gedek, M.; Suto, J.; Ishino, K.; Kasajima, R.; Matsuda, T.; Manirakiza, F.; et al. Non-CpG sites preference in G:C > A:T transition of TP53 in gastric cancer of Eastern Europe (Poland, Romania and Hungary) compared to East Asian countries (China and Japan). Genes. Environ. 2023, 45, 1. [Google Scholar] [CrossRef] [PubMed]
- Fedotova, E.Y.; Iakovenko, E.V.; Abramycheva, N.Y.; Illarioshkin, S.N. SNCA Gene Methylation in Parkinson’s Disease and Multiple System Atrophy. Epigenomes 2023, 7, 5. [Google Scholar] [CrossRef] [PubMed]
- Von Meyenn, F.; Reik, W. Forget the Parents: Epigenetic Reprogramming in Human Germ Cells. Cell 2015, 161, 1248–1251. [Google Scholar] [CrossRef]
- Miller, J.L.; Grant, P.A. The role of DNA methylation and histone modifications in transcriptional regulation in humans. Subcell. Biochem. 2013, 61, 289–317. [Google Scholar] [CrossRef]
- Zhu, L.; Marjani, S.L.; Jiang, Z. The Epigenetics of Gametes and Early Embryos and Potential Long-Range Consequences in Livestock Species-Filling in the Picture With Epigenomic Analyses. Front. Genet. 2021, 12, 557934. [Google Scholar] [CrossRef]
- McCarthy, R.L.; Zhang, J.; Zaret, K.S. Diverse heterochromatin states restricting cell identity and reprogramming. Trends Biochem. Sci. 2023, 48, 513–526. [Google Scholar] [CrossRef]
- Odroniec, A.; Olszewska, M.; Kurpisz, M. Epigenetic markers in the embryonal germ cell development and spermatogenesis. Basic. Clin. Androl. 2023, 33, 6. [Google Scholar] [CrossRef]
- Rossant, J. Stem cells and lineage development in the mammalian blastocyst. Reprod. Fertil. Dev. 2007, 19, 111–118. [Google Scholar] [CrossRef]
- Kunath, T.; Arnaud, D.; Uy, G.D.; Okamoto, I.; Chureau, C.; Yamanaka, Y.; Heard, E.; Gardner, R.L.; Avner, P.; Rossant, J. Imprinted X-inactivation in extra-embryonic endoderm cell lines from mouse blastocysts. Development 2005, 132, 1649–1661. [Google Scholar] [CrossRef]
- Harper, M.I.; Fosten, M.; Monk, M. Preferential paternal X inactivation in extraembryonic tissues of early mouse embryos. J. Embryol. Exp. Morphol. 1982, 67, 127–135. [Google Scholar] [CrossRef]
- Zuccotti, M.; Monk, M. Methylation of the mouse Xist gene in sperm and eggs correlates with imprinted Xist expression and paternal X-inactivation. Nat. Genet. 1995, 9, 316–320. [Google Scholar] [CrossRef]
- Wake, N.; Takagi, N.; Sasaki, M. Non-random inactivation of X chromosome in the rat yolk sac. Nature 1976, 262, 580–581. [Google Scholar] [CrossRef]
- Takagi, N.; Sasaki, M. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature 1975, 256, 640–642. [Google Scholar] [CrossRef]
- Huynh, K.D.; Lee, J.T. Imprinted X inactivation in eutherians: A model of gametic execution and zygotic relaxation. Curr. Opin. Cell Biol. 2001, 13, 690–697. [Google Scholar] [CrossRef]
- Huynh, K.D.; Lee, J.T. X-chromosome inactivation: A hypothesis linking ontogeny and phylogeny. Nat. Rev. Genet. 2005, 6, 410–418. [Google Scholar] [CrossRef]
- Wang, X.; Miller, D.C.; Clark, A.G.; Antczak, D.F. Random X inactivation in the mule and horse placenta. Genome Res. 2012, 22, 1855–1863. [Google Scholar] [CrossRef]
- Kalousek, D.K.; Vekemans, M. Confined placental mosaicism. J. Med. Genet. 1996, 33, 529–533. [Google Scholar] [CrossRef]
- Kalousek, D.K. Confined placental mosaicism and uniparental disomy. Funct. Dev. Morphol. 1994, 4, 93–98. [Google Scholar]
- Toutain, J.; Goutte-Gattat, D.; Horovitz, J.; Saura, R. Confined placental mosaicism revisited: Impact on pregnancy characteristics and outcome. PLoS ONE 2018, 13, e0195905. [Google Scholar] [CrossRef]
- Cheng, Z.L.; Zhang, M.L.; Lin, H.P.; Gao, C.; Song, J.B.; Zheng, Z.; Li, L.; Zhang, Y.; Shen, X.; Zhang, H.; et al. The Zscan4-Tet2 Transcription Nexus Regulates Metabolic Rewiring and Enhances Proteostasis to Promote Reprogramming. Cell Rep. 2020, 32, 107877. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Shih, J.; Hollern, D.P.; Wang, L.; Bowlby, R.; Tickoo, S.K.; Thorsson, V.; Mungall, A.J.; Newton, Y.; Hegde, A.M.; et al. Integrated Molecular Characterization of Testicular Germ Cell Tumors. Cell Rep. 2018, 23, 3392–3406. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Hu, B.; Yan, L.; Yong, J.; Wu, Y.; Gao, Y.; Guo, F.; Hou, Y.; Fan, X.; Dong, J.; et al. DNA methylation and chromatin accessibility profiling of mouse and human fetal germ cells. Cell Res. 2017, 27, 165–183. [Google Scholar] [CrossRef] [PubMed]
- Molaro, A.; Hodges, E.; Fang, F.; Song, Q.; McCombie, W.R.; Hannon, G.J.; Smith, A.D. Sperm methylation profiles reveal features of epigenetic inheritance and evolution in primates. Cell 2011, 146, 1029–1041. [Google Scholar] [CrossRef] [PubMed]
- Ehmcke, J.; Wistuba, J.; Schlatt, S. Spermatogonial stem cells: Questions, models and perspectives. Hum. Reprod. Update 2006, 12, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Okae, H.; Chiba, H.; Hiura, H.; Hamada, H.; Sato, A.; Utsunomiya, T.; Kikuchi, H.; Yoshida, H.; Tanaka, A.; Suyama, M.; et al. Genome-wide analysis of DNA methylation dynamics during early human development. PLoS Genet. 2014, 10, e1004868. [Google Scholar] [CrossRef]
- Bourc’his, D.; Xu, G.L.; Lin, C.S.; Bollman, B.; Bestor, T.H. Dnmt3L and the establishment of maternal genomic imprints. Science 2001, 294, 2536–2539. [Google Scholar] [CrossRef]
- Marques, C.J.; Joao Pinho, M.; Carvalho, F.; Bieche, I.; Barros, A.; Sousa, M. DNA methylation imprinting marks and DNA methyltransferase expression in human spermatogenic cell stages. Epigenetics 2011, 6, 1354–1361. [Google Scholar] [CrossRef]
- Brykczynska, U.; Hisano, M.; Erkek, S.; Ramos, L.; Oakeley, E.J.; Roloff, T.C.; Beisel, C.; Schubeler, D.; Stadler, M.B.; Peters, A.H. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat. Struct. Mol. Biol. 2010, 17, 679–687. [Google Scholar] [CrossRef]
- Hammoud, S.S.; Nix, D.A.; Zhang, H.; Purwar, J.; Carrell, D.T.; Cairns, B.R. Distinctive chromatin in human sperm packages genes for embryo development. Nature 2009, 460, 473–478. [Google Scholar] [CrossRef]
- Aoki, F.; Worrad, D.M.; Schultz, R.M. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev. Biol. 1997, 181, 296–307. [Google Scholar] [CrossRef] [PubMed]
- Braude, P.; Bolton, V.; Moore, S. Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature 1988, 332, 459–461. [Google Scholar] [CrossRef] [PubMed]
- Dahl, J.A.; Jung, I.; Aanes, H.; Greggains, G.D.; Manaf, A.; Lerdrup, M.; Li, G.; Kuan, S.; Li, B.; Lee, A.Y.; et al. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature 2016, 537, 548–552. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Xu, J.; Liu, B.; Yao, G.; Wang, P.; Lin, Z.; Huang, B.; Wang, X.; Li, T.; Shi, S.; et al. Publisher Correction: Chromatin analysis in human early development reveals epigenetic transition during ZGA. Nature 2018, 560, E27. [Google Scholar] [CrossRef]
- Xue, L.; Cai, J.Y.; Ma, J.; Huang, Z.; Guo, M.X.; Fu, L.Z.; Shi, Y.B.; Li, W.X. Global expression profiling reveals genetic programs underlying the developmental divergence between mouse and human embryogenesis. BMC Genomics 2013, 14, 568. [Google Scholar] [CrossRef]
- Amouroux, R.; Nashun, B.; Shirane, K.; Nakagawa, S.; Hill, P.W.; D’Souza, Z.; Nakayama, M.; Matsuda, M.; Turp, A.; Ndjetehe, E.; et al. De novo DNA methylation drives 5hmC accumulation in mouse zygotes. Nat. Cell Biol. 2016, 18, 225–233. [Google Scholar] [CrossRef]
- Zhu, P.; Guo, H.; Ren, Y.; Hou, Y.; Dong, J.; Li, R.; Lian, Y.; Fan, X.; Hu, B.; Gao, Y.; et al. Single-cell DNA methylome sequencing of human preimplantation embryos. Nat. Genet. 2018, 50, 12–19. [Google Scholar] [CrossRef]
- Hanna, C.W.; Demond, H.; Kelsey, G. Epigenetic regulation in development: Is the mouse a good model for the human? Hum. Reprod. Update 2018, 24, 556–576. [Google Scholar] [CrossRef]
- Auclair, G.; Borgel, J.; Sanz, L.A.; Vallet, J.; Guibert, S.; Dumas, M.; Cavelier, P.; Girardot, M.; Forne, T.; Feil, R.; et al. EHMT2 directs DNA methylation for efficient gene silencing in mouse embryos. Genome Res. 2016, 26, 192–202. [Google Scholar] [CrossRef]
- Andergassen, D.; Dotter, C.P.; Wenzel, D.; Sigl, V.; Bammer, P.C.; Muckenhuber, M.; Mayer, D.; Kulinski, T.M.; Theussl, H.C.; Penninger, J.M.; et al. Mapping the mouse Allelome reveals tissue-specific regulation of allelic expression. eLife 2017, 6, e25125. [Google Scholar] [CrossRef]
- Santoni, F.A.; Stamoulis, G.; Garieri, M.; Falconnet, E.; Ribaux, P.; Borel, C.; Antonarakis, S.E. Detection of Imprinted Genes by Single-Cell Allele-Specific Gene Expression. Am. J. Hum. Genet. 2017, 100, 444–453. [Google Scholar] [CrossRef]
- Babak, T.; DeVeale, B.; Tsang, E.K.; Zhou, Y.; Li, X.; Smith, K.S.; Kukurba, K.R.; Zhang, R.; Li, J.B.; van der Kooy, D.; et al. Genetic conflict reflected in tissue-specific maps of genomic imprinting in human and mouse. Nat. Genet. 2015, 47, 544–549. [Google Scholar] [CrossRef]
- Quenneville, S.; Verde, G.; Corsinotti, A.; Kapopoulou, A.; Jakobsson, J.; Offner, S.; Baglivo, I.; Pedone, P.V.; Grimaldi, G.; Riccio, A.; et al. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol. Cell 2011, 44, 361–372. [Google Scholar] [CrossRef]
- Inoue, A.; Jiang, L.; Lu, F.; Suzuki, T.; Zhang, Y. Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature 2017, 547, 419–424. [Google Scholar] [CrossRef]
- Hamada, H.; Okae, H.; Toh, H.; Chiba, H.; Hiura, H.; Shirane, K.; Sato, T.; Suyama, M.; Yaegashi, N.; Sasaki, H.; et al. Allele-Specific Methylome and Transcriptome Analysis Reveals Widespread Imprinting in the Human Placenta. Am. J. Hum. Genet. 2016, 99, 1045–1058. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, A.; Rappolee, D.A.; Ruden, D.M. Epigenetic Reprogramming in Mice and Humans: From Fertilization to Primordial Germ Cell Development. Cells 2023, 12, 1874. https://doi.org/10.3390/cells12141874
Singh A, Rappolee DA, Ruden DM. Epigenetic Reprogramming in Mice and Humans: From Fertilization to Primordial Germ Cell Development. Cells. 2023; 12(14):1874. https://doi.org/10.3390/cells12141874
Chicago/Turabian StyleSingh, Aditi, Daniel A. Rappolee, and Douglas M. Ruden. 2023. "Epigenetic Reprogramming in Mice and Humans: From Fertilization to Primordial Germ Cell Development" Cells 12, no. 14: 1874. https://doi.org/10.3390/cells12141874
APA StyleSingh, A., Rappolee, D. A., & Ruden, D. M. (2023). Epigenetic Reprogramming in Mice and Humans: From Fertilization to Primordial Germ Cell Development. Cells, 12(14), 1874. https://doi.org/10.3390/cells12141874







