Artificial Intelligence-Assisted Diagnostic Cytology and Genomic Testing for Hematologic Disorders
Abstract
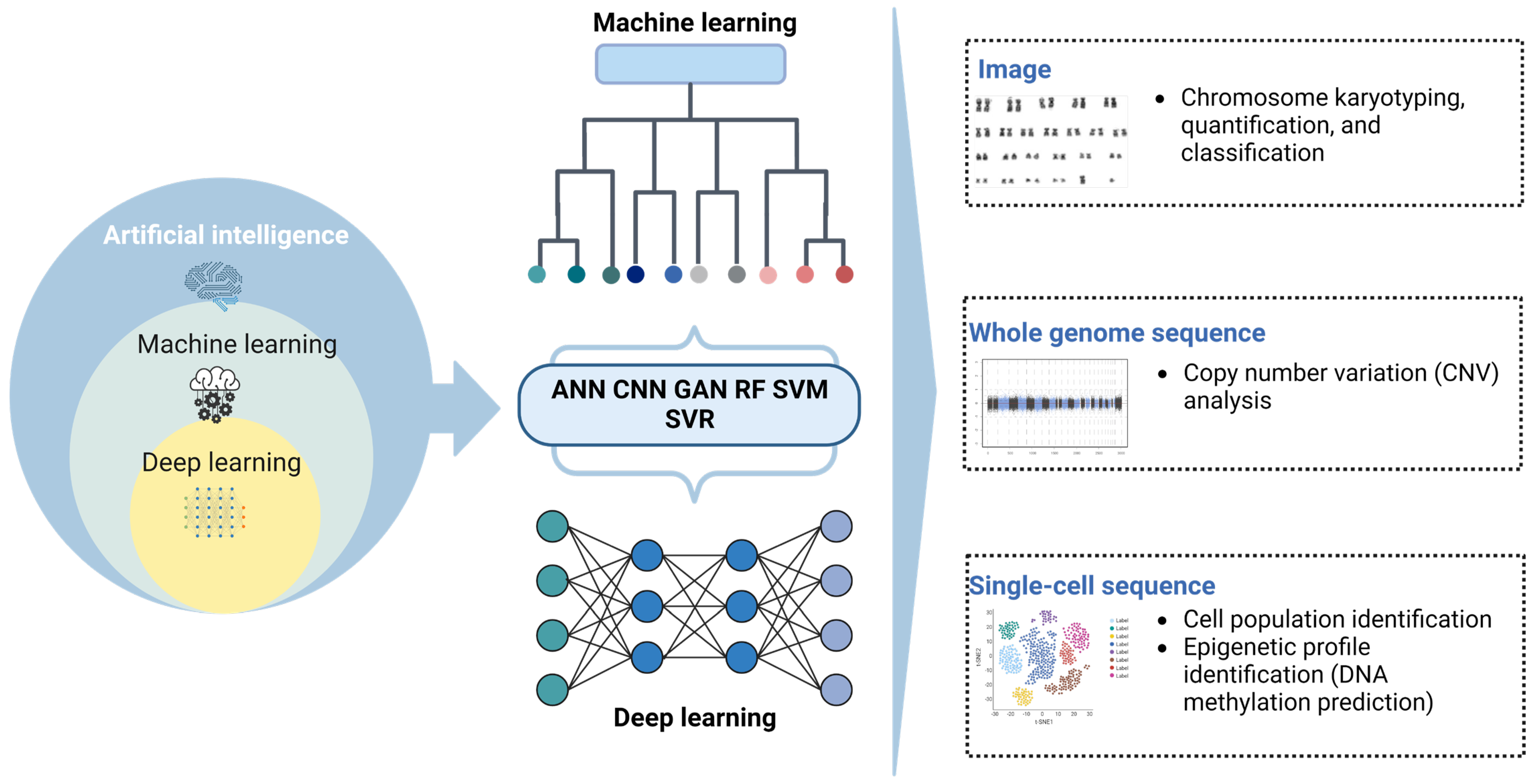
:1. Introduction
2. Current Applications of AI in Hematologic Cytology
2.1. Review of Blood/Marrow Smears
2.2. Detection of Hb-H Inclusion Bodies
2.3. Flow Cytometric Analysis
3. AI-Assisted Genomic Testing for Hematologic Disorders
3.1. Cytogenetic Karyotyping
3.2. Sequencing for Profiling of Genetic Markers
3.3. Whole-Genome Sequencing for Analysis of Copy Number Variations
3.4. Single-Cell Sequencing Analysis
3.5. Epigenetic Profiling to Identify Novel Biomarkers
| No. | Method/Device | AI Model | Function | Accuracy (%) | Year | References |
|---|---|---|---|---|---|---|
| 1 | Ikaros | CNN, DNN | Chromosome karyotyping myelofibrosis | 98 | 2023 | [80] |
| 2 | ChromoEnhancer method | CycleGAN | Bone marrow karyotyping | NA | 2022 | [90] |
| 3 | Softmax | CNN | Chromosome classification | 93.8 | 2019 | [91] |
| 4 | KaryoNet | MFIM, DAM | Chromosome quantitation and classification | 98.4–99.6 | 2023 | [133] |
| 5 | CNV-P | DL | CNV detection | 90 | 2021 | [104] |
| 6 | CUP-AI-Dx: | CNN | Predicts tumor primary site and molecular subtype | 98.5 | 2020 | [134] |
| 7 | scDCC | DL | Cell clustering | 90 | 2021 | [110] |
| 8 | DeepCpG: | DL | DNA methylation | NA | 2023 | [131] |
| 9 | EPiScore | DL | DNA methylation | NA | 2018 | [135] |
| 10 | MOM | ML | Therapeutic prediction | NA | 2022 | [117] |
| 11 | MulCNN | CNN | Cell clustering and batch effect removal | NA | 2023 | [118] |
| 12 | BERMUDA | DL | Cell clustering and batch effect removal | NA | 2019 | [119] |
| 13 | RCA2 | MEM, SNN | Cell clustering and batch effect removal | NA | 2021 | [120] |
| 14 | SpotLearn | CNN | DNA FISH detection | 98 | 2017 | [85] |
| 15 | DeepSpot | DNN | RNA FISH detection | 97 | 2022 | [86] |
| 16 | ChroSegNet | CNN, U-Net | Chromosome segmentation | 93.3 | 2023 | [92] |
4. AI-Assisted Clinical Prediction Models for Hematologic Disorders
5. Challenges in Developing Clinical AI Systems
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 2D | Two-dimentional |
| AA | Aplastic anemia |
| AI | Artificial intelligence |
| AIPSS-MF | Artificial intelligence prognostic scoring system for myelofibrosis |
| ALL | Acute lymphoblastic leukemia |
| ALPODS | Algorithmic population description approach |
| AML | Acute myeloid leukemia |
| ANN | Artificial neural network |
| APL | Acute promyelocytic leukemia |
| AUC | Area under the curve |
| BCB | Brilliant cresyl blue |
| BERMUDA | Batch effect removal using deep autoencoder |
| BMA | Bone marrow aspirate |
| BMS | Bone marrow smear |
| B-NHL | B- cell non-Hodgkin lymphoma |
| BNN | Bayesian neural network |
| CBC | Complete blood count |
| CD | Cluster of differentiation |
| CLL | Chronic lymphocytic leukemia |
| CML | Chronic myeloid leukemia |
| CNN | Convolution neural network |
| CNV | Copy number variation |
| CoAtNet | Convolution and attention network model |
| DNA | Deoxyribonucleic acid |
| DL | Deep learning |
| DLBCL | Diffuse large B-cell lymphoma |
| DMR | Differentially methylated region |
| DNN | Deep neural network |
| FCL | Follicular cell lymphoma |
| FDA | Food and Drug Administration |
| FDART | Federated gradient boosting trees with dropout |
| FGBT | Federated gradient boosting tree |
| FISH | Fluorescence in situ hybridization |
| FMLP | Federated multi-layer perceptron |
| FMNB | Federated multinomial naive bayes |
| H&E | Hematoxylin and eosin |
| Hb | Hemoglobin |
| Hb-H | Hemoglobin H |
| HPLC | High-performance liquid chromatography |
| IHC | Immunohistochemistry |
| LPL | Lymphoplasmacytic lymphoma |
| MALT | Mucosa associated lymphoma tissue |
| MCL | Mantle cell lymphoma |
| MDS | Myelodysplastic syndrome |
| MF | Myelofibrosis |
| MFC | Multiparameter flow cytometry |
| ML | Machine learning |
| MLP | Multi-layer perception |
| MM | Multiple myeloma |
| MOM | Multi-dimensional module optimization |
| MPN | Myeloproliferative neoplasm |
| MRD | Minimal residual disease |
| MulCNN | Multi-level convolutional neural network |
| MZL | Marginal zone lymphoma |
| PBMC | Peripheral blood mononuclear cell |
| PBS | Peripheral blood smear |
| PCT | Prediction confidence score |
| RBC | Red blood cell |
| RCA2 | Reference component analysis 2 |
| RGS1 | Regulator of G-protein signaling 1 |
| RNN | Recurrent neural network |
| ScRNA-seq | Single-cell RNA sequencing |
| SOM | Self-organized map |
| SVM | Support vector machine |
| WBC | White blood cell |
| WGS | Whole-genome sequencing |
| WSI | Whole-slide image |
| XAI | Explainable artificial intelligence |
References
- Shouval, R.; Fein, J.A.; Savani, B.; Mohty, M.; Nagler, A. Machine learning and artificial intelligence in hematology. Br. J. Hematol. 2021, 192, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Briganti, G.; Le Moine, O. Artificial intelligence in medicine: Today and tomorrow. Front. Med. 2020, 7, 27. [Google Scholar] [CrossRef] [PubMed]
- Kazancı, E.G.; Güven, D. Artificial intelligence applications in hematology. AITA 2021, 1, 1–7. [Google Scholar]
- Zini, G. Artificial intelligence in hematology. Hematology 2005, 10, 393–400. [Google Scholar] [CrossRef]
- Diamond, L.W.; Mishka, V.G.; Seal, A.H.; Nguyen, D.T. Multiparameter interpretative reporting hematology in diagnostic laboratory. Int. J. Biomed. Comput. 1994, 37, 211–224. [Google Scholar] [CrossRef]
- Wong, A.N.N.; He, Z.; Leung, K.L.; To, C.C.K.; Wong, C.Y.; Wong, S.C.C.; Yoo, J.S.; Chan, C.K.R.; Chan, A.Z.; Lacambra, M.D. Current developments of artificial intelligence in digital pathology and its future clinical applications in gastrointestinal cancers. Cancers 2022, 14, 3780. [Google Scholar] [CrossRef]
- Allegra, A.; Tonacci, A.; Sciaccotta, R.; Genovese, S.; Musolino, C.; Pioggia, G.; Gangemi, S. Machine learning and deep learning applications in multiple myeloma diagnosis, prognosis, and treatment selection. Cancers 2022, 14, 606. [Google Scholar] [CrossRef]
- Muhsen, I.N.; Shyr, D.; Sung, A.D.; Hashmi, S.K. Machine learning applications in the diagnosis of benign and malignant hematological diseases. Clin. Hematol. Int. 2021, 3, 13–20. [Google Scholar] [CrossRef]
- Khouani, A.; El Habib Daho, M.; Mahmoudi, S.A.; Chikh, M.A.; Benzineb, B. Automated recognition of white blood cells using deep learning. Biomed. Eng. Lett. 2020, 10, 359–367. [Google Scholar] [CrossRef]
- Ryan, L.; Mataraso, S.; Siefkas, A.; Pellegrini, E.; Barnes, G.; Green-Saxena, A.; Hoffman, J.; Calvert, J.; Das, R.A. Machine learning approach to predict deep venous thrombosis among hospitalized patients. Clin. Appl. Thromb. Hemost. 2021, 27, 1076029621991185. [Google Scholar] [CrossRef]
- Dimitriou, N.; Arandjelovic, O.; Caie, P.D. Deep learning for whole slide image analysis: An overview. Front. Med. 2019, 6, 264. [Google Scholar] [CrossRef]
- WHO. World Health Organization: Non-Communicable Diseases. 2022. Available online: https://www.who.int/news-room/fact-sheets/detail/noncommunicable-diseases (accessed on 29 April 2023).
- GBD 2019 Cancer Risk Factors Collaborators. The global burden of cancer attributable to risk factors, 2010–2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet 2022, 400, 563–591. [Google Scholar] [CrossRef]
- Taylor, J.; Xiao, W.; Abdel-Wahab, O. Diagnosis and classification of hematologic malignancies on the basis of genetics. Blood 2017, 130, 410–423. [Google Scholar] [CrossRef]
- Sochacka-Cwikla, A.; Maczynski, M.; Regiec, A. FDA-approved drugs for hematological malignancies-The last decade review. Cancers 2021, 14, 87. [Google Scholar] [CrossRef]
- Bersabe, A.R.; Aden, J.K.; Shumway, N.M.; Osswald, M.B. Peripheral smear review and bone marrow biopsy correlation. J. Clin. Diagn. Res. 2017, 11, JC01–JC03. [Google Scholar] [CrossRef]
- Agale, D.; Mishra, D.; Kinake, D.; Rathod, D.; Bijwe, D.; Dcunha, D.; Sharma, D. Utility and efficacy of bone marrow examination in the diagnosis of hematological and non-hematological disorders. Int. J. Med. Rev. Case Rep. 2021, 5, 24–30. [Google Scholar]
- Kaur, M.; Singh Rana, A.P.; Kapoor, S.; Puri, A. Diagnostic value of bone marrow aspiration and biopsy in routine hematology practice. J. Clin. Diagn. Res. 2014, 8, FC13–FC16. [Google Scholar]
- Park, S.J.; Yoon, J.; Kwon, J.A.; Yoon, S.Y. Evaluation of the CellaVision advanced RBC application for detecting red blood cell morphological abnormalities. Ann. Lab. Med. 2021, 41, 44–50. [Google Scholar] [CrossRef]
- Fu, X.; Fu, M.; Li, Q.; Peng, X.; Lu, J.; Fang, F.; Chen, M. Morphogo: An automatic bone marrow cell classification system on digital images analyzed by artificial intelligence. Acta Cytol. 2020, 64, 588–596. [Google Scholar] [CrossRef]
- Arya, P.; Modani, U.S. Applications of artificial neural network in image processing: A survey. IJSER 2019, 10, 329–333. [Google Scholar]
- Lin, E.; Fuda, F.; Luu, H.S.; Cox, A.M.; Fang, F.; Feng, J.; Chen, M. Digital pathology and artificial intelligence as the next chapter in diagnostic hematopathology. Semin. Diagn. Pathol. 2023, 40, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Cornet, E.; Perol, J.P.; Troussard, X. Performance evaluation and relevance of the CellaVision DM96 system in routine analysis and in patients with malignant hematological diseases. Int. J. Lab. Hematol. 2008, 30, 536–542. [Google Scholar] [CrossRef] [PubMed]
- Kratz, A.; Bengtsson, H.I.; Casey, J.E.; Keefe, J.M.; Beatrice, G.H.; Grzybek, D.Y.; Lewandrowski, K.B.; Van Cott, E.M. Performance evaluation of the CellaVision DM96 System: WBC differentials by automated digital image analysis supported by an artificial neural network. Am. J. Clin. Pathol. 2005, 124, 770–781. [Google Scholar] [CrossRef]
- Horn, C.L.; Mansoor, A.; Wood, B.; Nelson, H.; Higa, D.; Lee, L.H.; Naugler, C. Performance of the CellaVision (R) DM96 system for detecting red blood cell morphologic abnormalities. J. Pathol. Inform. 2015, 6, 11. [Google Scholar] [CrossRef] [PubMed]
- Florin, L.; Maelegheer, K.; Muyldermans, A.; Van Esbroeck, M.; Nulens, E.; Emmerechts, J. Evaluation of the CellaVision DM96 advanced RBC application for screening and follow-up of malaria infection. Diagn. Microbiol. Infect. Dis. 2018, 90, 253–256. [Google Scholar] [CrossRef] [PubMed]
- Billard, M.; Lainey, E.; Armoogum, P.; Alberti, C.; Fenneteau, O.; Da Costa, L. Evaluation of the CellaVision DM automated microscope in pediatrics. Int. J. Lab. Hematol. 2010, 32, 530–538. [Google Scholar] [CrossRef]
- McCulloch, W.S.; Pitts, W. A logical calculus of the ideas immanent in nervous activity.1943. Bull. Math. Biol. 1990, 52, 9–115. [Google Scholar] [CrossRef]
- Chen, P.; Chen Xu, R.; Chen, N.; Zhang, L.; Zhang, L.; Zhu, J.; Pan, B.; Wang, B.; Guo, W. Detection of metastatic tumor cells in the bone marrow aspirate smears by artificial intelligence (AI)-based Morphogo system. Front. Oncol. 2021, 11, 742395. [Google Scholar] [CrossRef]
- Jin, H.; Fu, X.; Cao, X.; Sun, M.; Wang, X.; Zhong, Y.; Yang, S.; Qi, C.; Peng, B.; He, X.; et al. Developing and preliminary validating an automatic cell classification system for bone marrow smears: A Pilot study. J. Med. Syst. 2020, 44, 184. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Y.; Qi, C.; Qiao, S.; Yang, S.; Wang, R.; Jin, H.; Zhang, J. The application of Morphogo in the detection of megakaryocytes from bone marrow digital images with convolutional neural networks. Technol. Cancer Res. Treat. 2023, 22, 15330338221150069. [Google Scholar] [CrossRef]
- Katz, B.Z.; Feldman, M.D.; Tessema, M.; Benisty, D.; Toles, G.S.; Andre, A.; Shtreker, B.; Paz, F.M.; Edwards, J.; Jengehino, D.; et al. Evaluation of Scopio Labs X100 full field PBS: The first high-resolution full field viewing of peripheral blood specimens combined with artificial intelligence-based morphological analysis. Int. J. Lab. Hematol. 2021, 43, 1408–1416. [Google Scholar] [CrossRef]
- Scopio. Full Field Bone Marrow Aspirate. 2023. Available online: https://scopiolabs.com/hematology (accessed on 29 April 2023).
- Mantiscope. Mantiscope: AI Assistant Blood Cell Analysis. 2023. Available online: https://www.mantiscope.com (accessed on 29 April 2023).
- Kratz, A.; Lee, S.H.; Zini, G.; Riedl, J.A.; Hur, M.; Machin, S.; International council for standardization in hematology. Digital morphology analyzers in hematology: ICSH review and recommendations. Int. J. Lab. Hematol. 2019, 41, 437–447. [Google Scholar] [CrossRef]
- Kimura, K.; Tabe, Y.; Ai, T.; Takehara, I.; Fukuda, H.; Takahashi, H.; Naito, T.; Komatsu, N.; Uchihashi, K.; Ohsaka, A. A novel automated image analysis system using deep convolutional neural networks can assist to differentiate MDS and AA. Sci. Rep. 2019, 9, 13385. [Google Scholar] [CrossRef]
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef]
- Zhang, X.; Guo, X. Risk factors of thrombosis in Chinese subjects with acute promyelocytic leukemia. Thromb. J. 2021, 19, 42. [Google Scholar] [CrossRef]
- Eckardt, J.N.; Schmittmann, T.; Riechert, S.; Kramer, M.; Sulaiman, A.S.; Sockel, K.; Kroschinsky, F.; Schetelig, J.; Wagenfuhr, L.; Schuler, U.; et al. Deep learning identifies acute promyelocytic leukemia in bone marrow smears. BMC Cancer 2022, 22, 201. [Google Scholar] [CrossRef]
- Li, D.; Bledsoe, J.R.; Zeng, Y.; Liu, W.; Hu, Y.; Bi, K.; Liang, A.; Li, S. A deep learning diagnostic platform for diffuse large B-cell lymphoma with high accuracy across multiple hospitals. Nat. Commun. 2020, 11, 6004. [Google Scholar] [CrossRef]
- Tripathi, S.; Augustin, A.I.; Sukumaranc, R.; Dheer, S.; Kime, E. HematoNet: Expert level classification of bone marrow cytology morphology in hematological malignancy with deep learning. Artif. Intell. Life Sci. 2022, 2, 100043. [Google Scholar]
- Eckardt, J.N.; Middeke, J.M.; Riechert, S.; Schmittmann, T.; Sulaiman, A.S.; Kramer, M.; Sockel, K.; Kroschinsky, F.; Schuler, U.; Schetelig, J.; et al. Deep learning detects acute myeloid leukemia and predicts NPM1 mutation status from bone marrow smears. Leukemia 2022, 36, 111–118. [Google Scholar] [CrossRef]
- Orlov, N.V.; Chen, W.W.; Eckley, D.M.; Macura, T.J.; Shamir, L.; Jaffe, E.S.; Goldberg, I.G. Automatic classification of lymphoma images with transform-based global features. IEEE Trans. Inf. Technol. Biomed. 2010, 14, 1003–1013. [Google Scholar] [CrossRef]
- Steinbuss, G.; Kriegsmann, M.; Zgorzelski, C.; Brobeil, A.; Goeppert, B.; Dietrich, S.; Mechtersheimer, G.; Kriegsmann, K. Deep learning for the classification of non-Hodgkin lymphoma on histopathological images. Cancers 2021, 13, 2419. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, H.; Sato, K.; Kabeya, Y.; Yonezawa, S.; Nakano, H.; Takeuchi, Y.; Ozawa, I.; Higo, S.; Yanagida, E.; Yamada, K.; et al. Deep learning shows the capability of high-level computer-aided diagnosis in malignant lymphoma. Lab. Investig. 2020, 100, 1300–1310. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.H.; Li, C.H.; Wang, R.C.; Yeh, C.Y.; Chuang, S.S. Machine learning based on morphological features enables classification of primary intestinal T-cell lymphomas. Cancers 2021, 13, 5463. [Google Scholar] [CrossRef] [PubMed]
- Achi, H.E.; Belousova, T.; Chen, L.; Wahed, A.; Wang, I.; Hu, Z.; Kanaan, Z.; Rios, A.; Nguyen, A.N.D. Automated diagnosis of lymphoma with digital pathology images using deep learning. Ann. Clin. Lab. Sci. 2019, 49, 153–160. [Google Scholar] [PubMed]
- Syrykh, C.; Abreu, A.; Amara, N.; Siegfried, A.; Maisongrosse, V.; Frenois, F.X.; Martin, L.; Rossi, C.; Laurent, C.; Brousset, P. Accurate diagnosis of lymphoma on whole-slide histopathology images using deep learning. NPJ Digit. Med. 2020, 3, 63. [Google Scholar] [CrossRef] [PubMed]
- Pezoulas, V.C.; Kalatzis, F.; Exarchos, T.P.; Chatzis, L.; Gandolfo, S.; Goules, A.; De Vita, S.; Tzioufas, A.G.; Fotiadis, D.I. A federated AI strategy for the classification of patients with mucosa associated lymphoma tissue (MALT) lymphoma across multiple harmonized cohorts. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2021, 2021, 1666–1669. [Google Scholar]
- Shankar, V.; Yang, X.; Krishna, V.; Tan, B.T.; Silva, O.; Rojansky, R.; Ng, A.Y.; Valvert, F.; Briercheck, E.L.; Weinstock, D.M.; et al. LymphoML: An Interpretable Artificial Intelligence-Based Method Identifies Morphologic Features That Correlate with Lymphoma Subtype. MedRxiv Preprint. Available online: https://www.medrxiv.org/content/10.1101/2023.03.14.23287143v1 (accessed on 17 May 2023).
- Magaki, S.; Hojat, S.A.; Wei, B.; So, A.; Yong, W.H. An introduction to the performance of immunohistochemistry. Methods Mol. Biol. 2019, 1897, 289–298. [Google Scholar]
- Abdul-Ghafar, J.; Seo, K.J.; Jung, H.R.; Park, G.; Lee, S.S.; Chong, Y. Validation of a machine learning expert supporting system, ImmunoGenius, using immunohistochemistry results of 3000 patients with lymphoid neoplasms. Diagnostics 2023, 13, 1308. [Google Scholar] [CrossRef]
- Costa, C.B.T. Machine learning provides an accurate classification of diffuse large B-cell lymphoma from immunohistochemical data. J. Pathol. Inform. 2018, 9, 21. [Google Scholar] [CrossRef]
- Carreras, J.; Nakamura, N.; Hamoudi, R. Artificial intelligence analysis of gene expression predicted the overall survival of mantle cell lymphoma and a large pan-cancer Series. Healthcare 2022, 10, 155. [Google Scholar] [CrossRef]
- Saputra, D.C.E.; Sunat, K.; Ratnaningsih, T. A new artificial intelligence approach using extreme learning machine as the potentially effective model to predict and analyze the diagnosis of anemia. Healthcare 2023, 11, 697. [Google Scholar] [CrossRef]
- Ferih, K.; Elsayed, B.; Elshoeibi, A.M.; Elsabagh, A.A.; Elhadary, M.; Soliman, A.; Abdalgayoom, M.; Yassin, M. Applications of artificial intelligence in thalassemia: A comprehensive review. Diagnostics 2023, 13, 1551. [Google Scholar] [CrossRef]
- Chan, A.Y.; So, C.K.; Chan, L.C. Comparison of the HbH inclusion test and a PCR test in routine screening for alpha thalassemia in Hong Kong. J. Clin. Pathol. 1996, 49, 411–413. [Google Scholar] [CrossRef]
- Pan, L.L.; Eng, H.L.; Kuo, C.Y.; Chen, W.J.; Huang, H.Y. Usefulness of brilliant cresyl blue staining as an auxiliary method of screening for alpha-thalassemia. J. Lab. Clin. Med. 2005, 145, 94–97. [Google Scholar] [CrossRef]
- Lee, S.Y.; Chen, C.M.E.; Lim, E.Y.P.; Shen, L.; Sathe, A.; Singh, A.; Sauer, J.; Taghipour, K.; Yip, C.Y.C. Image analysis using machine learning for automated detection of hemoglobin H inclusions in blood smears–A method for morphologic detection of rare cells. J. Pathol. Inform. 2021, 12, 18. [Google Scholar] [CrossRef]
- Kensert, A.; Bouwmeester, R.; Efthymiadis, K.; Van Broeck, P.; Desmet, G.; Cabooter, D. Graph convolutional networks for improved prediction and interpretability of chromatographic retention data. Anal. Chem. 2021, 93, 15633–15641. [Google Scholar] [CrossRef]
- Uçucu, S.; Karabıyık, T.; Azik, F.M. Machine learning models can predict the presence of variants in hemoglobin: Artificial neural network-based recognition of human hemoglobin variants by HPLC. TJB 2023, 48, 5–11. [Google Scholar] [CrossRef]
- Michael Brown, C.W. Flow cytometry: Principles and clinical applications in hematology. Clin. Chem. 2000, 46, 1221–1229. [Google Scholar] [CrossRef]
- Salama, M.E.; Otteson, G.E.; Camp, J.J.; Seheult, J.N.; Jevremovic, D.; Holmes, D.R., 3rd; Olteanu, H.; Shi, M. Artificial intelligence enhances diagnostic flow cytometry workflow in the detection of minimal residual disease of chronic lymphocytic leukemia. Cancers 2022, 14, 2537. [Google Scholar] [CrossRef]
- Chulian, S.; Martinez-Rubio, A.; Perez-Garcia, V.M.; Rosa, M.; Blazquez Goni, C.; Rodriguez Gutierrez, J.F.; Hermosin-Ramos, L.; Molinos Quintana, A.; Caballero-Velazquez, T.; Ramirez-Orellana, M. High-dimensional analysis of single-cell flow cytometry data predicts relapse in childhood acute lymphoblastic leukemia. Cancers 2020, 13, 17. [Google Scholar] [CrossRef] [PubMed]
- Vial, J.P.; Lechevalier, N.; Lacombe, F.; Dumas, P.Y.; Bidet, A.; Leguay, T.; Vergez, F.; Pigneux, A.; Bene, M.C. Unsupervised flow cytometry analysis allows for an accurate identification of minimal residual disease assessment in acute myeloid leukemia. Cancers 2021, 13, 629. [Google Scholar] [CrossRef] [PubMed]
- Gaidano, V.; Tenace, V.; Santoro, N.; Varvello, S.; Cignetti, A.; Prato, G.; Saglio, G.; De Rosa, G.; Geuna, M. A clinically applicable approach to the classification of B-cell non-Hodgkin lymphomas with flow cytometry and machine learning. Cancers 2020, 12, 1684. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Mallesh, N.; Hollein, A.; Schabath, R.; Haferlach, C.; Haferlach, T.; Elsner, F.; Luling, H.; Krawitz, P.; Kern, W. Hematologist-level classification of mature B-cell neoplasm using deep learning on multiparameter flow cytometry data. Cytometry A 2020, 97, 1073–1080. [Google Scholar] [CrossRef]
- Ji, D.; Putzel, P.; Qian, Y.; Chang, I.; Mandava, A.; Scheuermann, R.H.; Bui, J.D.; Wang, H.Y.; Smyth, P. Machine learning of discriminative gate locations for clinical diagnosis. Cytometry A 2020, 97, 296–307. [Google Scholar] [CrossRef]
- Arvaniti, E.; Claassen, M. Sensitive detection of rare disease-associated cell subsets via representation learning. Nat. Commun. 2017, 8, 14825. [Google Scholar] [CrossRef]
- Abir, W.H.; Uddin, M.F.; Khanam, F.R.; Tazin, T.; Khan, M.M.; Masud, M.; Aljahdali, S. Explainable AI in diagnosing and anticipating leukemia using transfer learning method. Comput. Intell. Neurosci. 2022, 2022, 5140148. [Google Scholar] [CrossRef]
- Herishanu, S.A. AI-based clinical decision support system for treatment of chronic lymphocytic leukemia/small lymphocytic lymphoma (CLL/SLL) and prediction of treatment efficiency. Blood 2022, 140, 2393–12394. [Google Scholar] [CrossRef]
- Lee, S.Y.; Lee, H.T.; Kwoun, W.-J.; Lee, Y.S.; Ahn, J.-Y. Evaluating the performance of the Sysmex DI-60 automated cell image analyzer for the differential analysis of leukocytes. Lab. Med. Qual. Assur. 2020, 42, 70–76. [Google Scholar] [CrossRef]
- Çelebi, S.; Burkay Çöteli, M. Red and white blood cell classification using artificial neural networks. AIMS Bioeng. 2018, 5, 179–191. [Google Scholar] [CrossRef]
- Kim, H.; Lee, G.H.; Yoon, S.; Hur, M.; Kim, H.N.; Park, M.; Kim, S.W. Performance of digital morphology analyzer Medica EasyCell assistant. Clin. Chem. Lab. Med. 2023. online ahead of print. [Google Scholar] [CrossRef]
- Wang, W.; Luo, M.; Guo, P.; Wei, Y.; Tan, Y.; Shi, H. Artificial intelligence-assisted diagnosis of hematologic diseases based on bone marrow smears using deep neural networks. Comput. Methods Programs Biomed. 2023, 231, 107343. [Google Scholar] [CrossRef]
- Yu, H.; Ok, C.Y.; Hesse, A.; Nordell, P.; Connor, D.; Sjostedt, E.; Pechet, L.; Snyder, L.M. Evaluation of an automated digital imaging system, Nextslide digital review network, for examination of peripheral blood smears. Arch. Pathol. Lab. Med. 2012, 136, 660–667. [Google Scholar] [CrossRef]
- Kimura, K.; Ai, T.; Horiuchi, Y.; Matsuzaki, A.; Nishibe, K.; Marutani, S.; Saito, K.; Kaniyu, K.; Takehara, I.; Uchihashi, K.; et al. Automated diagnostic support system with deep learning algorithms for distinction of Philadelphia chromosome-negative myeloproliferative neoplasms using peripheral blood specimen. Sci. Rep. 2021, 11, 3367. [Google Scholar] [CrossRef]
- Li, M.; Lin, C.; Ge, P.; Li, L.; Song, S.; Zhang, H.; Lu, L.; Liu, X.; Zheng, F.; Zhang, S.; et al. A deep learning model for detection of leukocytes under various interference factors. Sci. Rep. 2023, 13, 2160. [Google Scholar] [CrossRef]
- Sirinukunwattana, K.; Aberdeen, A.; Theissen, H.; Sousos, N.; Psaila, B.; Mead, A.J.; Turner, G.D.H.; Rees, G.; Rittscher, J.; Royston, D. Artificial intelligence-based morphological fingerprinting of megakaryocytes: A new tool for assessing disease in MPN patients. Blood. Adv. 2020, 4, 3284–3294. [Google Scholar] [CrossRef]
- Mosquera-Orgueira, A.; Perez-Encinas, M.; Hernandez-Sanchez, A.; Gonzalez-Martinez, T.; Arellano-Rodrigo, E.; Martinez-Elicegui, J.; Villaverde-Ramiro, A.; Raya, J.M.; Ayala, R.; Ferrer-Marin, F.; et al. Machine learning improves risk stratification in myelofibrosis: An analysis of the Spanish registry of myelofibrosis. Hemasphere 2023, 7, e818. [Google Scholar] [CrossRef]
- Lippeveld, M.; Knill, C.; Ladlow, E.; Fuller, A.; Michaelis, L.J.; Saeys, Y.; Filby, A.; Peralta, D. Classification of human white blood cells using machine learning for stain-free imaging flow cytometry. Cytometry A 2020, 97, 308–319. [Google Scholar] [CrossRef]
- Flow Cytometry. Automated Image Analysis Reduces User-To-User Variability in Flow Cytometry Gating Strategies. 2022. Available online: https://www.thermofisher.com/hk/en/home/life-science/cell-analysis/flow-cytometry/flow-cytometers/attune-nxt-flow-cytometer/models (accessed on 4 May 2023).
- Luo, S.; Shi, Y.; Chin, L.K.; Hutchinson, P.E.; Zhang, Y.; Chierchia, G.; Talbot, H.; Jiang, X.; Bourouina, T.; Liu, A.-Q. Machine-learning-assisted intelligent imaging flow cytometry: A review. AdvIntell. Syst. 2021, 3, 2100073. [Google Scholar] [CrossRef]
- Shawly, T.; Alsheikhy, A.A. Biomedical diagnosis of leukemia using a deep learner classifier. Comput. Intell. Neurosci. 2022, 2022, 1568375. [Google Scholar] [CrossRef]
- Gudla, P.R.; Nakayama, K.; Pegoraro, G.; Misteli, T. SpotLearn: Convolutional neural network for detection of fluorescence in situ hybridization (FISH) signals in high-throughput imaging approaches. Cold Spring Harb. Symp. Quant. Biol. 2017, 82, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Bouilhol, E.; Savulescu, A.F.; Lefevre, E.; Dartigues, B.; Brackin, R.; Nikolski, M. DeepSpot: A deep neural network for RNA spot enhancement in single-molecule fluorescence in-situ hybridization microscopy images. Biol. Imaging 2022, 2, E4. [Google Scholar] [CrossRef]
- Yenamandra, A.K.; Hughes, C.; Maris, A.S. Artificial intelligence in plasma cell myeloma: Neural networks and support vector machines in the classification of plasma cell myeloma data at diagnosis. J. Pathol. Inform. 2021, 12, 35. [Google Scholar] [CrossRef] [PubMed]
- Bates, S.E. Classical cytogenetics: Karyotyping techniques. Hum. Pluripotent Stem Cells 2011, 767, 177–190. [Google Scholar]
- Xie, N.; Li, X.; Li, K.; Yang, Y.; Shen, H.T. Statistical karyotype analysis using CNN and geometric optimization. IEEE Access 2019, 7, 179445–179453. [Google Scholar] [CrossRef]
- Bokhari, Y.; Alhareeri, A.; Aljouie, A.; Alkhaldi, A.; Rashid, M.; Alawad, M.; Alhassnan, R.; Samargandy, S.; Panahi, A.; Heidrich, W.; et al. ChromoEnhancer: An artificial-intelligence-based tool to enhance neoplastic karyograms as an aid for effective analysis. Cells 2022, 11, 2244. [Google Scholar] [CrossRef]
- Hu, X.; Yi, W.; Jiang, L.; Wu, S.; Zhang, Y.; Du, J.; Ma, T.; Wang, T.; Wu, X. Classification of metaphase chromosomes using deep convolutional neural network. J. Comput. Biol. 2019, 26, 473–484. [Google Scholar] [CrossRef]
- Chen, X.; Cai, Q.; Ma, N.; Li, H. ChroSegNet: An Attention-based model for chromosome segmentation with enhanced processing. Appl. Sci. 2023, 13, 2308. [Google Scholar] [CrossRef]
- Vajen, B.; Hanselmann, S.; Lutterloh, F.; Kafer, S.; Espenkotter, J.; Beening, A.; Bogin, J.; Schlegelberger, B.; Gohring, G. Classification of fluorescent R-Band metaphase chromosomes using a convolutional neural network is precise and fast in generating karyograms of hematologic neoplastic cells. Cancer Genet. 2022, 260–261, 23–29. [Google Scholar] [CrossRef]
- Bobee, V.; Drieux, F.; Marchand, V.; Sater, V.; Veresezan, L.; Picquenot, J.M.; Viailly, P.J.; Lanic, M.D.; Viennot, M.; Bohers, E.; et al. Combining gene expression profiling and machine learning to diagnose B-cell non-Hodgkin lymphoma. Blood Cancer J. 2020, 10, 59. [Google Scholar] [CrossRef]
- Prakash, G.; Kaur, A.; Malhotra, P.; Khadwal, A.; Sharma, P.; Suri, V.; Varma, N.; Varma, S. Current role of genetics in hematologic malignancies. Indian J. Hematol. Blood Transfus. 2016, 32, 18–31. [Google Scholar] [CrossRef]
- Awada, H.; Durmaz, A.; Gurnari, C.; Kishtagari, A.; Meggendorfer, M.; Kerr, C.M.; Kuzmanovic, T.; Durrani, J.; Shreve, J.; Nagata, Y. Machine learning integrates genomic signatures for subclassification beyond primary and secondary acute myeloid leukemia. Blood 2021, 138, 1885–1895. [Google Scholar] [CrossRef]
- Warnat-Herresthal, S.; Oestreich, M.; Schultze, J.L.; Becker, M. Artificial intelligence in blood transcriptomics. In Artificial Intelligence in Medicine; Springer: Cham, Switzerland, 2022; pp. 1109–1123. [Google Scholar]
- Wan, J.; Gao, Y.; Zhao, X.; Wu, Q.; Fu, X.; Shao, Y.; Yang, H.; Guan, M.; Yu, B.; Zhang, W. The association between the copy-number variations of ZMAT4 and hematological malignancy. Hematology 2011, 16, 20–23. [Google Scholar] [CrossRef]
- Laurent, A.P.; Kotecha, R.S.; Malinge, S. Gain of chromosome 21 in hematological malignancies: Lessons from studying leukemia in children with Down syndrome. Leukemia 2020, 34, 1984–1999. [Google Scholar] [CrossRef]
- Schmidt, T.M.; Fonseca, R.; Usmani, S.Z. Chromosome 1q21 abnormalities in multiple myeloma. Blood Cancer J. 2021, 11, 83. [Google Scholar] [CrossRef]
- Song, Y.; Fang, Q.; Mi, Y. Prognostic significance of copy number variation in B-cell acute lymphoblastic leukemia. Front. Oncol. 2022, 12, 981036. [Google Scholar] [CrossRef]
- Chicano, M.; Carbonell, D.; Suarez-Gonzalez, J.; Lois, S.; Ballesteros-Culebras, M.; Andres-Zayas, C.; Muniz, P.; Rodriguez-Macias, G.; Bastos-Oreiro, M.; Font, P.; et al. Next generation cytogenetics in myeloid hematological neoplasms: Detection of CNVs and translocations. Cancers 2021, 13, 3001. [Google Scholar] [CrossRef]
- Haferlach, C.; Hänselmann, S.; Walter, W.; Volkert, S.; Zenger, M.; Kern, W.; Stengel, A.; Lörch, T.; Haferlach, T. Artificial intelligence substantially supports chromosome banding analysis maintaining its strengths in hematologic diagnostics even in the era of newer technologies. Blood 2020, 136, 47–48. [Google Scholar] [CrossRef]
- Wang, T.; Sun, J.; Zhang, X.; Wang, W.J.; Zhou, Q. CNV-P: A machine-learning framework for predicting high confident copy number variations. Peer J. 2021, 9, e12564. [Google Scholar] [CrossRef]
- Zhang, L.; Shi, J.; Ouyang, J.; Zhang, R.; Tao, Y.; Yuan, D.; Lv, C.; Wang, R.; Ning, B.; Roberts, R.; et al. X-CNV: Genome-wide prediction of the pathogenicity of copy number variations. Genome Med. 2021, 13, 132. [Google Scholar] [CrossRef]
- Shalek, A.K.; Benson, M. Single-cell analyses to tailor treatments. Sci. Transl. Med. 2017, 9, eaan4730. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Tang, R.; Xu, J.; Wang, W.; Zhang, B.; Liu, J.; Yu, X.; Shi, S. Applications of single-cell sequencing in cancer research: Progress and perspectives. J. Hematol. Oncol. 2021, 14, 91. [Google Scholar] [CrossRef] [PubMed]
- Syed-Abdul, S.; Firdani, R.P.; Chung, H.J.; Uddin, M.; Hur, M.; Park, J.H.; Kim, H.W.; Gradisek, A.; Dovgan, E. Artificial intelligence based models for screening of hematologic malignancies using cell population data. Sci. Rep. 2020, 10, 4583. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, K.; Lyu, Y.; Pan, H.; Zhang, J.; Stambolian, D.; Susztak, K.; Reilly, M.P.; Hu, G.; Li, M. Deep learning enables accurate clustering with batch effect removal in single-cell RNA-seq analysis. Nat. Commun. 2020, 11, 2338. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Zhang, J.; Lin, X.; Wei, Z.; Hakonarson, H. Model-based deep embedding for constrained clustering analysis of single cell RNA-seq data. Nat. Commun. 2021, 12, 1873. [Google Scholar] [CrossRef]
- Tian, T.; Wan, J.; Song, Q.; Wei, Z. Clustering single-cell RNA-seq data with a model-based deep learning approach. Nat. Mach. Intell. 2019, 1, 191–198. [Google Scholar] [CrossRef]
- Awada, H.; Gurnari, C.; Durmaz, A.; Awada, H.; Pagliuca, S.; Visconte, V. Personalized risk schemes and machine learning to empower genomic prognostication models in myelodysplastic syndromes. Int. J. Mol. Sci. 2022, 23, 2802. [Google Scholar] [CrossRef]
- Radakovich, N.; Cortese, M.; Nazha, A. Acute myeloid leukemia and artificial intelligence, algorithms and new scores. Best. Pract. Res. Clin. Hematol. 2020, 33, 101192. [Google Scholar] [CrossRef]
- Giovanna Nicora, R.B. A reliable machine learning approach applied to single-cell classification in acute myeloid leukemia. AMIA. Annu. Symp. Proc. 2020, 2020, 925–932. [Google Scholar]
- Moreno-Sanchez, F.; Gomez-Gomez, B. Antibiotic management of patients with hematologic malignancies: From prophylaxis to unusual infections. Curr. Oncol. Rep. 2022, 24, 835–842. [Google Scholar] [CrossRef]
- Chen, X.; Chen, H.Y.; Chen, Z.D.; Gong, J.N.; Chen, C.Y. A novel artificial intelligence protocol for finding potential inhibitors of acute myeloid leukemia. J. Mater. Chem. B 2020, 8, 2063–2081. [Google Scholar] [CrossRef]
- Gimeno, M.; San Jose-Eneriz, E.; Villar, S.; Agirre, X.; Prosper, F.; Rubio, A.; Carazo, F. Explainable artificial intelligence for precision medicine in acute myeloid leukemia. Front. Immunol. 2022, 13, 977358. [Google Scholar] [CrossRef]
- Jiao, L.; Ren, Y.; Wang, L.; Gao, C.; Wang, S.; Song, T. MulCNN: An efficient and accurate deep learning method based on gene embedding for cell type identification in single-cell RNA-seq data. Front. Genet. 2023, 14, 1179859. [Google Scholar] [CrossRef]
- Wang, T.; Johnson, T.S.; Shao, W.; Lu, Z.; Helm, B.R.; Zhang, J.; Huang, K. BERMUDA: A novel deep transfer learning method for single-cell RNA sequencing batch correction reveals hidden high-resolution cellular subtypes. Genome Biol. 2019, 20, 165. [Google Scholar] [CrossRef]
- Schmidt, F.; Ranjan, B.; Lin, Q.X.X.; Krishnan, V.; Joanito, I.; Honardoost, M.A.; Nawaz, Z.; Venkatesh, P.N.; Tan, J.; Rayan, N.A.; et al. RCA2: A scalable supervised clustering algorithm that reduces batch effects in scRNA-seq data. Nucleic Acids Res. 2021, 49, 8505–8519. [Google Scholar] [CrossRef]
- Lahnemann, D.; Koster, J.; Szczurek, E.; McCarthy, D.J.; Hicks, S.C.; Robinson, M.D.; Vallejos, C.A.; Campbell, K.R.; Beerenwinkel, N.; Mahfouz, A.; et al. Eleven grand challenges in single-cell data science. Genome Biol. 2020, 21, 31. [Google Scholar] [CrossRef]
- Sapoval, N.; Aghazadeh, A.; Nute, M.G.; Antunes, D.A.; Balaji, A.; Baraniuk, R.; Barberan, C.J.; Dannenfelser, R.; Dun, C.; Edrisi, M.; et al. Current progress and open challenges for applying deep learning across the biosciences. Nat. Commun. 2022, 13, 1728. [Google Scholar] [CrossRef]
- Teschendorff, A.E.; Zhu, T.; Breeze, C.E.; Beck, S. EPISCORE: Cell type deconvolution of bulk tissue DNA methylomes from single-cell RNA-seq data. Genome Biol. 2020, 21, 221. [Google Scholar] [CrossRef]
- Koldobskiy, M.A.; Jenkinson, G.; Abante, J.; Rodriguez DiBlasi, V.A.; Zhou, W.; Pujadas, E.; Idrizi, A.; Tryggvadottir, R.; Callahan, C.; Bonifant, C.L.; et al. Converging genetic and epigenetic drivers of pediatric acute lymphoblastic leukemia identified by an information-theoretic analysis. Nat. Biomed. Eng. 2021, 5, 360–376. [Google Scholar] [CrossRef]
- Levy, J.J.; Titus, A.J.; Petersen, C.L.; Chen, Y.; Salas, L.A.; Christensen, B.C. MethylNet: An automated and modular deep learning approach for DNA methylation analysis. BMC Bioinform. 2020, 21, 108. [Google Scholar] [CrossRef]
- Reilly, B.; Tanaka, T.N.; Diep, D.; Yeerna, H.; Tamayo, P.; Zhang, K.; Bejar, R. DNA methylation identifies genetically and prognostically distinct subtypes of myelodysplastic syndromes. Blood Adv. 2019, 3, 2845–2858. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Xu, B.; Xu, J.; Li, J.; Jiang, J.; Ren, Y.; Liu, P. A machine learning model to predict survival and therapeutic responses in multiple myeloma. Int. J. Mol. Sci. 2023, 24, 6683. [Google Scholar] [CrossRef] [PubMed]
- Derrien, J.; Guerin-Charbonnel, C.; Gaborit, V.; Campion, L.; Devic, M.; Douillard, E.; Roi, N.; Avet-Loiseau, H.; Decaux, O.; Facon, T.; et al. The DNA methylation landscape of multiple myeloma shows extensive inter- and intrapatient heterogeneity that fuels transcriptomic variability. Genome Med. 2021, 13, 127. [Google Scholar] [CrossRef] [PubMed]
- Saleem, M.M.; Mendoza-Parra, M.A.; Cholley, P.E.; Blum, M.; Gronemeyer, H. Epimetheus–A multi-profile normalizer for epigenomic sequencing data. BMC Bioinform. 2017, 18, 259. [Google Scholar] [CrossRef]
- Ovejero, S.; Moreaux, J. Multi-omics tumor profiling technologies to develop precision medicine in multiple myeloma. Explor. Target. Antitumor. Ther. 2021, 2, 65–106. [Google Scholar]
- Angermueller, C.; Lee, H.J.; Reik, W.; Stegle, O. DeepCpG: Accurate prediction of single-cell DNA methylation states using deep learning. Genome Biol. 2017, 18, 67. [Google Scholar] [CrossRef]
- Rauschert, S.; Raubenheimer, K.; Melton, P.E.; Huang, R.C. Machine learning and clinical epigenetics: A review of challenges for diagnosis and classification. Clin. Epigenet. 2020, 12, 51. [Google Scholar] [CrossRef]
- Xia, C.; Wang, J.; Qin, Y.; Wen, J.; Liu, Z.; Song, N.; Wu, L.; Chen, B.; Gu, Y.; Yang, J. KaryoNet: Chromosome recognition with end-to-end combinatorial optimization network. IEEE Trans. Med. Imaging 2023. Early Access. [Google Scholar] [CrossRef]
- Zhao, Y.; Pan, Z.; Namburi, S.; Pattison, A.; Posner, A.; Balachander, S.; Paisie, C.A.; Reddi, H.V.; Rueter, J.; Gill, A.J.; et al. CUP-AI-Dx: A tool for inferring cancer tissue of origin and molecular subtype using RNA gene-expression data and artificial intelligence. EBioMedicine 2020, 61, 103030. [Google Scholar] [CrossRef]
- Szablewski, V.; Bret, C.; Kassambara, A.; Devin, J.; Cartron, G.; Martineau, V.C.; Moreaux, J. An epigenetic regulator-related score (EpiScore) predicts survival in patients with diffuse large B cell lymphoma and identifies patients who may benefit from epigenetic therapy. Oncotarget 2018, 9, 19079–19099. [Google Scholar] [CrossRef]
- Pan, L.; Liu, G.; Lin, F.; Zhong, S.; Xia, H.; Sun, X.; Liang, H. Machine learning applications for prediction of relapse in childhood acute lymphoblastic leukemia. Sci. Rep. 2017, 7, 7402. [Google Scholar] [CrossRef]
- Chen, L. Overview of clinical prediction models. Ann. Transl. Med. 2020, 8, 71. [Google Scholar] [CrossRef]
- Mosquera Orgueira, A.; Gonzalez Perez, M.S.; Diaz Arias, J.A.; Antelo Rodriguez, B.; Alonso Vence, N.; Bendana Lopez, A.; Abuin Blanco, A.; Bao Perez, L.; Peleteiro Raindo, A.; Cid Lopez, M.; et al. Survival prediction and treatment optimization of multiple myeloma patients using machine-learning models based on clinical and gene expression data. Leukemia 2021, 35, 2924–2935. [Google Scholar] [CrossRef]
- Wallington-Beddoe, C.T.; Mynott, R.L. Prognostic and predictive biomarker developments in multiple myeloma. J. Hematol. Oncol. 2021, 14, 151. [Google Scholar] [CrossRef]
- Siddiqui, N.S.; Klein, A.; Godara, A.; Buchsbaum, R.J.; Hughes, M.C. Predicting in-hospital mortality after acute myeloid leukemia therapy: Through supervised machine learning algorithms. JCO Clin. Cancer Inform. 2022, 6, e2200044. [Google Scholar] [CrossRef]
- Li, J.L.; Wang, Y.F.; Ko, B.S.; Li, C.C.; Tang, J.L.; Lee, C.C. Learning a cytometric deep phenotype embedding for automatic hematological malignancies classification. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2019, 2019, 1733–1736. [Google Scholar]
- Hoffmann, J.; Eminovic, S.; Wilhelm, C.; Krause, S.W.; Neubauer, A.; Thrun, M.C.; Ultsch, A.; Brendel, C. Prediction of clinical outcomes with explainable artificial intelligence in patients with chronic lymphocytic leukemia. Curr. Oncol. 2023, 30, 1903–1915. [Google Scholar] [CrossRef]
- Agius, R.; Brieghel, C.; Andersen, M.A.; Pearson, A.T.; Ledergerber, B.; Cozzi-Lepri, A.; Louzoun, Y.; Andersen, C.L.; Bergstedt, J.; von Stemann, J.H.; et al. Machine learning can identify newly diagnosed patients with CLL at high risk of infection. Nat. Commun. 2020, 11, 363. [Google Scholar] [CrossRef]
- Shanbehzadeh, M.; Afrash, M.R.; Mirani, N.; Kazemi-Arpanahi, H. Comparing machine learning algorithms to predict 5-year survival in patients with chronic myeloid leukemia. BMC Med. Inform. Decis. Mak. 2022, 22, 236. [Google Scholar] [CrossRef]
- Yang, C.C. Explainable artificial intelligence for predictive modeling in healthcare. J. Healthc. Inform. Res. 2022, 6, 228–239. [Google Scholar] [CrossRef]
- Cadamuro, J. Rise of the Machines: The inevitable evolution of medicine and medical laboratories intertwining with artificial intelligence-A narrative review. Diagnostics 2021, 11, 1399. [Google Scholar] [CrossRef] [PubMed]
- Morley, J.; Machado, C.C.V.; Burr, C.; Cowls, J.; Joshi, I.; Taddeo, M.; Floridi, L. The ethics of AI in health care: A mapping review. Soc. Sci. Med. 2020, 260, 113172. [Google Scholar] [CrossRef]
- Murdoch, B. Privacy and artificial intelligence: Challenges for protecting health information in a new era. BMC. Med. Ethics 2021, 22, 122. [Google Scholar] [CrossRef] [PubMed]
- Bashshur, R.L.; Krupinski, E.A.; Doarn, C.R.; Merrell, R.C.; Woolliscroft, J.O.; Frenk, J. Telemedicine across time: Integrated health system of the future-A prelude. Telemed. J. E Health 2020, 26, 128–130. [Google Scholar] [CrossRef] [PubMed]
- Kelly, C.J.; Karthikesalingam, A.; Suleyman, M.; Corrado, G.; King, D. Key challenges for delivering clinical impact with artificial intelligence. BMC Med. 2019, 17, 195. [Google Scholar] [CrossRef]
- Obermeyer, Z.; Weinstein, B.J. Adoption of artificial intelligence and machine learning is increasing, but irrational exuberance remains. NEJM. Catal. Innov. Care. Deliv. 2019, 1, 1–18. [Google Scholar] [CrossRef]
- Noorbakhsh-Sabet, N.; Zand, R.; Zhang, Y.; Abedi, V. Artificial intelligence transforms the future of health care. Am. J. Med. 2019, 132, 795–801. [Google Scholar] [CrossRef]
- Chai, S.Y.; Hayat, A.; Flaherty, G.T. Integrating artificial intelligence into hematology training and practice: Opportunities, threats and proposed solutions. Br. J. Hematol. 2022, 198, 807–811. [Google Scholar] [CrossRef]



| No. | Methods | Function | Model Used | Accuracy (%) | Year | References |
|---|---|---|---|---|---|---|
| 1 | WSI analysis | Automated detection of HB-H inclusions in RBCs | ML, CNN | 97.6 | 2021 | [59] |
| 2 | CellaVision | Blood and marrow smear image analysis Advanced RBC morphology analysis | AI, ANN | 98 | 2020 | [19,23] |
| 3 | DI-60 | Automated cell image analyzer | 91 | 2022 | [72] | |
| 4 | Morphogo | Blood and marrow smear image analysis | AI, CNN | 85.7–91 | 2021 | [20,30] |
| 5 | Scorpio | Fulfilled PBS and BMA image analysis | AI | 94.9 | 2020 | [33] |
| 6 | Mantiscope | Digital PBS and BMS preparation and image analysis | ANN, CNN | NA | 2018 | [73] |
| 7 | Vision Hema | Blood cell identification and pre-classification | AI | NA | 2019 | [35] |
| 8 | EasyCell Assistant | Automatic detection and classification of cell morphology | ML | NA | 2023 | [74] |
| 9 | YOLOX-s model | BM cell classification | DNN | 92.5 | 2023 | [75] |
| 10 | Nextslide | Automated digital imaging | AI | 99.7 | 2012 | [76] |
| 11 | XGBoost | Differentiate PV, ET, and MF | CNN, DL | 90 | 2021 | [77] |
| 12 | HematoNet | BM cell detection and classification | DL, CoAtNet | 95 | 2022 | [41] |
| 13 | Ensemble model | WBC detection | DL | 98.8 | 2023 | [78] |
| 14 | Automated BMT phenotyping | Morphological identification of megakaryocytes | AI, ML | 95 | 2020 | [79] |
| 15 | AIPSS-MF | Risk stratification of MF patients | ML, random forest | 82 | 2023 | [80] |
| 16 | ImageStream (Amnis) | Identification of white blood cells | ML, SVM | 99 | 2020 | [81] |
| 17 | Attune CytPix | High-resolution, real-time imaging of cells in flow cytometry | AI | NA | 2023 | [82] |
| 18 | ImageStream (Amnis) | Leukemia monitoring | CNN, linear SVM | 98.2 | 2021 | [83] |
| 19 | AlexNet | Detection of ALL and AML | ML, CNN | 98 | 2022 | [84] |
| 20 | DNN-FC | Detection of CLL-MRD | AI, DNN | 97.1 | 2022 | [63] |
| 21 | XAI | Translate AI data in ALL | AI, DL | 98.4 | 2022 | [70] |
| 22 | EfficientNet | Differentiate NHL | CNN | 95.6 | 2021 | [44] |
| 23 | LymphoML | Predict lymphoma types | ML, LightGBM | 85 | 2023 | [50] |
| 24 | ImmunoGenius | Predict and differentiate lymphoma subtypes | ML, decision tree algorithm | 91.8 | 2023 | [52] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gedefaw, L.; Liu, C.-F.; Ip, R.K.L.; Tse, H.-F.; Yeung, M.H.Y.; Yip, S.P.; Huang, C.-L. Artificial Intelligence-Assisted Diagnostic Cytology and Genomic Testing for Hematologic Disorders. Cells 2023, 12, 1755. https://doi.org/10.3390/cells12131755
Gedefaw L, Liu C-F, Ip RKL, Tse H-F, Yeung MHY, Yip SP, Huang C-L. Artificial Intelligence-Assisted Diagnostic Cytology and Genomic Testing for Hematologic Disorders. Cells. 2023; 12(13):1755. https://doi.org/10.3390/cells12131755
Chicago/Turabian StyleGedefaw, Lealem, Chia-Fei Liu, Rosalina Ka Ling Ip, Hing-Fung Tse, Martin Ho Yin Yeung, Shea Ping Yip, and Chien-Ling Huang. 2023. "Artificial Intelligence-Assisted Diagnostic Cytology and Genomic Testing for Hematologic Disorders" Cells 12, no. 13: 1755. https://doi.org/10.3390/cells12131755
APA StyleGedefaw, L., Liu, C.-F., Ip, R. K. L., Tse, H.-F., Yeung, M. H. Y., Yip, S. P., & Huang, C.-L. (2023). Artificial Intelligence-Assisted Diagnostic Cytology and Genomic Testing for Hematologic Disorders. Cells, 12(13), 1755. https://doi.org/10.3390/cells12131755






