The Prodigious Potential of mRNA Electrotransfer as a Substitute to Conventional DNA-Based Transient Transfection
Abstract
:1. Introduction
2. Results
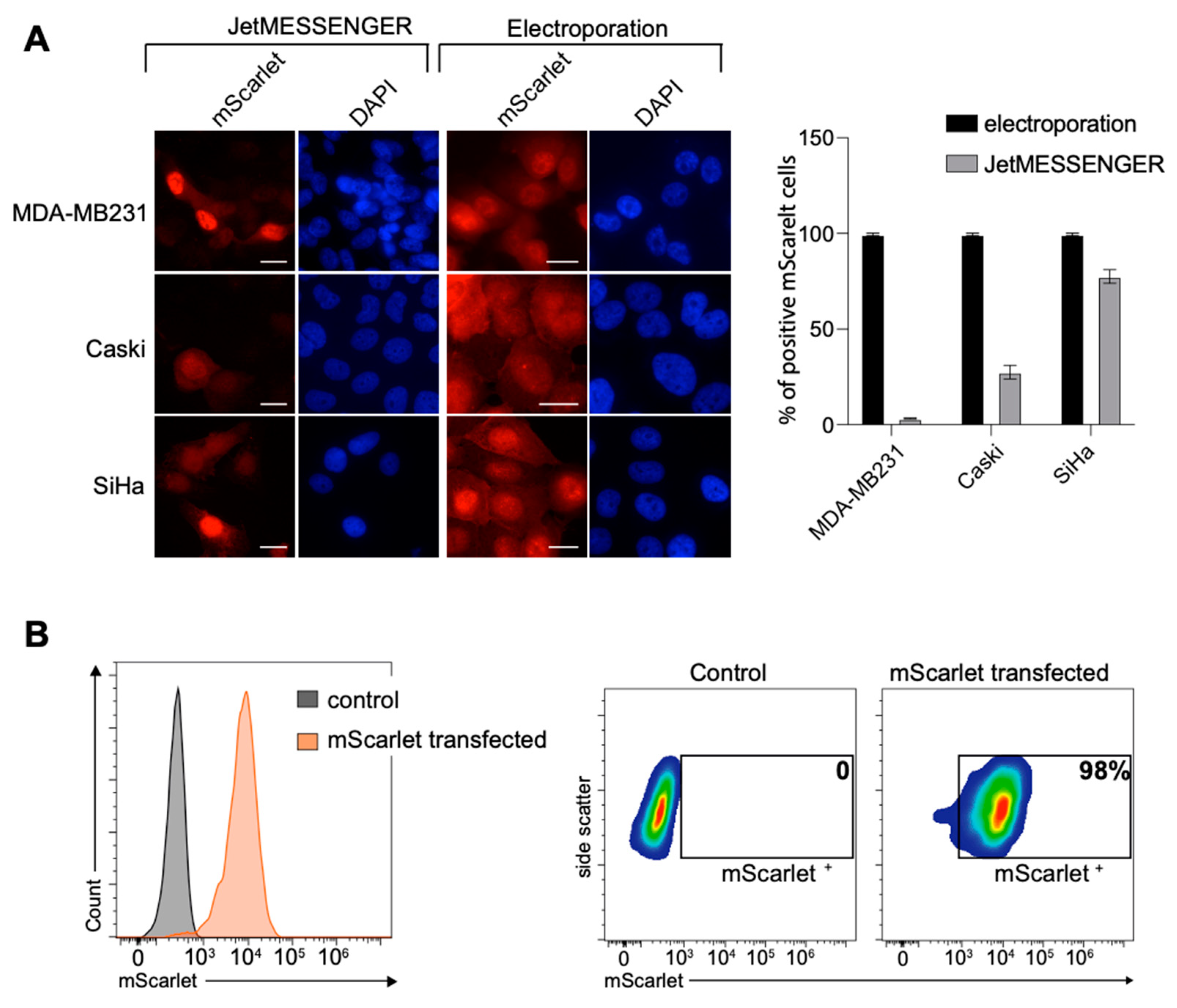
2.1. High Transfection Efficiency for Various Cell Lines
2.2. E6 Oncoprotein from HPV16
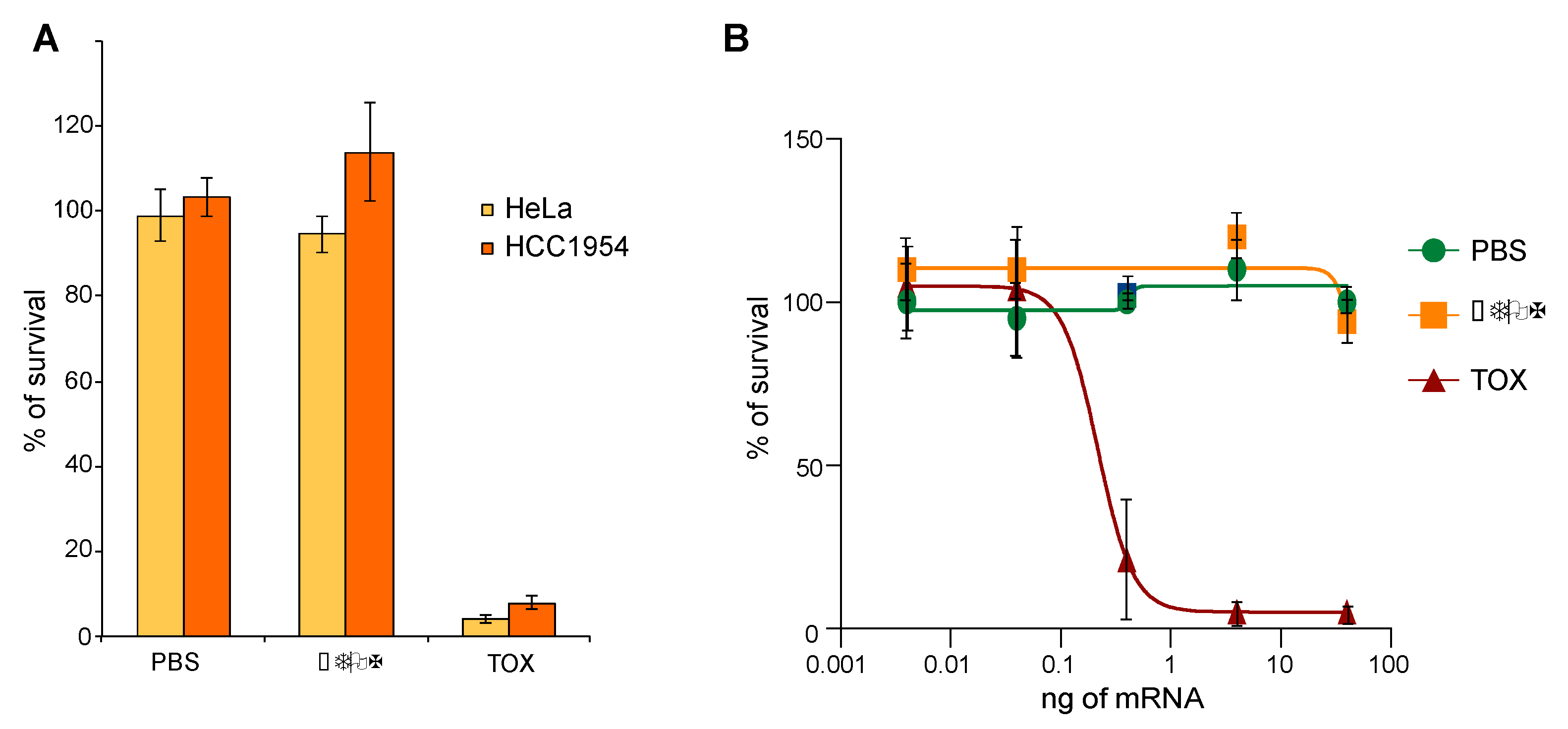
2.3. TOX Fragment from Pseudomonase aeruginosa
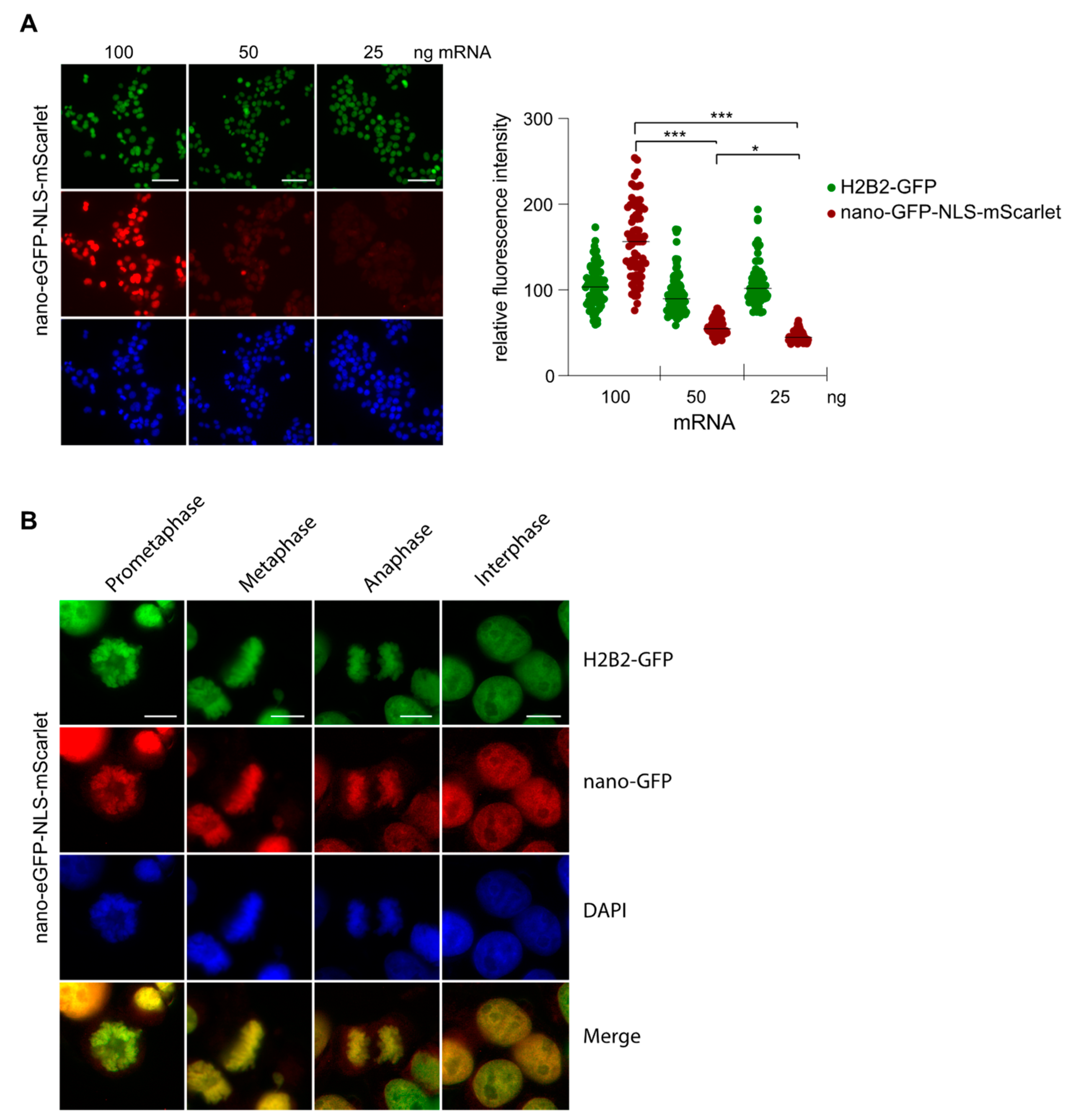
2.4. Nanobody Anti-GFP
2.5. Interplay between PCNA, Con1 and DNA Polymerase η
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Fus-Kujawa, A.; Prus, P.; Bajdak-Rusinek, K.; Teper, P.; Gawron, K.; Kowalczuk, A.; Sieron, A.L. An Overview of Methods and Tools for Transfection of Eukaryotic Cells in Vitro. Front. Bioeng. Biotechnol. 2021, 9, 701031. [Google Scholar] [CrossRef]
- Durocher, Y.; Perret, S.; Kamen, A. High-Level and High-Throughput Recombinant Protein Production by Transient Transfection of Suspension-Growing Human 293-EBNA1 Cells. Nucleic Acids Res. 2002, 30, e9. [Google Scholar] [CrossRef]
- Colosimo, A.; Goncz, K.K.; Holmes, A.R.; Kunzelmann, K.; Novelli, G.; Malones, R.W.; Bennett, M.J.; Gruenert, D.C. Transfer and Expression of Foreign Genes in Mammalian Cells. Biotechniques 2000, 29, 314–331. [Google Scholar] [CrossRef] [Green Version]
- Schenborn, E.T.; Oler, J. Liposome-Mediated Transfection of Mammalian Cells. Methods Mol. Biol. 2000, 130, 155–164. [Google Scholar] [CrossRef]
- Shi, J.; Ma, Y.; Zhu, J.; Chen, Y.; Sun, Y.; Yao, Y.; Yang, Z.; Xie, J. A Review on Electroporation-Based Intracellular Delivery. Molecules 2018, 23, 3044. [Google Scholar] [CrossRef] [Green Version]
- Tsong, T.Y. Electroporation of Cell Membranes. Biophys. J. 1991, 60, 297–306. [Google Scholar] [CrossRef] [Green Version]
- Butash, K.A.; Young, A.; Fox, D.K. Reexamination of the Effect of Endotoxin on Cell Proliferation and Transfection Efficiency. BioTechniques 2000, 29, 610–614. [Google Scholar] [CrossRef]
- Nomura, Y.; Fukui, C.; Morishita, Y.; Haishima, Y. A Biological Study Establishing the Endotoxin Limit for in Vitro Proliferation of Human Mesenchymal Stem Cells. Regen. Ther. 2017, 7, 45–51. [Google Scholar] [CrossRef]
- Faust, C.; Beil, C.; Dittrich, W.; Rao, E.; Langer, T. Impact of Lipopolysaccharides on Cultivation and Recombinant Protein Expression in Human Embryonal Kidney (HEK-293) Cells. Eng. Life Sci. 2021, 21, 778–785. [Google Scholar] [CrossRef]
- Vigneron, M.; Dietsch, F.; Bianchetti, L.; Dejaegere, A.; Nominé, Y.; Cordonnier, A.; Zuber, G.; Chatton, B.; Donzeau, M. Self-Associating Peptides for Modular Bifunctional Conjugation of Tetramer Macromolecules in Living Cells. Bioconjug. Chem. 2019, 30, 1734–1744. [Google Scholar] [CrossRef]
- Maurisse, R.; De Semir, D.; Emamekhoo, H.; Bedayat, B.; Abdolmohammadi, A.; Parsi, H.; Gruenert, D.C. Comparative Transfection of DNA into Primary and Transformed Mammalian Cells from Different Lineages. BMC Biotechnol. 2010, 10, 9. [Google Scholar] [CrossRef] [Green Version]
- Avci-Adali, M.; Behring, A.; Steinle, H.; Keller, T.; Krajeweski, S.; Schlensak, C.; Wendel, H.P. In Vitro Synthesis of Modified MRNA for Induction of Protein Expression in Human Cells. J. Vis. Exp. 2014, 93, e51943. [Google Scholar] [CrossRef] [Green Version]
- McLenachan, S.; Zhang, D.; Palomo, A.B.A.; Edel, M.J.; Chen, F.K. MRNA Transfection of Mouse and Human Neural Stem Cell Cultures. PLoS ONE 2013, 8, e83596. [Google Scholar] [CrossRef] [Green Version]
- Chandra, P.; Philips, J.A. Ectopic Gene Expression in Macrophages Using in Vitro Transcribed MRNA. Bio-Protocol 2018, 8, e2857. [Google Scholar] [CrossRef]
- Zhou, X.; Hao, R.; Chen, C.; Su, Z.; Zhao, L.; Luo, Z.; Xie, W. Rapid Delivery of Nanobodies/VHHs into Living Cells via Expressing In Vitro-Transcribed MRNA. Mol. Ther. Methods Clin. Dev. 2020, 17, 401–408. [Google Scholar] [CrossRef]
- Howley, P.M. Warts, Cancer and Ubiquitylation: Lessons from the Papillomaviruses. Trans. Am. Clin. Climatol. Assoc. 2006, 117, 113–127. [Google Scholar]
- Moldovan, G.-L.L.; Pfander, B.; Jentsch, S. PCNA, the Maestro of the Replication Fork. Cell 2007, 129, 665–679. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.C. PCNA: A Silent Housekeeper or a Potential Therapeutic Target? Trends Pharmacol. Sci. 2014, 35, 178–186. [Google Scholar] [CrossRef]
- Michalska, M.; Wolf, P. Pseudomonas Exotoxin A: Optimized by Evolution for Effective Killing. Front. Microbiol. 2015, 6, 963. [Google Scholar] [CrossRef] [Green Version]
- Stoessel, A.; Groysbeck, N.; Guyot, L.; Barret, L.; Nominé, Y.; Nguekeu-Zebaze, L.; Bender, A.; Voilquin, L.; Lutz, T.; Pallaoro, N.; et al. Modular Conjugation of a Potent Anti-HER2 Immunotoxin Using Coassociating Peptides. Bioconjug. Chem. 2020, 31, 2421–2430. [Google Scholar] [CrossRef]
- Rothbauer, U.; Zolghadr, K.; Tillib, S.; Nowak, D.; Schermelleh, L.; Gahl, A.; Backmann, N.; Conrath, K.; Muyldermans, S.; Cardoso, M.C.; et al. Targeting and Tracing Antigens in Live Cells with Fluorescent Nanobodies. Nat. Methods 2006, 3, 887–889. [Google Scholar] [CrossRef]
- Boissel, L.; Betancur, M.; Wels, W.S.; Tuncer, H.; Klingemann, H. Transfection with MRNA for CD19 Specific Chimeric Antigen Receptor Restores NK Cell Mediated Killing of CLL Cells. Leuk. Res. 2009, 33, 1255–1259. [Google Scholar] [CrossRef] [Green Version]
- Gharaati-Far, N.; Tohidkia, M.R.; Dehnad, A.; Omidi, Y. Efficiency and Cytotoxicity Analysis of Cationic Lipids-Mediated Gene Transfection into AGS Gastric Cancer Cells. Artif. Cells Nanomed. Biotechnol. 2018, 46, 1001–1008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Travé, G.; Zanier, K. HPV-Mediated Inactivation of Tumor Suppressor P53. Cell Cycle 2016, 15, 2231–2232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez-Zapien, D.; Ruiz, F.X.; Poirson, J.; Mitschler, A.; Ramirez, J.; Forster, A.; Cousido-Siah, A.; Masson, M.; Vande Pol, S.; Podjarny, A.; et al. Structure of the E6/E6AP/P53 Complex Required for HPV-Mediated Degradation of P53. Nature 2016, 529, 541–545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaisman, A.; Woodgate, R. Translesion DNA Polymerases in Eukaryotes: What Makes Them Tick? Crit. Rev. Biochem. Mol. Biol. 2017, 52, 274–303. [Google Scholar] [CrossRef] [Green Version]
- Nettersheim, J.; Janel-bintz, R.; Kuhn, L. DNA Polymerase η Is a Substrate for Calpain: A Possible Mechanism for Pol η Retention in UV-Induced Replication Foci. J. Cell Sci. 2021, 134, 258637. [Google Scholar] [CrossRef] [PubMed]
- Acharya, N.; Patel, S.K.; Sahu, S.R.; Kumari, P. ‘PIPs’ in DNA Polymerase: PCNA Interaction Affairs. Biochem. Soc. Trans. 2020, 48, 2811–2822. [Google Scholar] [CrossRef]
- Prives, C.; Gottifredi, V. The P21 and PCNA Partnership. Cell Cycle 2008, 7, 3840–3846. [Google Scholar] [CrossRef] [Green Version]
- Zheleva, D.I.; Zhelev, N.Z.; Fischer, P.M.; Duff, S.V.; Warbrick, E.; Blake, D.G.; Lane, D.P. A Quantitative Study of the in Vitro Binding of the C-Terminal Domain of P21 to PCNA: Affinity, Stoichiometry, and Thermodynamics. Biochemistry 2000, 39, 7388–7397. [Google Scholar] [CrossRef]
- Dietsch, F.; Nominé, Y.; Stoessel, A.; Kostmann, C.; Bonhoure, A.; Chatton, B.; Donzeau, M. Small P53 Derived Peptide Suitable for Robust Nanobodies Dimerization. J. Immunol. Methods 2021, 498, 113144. [Google Scholar] [CrossRef] [PubMed]
- Moeglin, E.; Barret, L.; Chatton, B.; Donzeau, M. Modular Site-Specific Conjugation of Nanobodies Using Two Co-Associating Tags. Int. J. Mol. Sci. 2022, 23, 14405. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Juncker, T.; Chatton, B.; Donzeau, M. The Prodigious Potential of mRNA Electrotransfer as a Substitute to Conventional DNA-Based Transient Transfection. Cells 2023, 12, 1591. https://doi.org/10.3390/cells12121591
Juncker T, Chatton B, Donzeau M. The Prodigious Potential of mRNA Electrotransfer as a Substitute to Conventional DNA-Based Transient Transfection. Cells. 2023; 12(12):1591. https://doi.org/10.3390/cells12121591
Chicago/Turabian StyleJuncker, Théo, Bruno Chatton, and Mariel Donzeau. 2023. "The Prodigious Potential of mRNA Electrotransfer as a Substitute to Conventional DNA-Based Transient Transfection" Cells 12, no. 12: 1591. https://doi.org/10.3390/cells12121591
APA StyleJuncker, T., Chatton, B., & Donzeau, M. (2023). The Prodigious Potential of mRNA Electrotransfer as a Substitute to Conventional DNA-Based Transient Transfection. Cells, 12(12), 1591. https://doi.org/10.3390/cells12121591





