Relevance of Bile Acids in Cholangiocarcinoma Pathogenesis: Critical Revision and Future Directions
Abstract
:1. Introduction
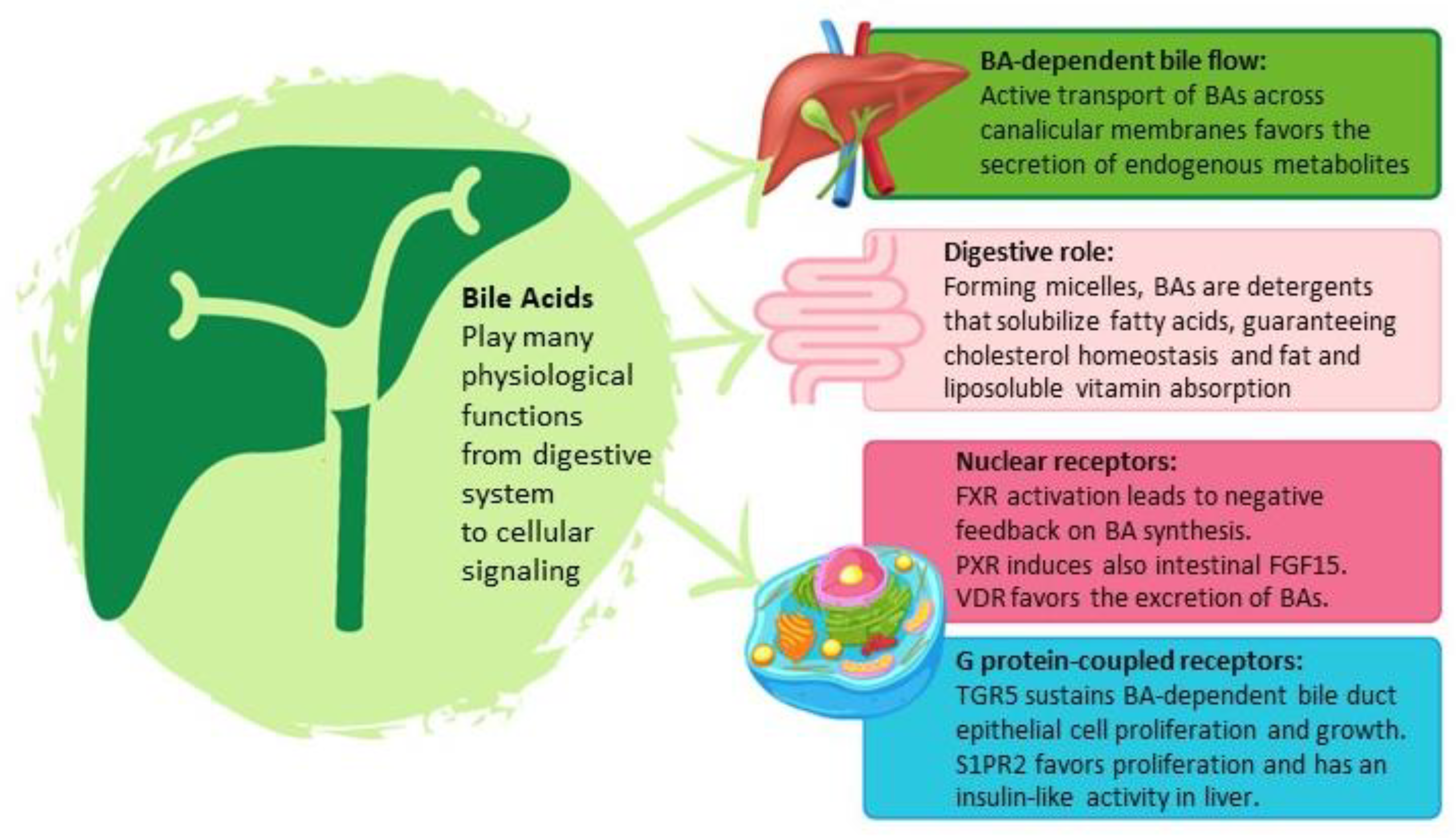
2. Bile Acid Physiology
2.1. Bile Acid Synthesis
2.2. Physiological Bile Acid Functions
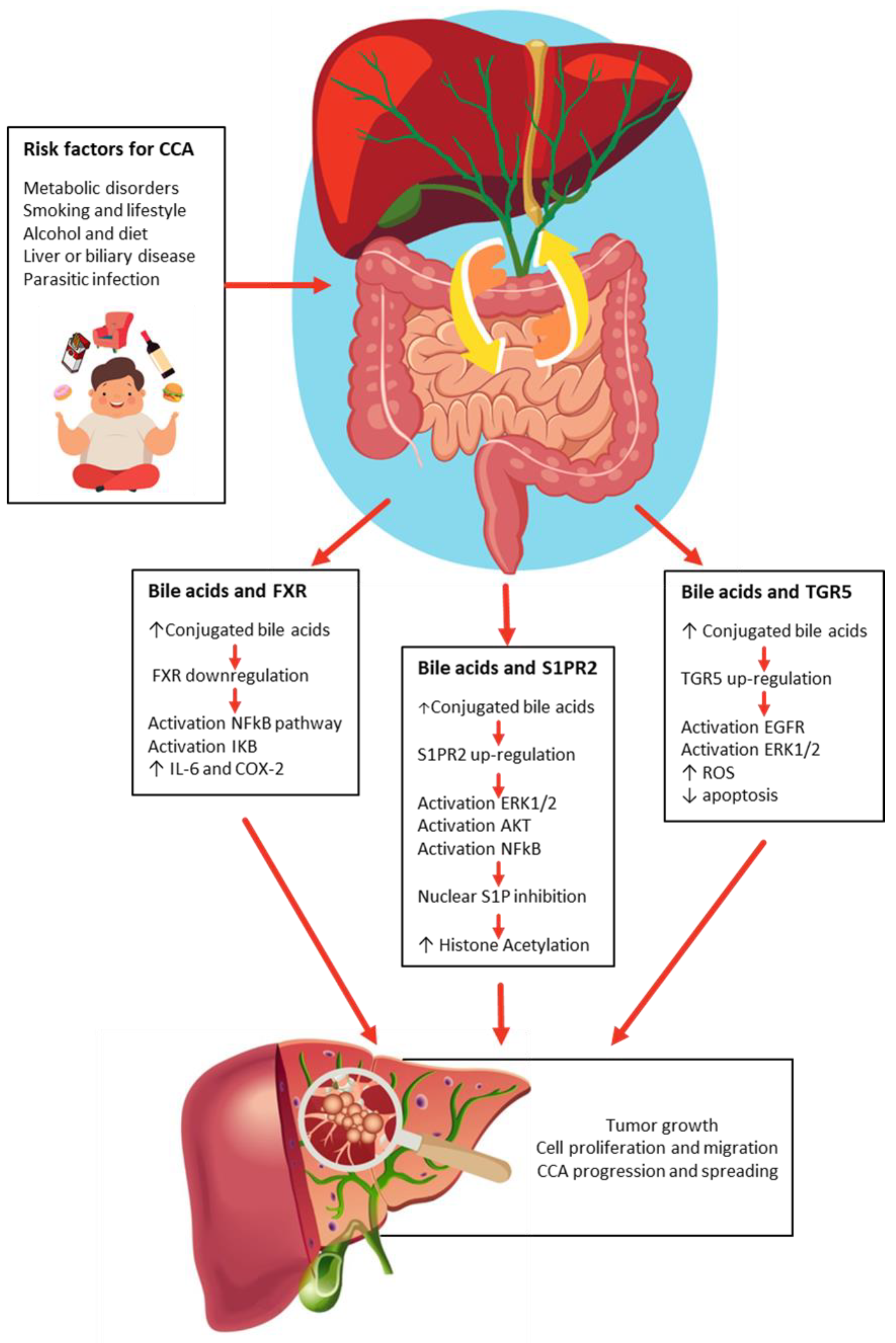
3. Bile Acids Act like a Double-Edged Sword in Cholangiocarcinogenesis
3.1. FXR and Bile Acids
3.2. SIPR2 and Bile Acids
3.3. TGR5 and Bile Acids
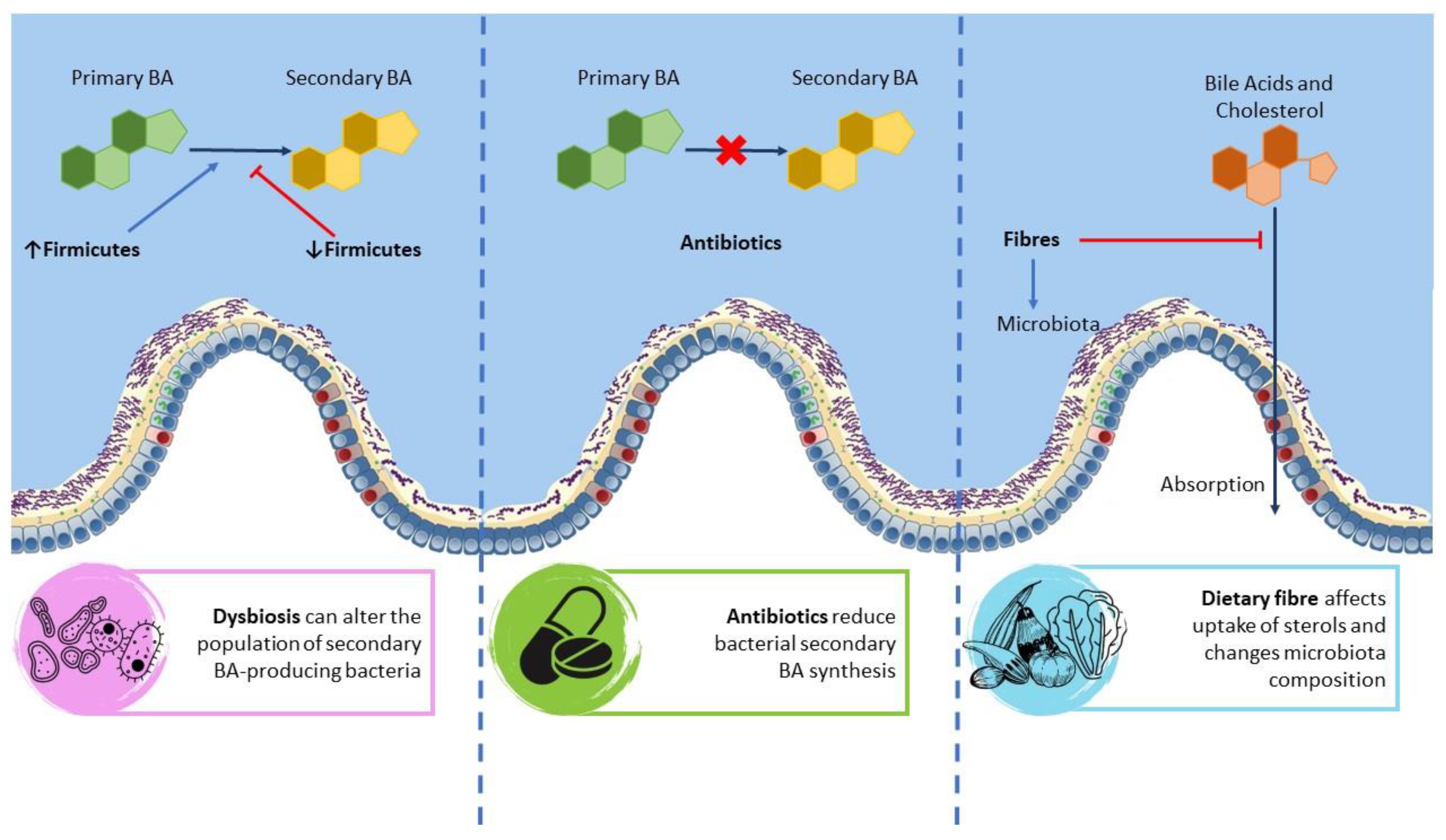
4. The Mutual Interaction between BA and Microbiota
4.1. Gut Microbiota
4.2. Biliary Microbiota
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Valle, J.W.; Kelley, R.K.; Nervi, B.; Oh, D.Y.; Zhu, A.X. Biliary tract cancer. Lancet 2021, 397, 428–444. [Google Scholar] [CrossRef]
- Banales, J.M.; Marin, J.J.G.; Lamarca, A.; Rodrigues, P.M.; Khan, S.A.; Roberts, L.R.; Cardinale, V.; Carpino, G.; Andersen, J.B.; Braconi, C.; et al. Cholangiocarcinoma 2020: The next horizon in mechanisms and management. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 557–588. [Google Scholar] [CrossRef]
- Rizvi, S.; Khan, S.A.; Hallemeier, C.L.; Kelley, R.K.; Gores, G.J. Cholangiocarcinoma—Evolving concepts and therapeutic strategies. Nat. Rev. Clin. Oncol. 2018, 15, 95–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Izquierdo-Sanchez, L.; Lamarca, A.; La Casta, A.; Buettner, S.; Utpatel, K.; Klümpen, H.J.; Adeva, J.; Vogel, A.; Lleo, A.; Fabris, L.; et al. Cholangiocarcinoma landscape in Europe: Diagnostic, prognostic and therapeutic insights from the ENSCCA Registry. J. Hepatol. 2022, 76, 1109–1121. [Google Scholar] [CrossRef] [PubMed]
- Alabraba, E.; Joshi, H.; Bird, N.; Griffin, R.; Sturgess, R.; Stern, N.; Sieberhagen, C.; Cross, T.; Camenzuli, A.; Davis, R.; et al. Increased multimodality treatment options has improved survival for Hepatocellular carcinoma but poor survival for biliary tract cancers remains unchanged. Eur. J. Surg. Oncol. 2019, 45, 1660–1667. [Google Scholar] [CrossRef] [PubMed]
- Cook, J.W. Cancer-Producing Chemical Compounds. Nature 1940, 145, 335–338. [Google Scholar] [CrossRef] [Green Version]
- Volle, D.H. Bile acids, roles in integrative physiology and pathophysiology. Mol. Aspects Med. 2017, 56, 1. [Google Scholar] [CrossRef]
- Chiang, J.Y. Bile acids: Regulation of synthesis. J. Lipid Res. 2009, 50, 1955–1966. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Chiang, J.Y. Bile acid signaling in metabolic disease and drug therapy. Pharmacol. Rev. 2014, 66, 948–983. [Google Scholar] [CrossRef] [Green Version]
- Chiang, J.Y. Regulation of bile acid synthesis: Pathways, nuclear receptors, and mechanisms. J. Hepatol. 2004, 40, 539–551. [Google Scholar] [CrossRef]
- Russell, D.W. The enzymes, regulation, and genetics of bile acid synthesis. Annu. Rev. Biochem. 2003, 72, 137–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keppler, D. Progress in the Molecular Characterization of Hepatobiliary Transporters. Dig Dis. 2017, 35, 197–202. [Google Scholar] [CrossRef]
- Dawson, P.A.; Lan, T.; Rao, A. Bile acid transporters. J. Lipid Res. 2009, 50, 2340–2357. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Apte, U. Bile Acid Metabolism and Signaling in Cholestasis, Inflammation, and Cancer. Adv. Pharmacol. 2015, 74, 263–302. [Google Scholar]
- Nguyen, J.T.; Shaw, R.P.H.; Anakk, S. Bile Acids-A Peek into Their History and Signaling. Endocrinology 2022, 163, bqac155. [Google Scholar] [CrossRef] [PubMed]
- Režen, T.; Rozman, D.; Kovács, T.; Kovács, P.; Sipos, A.; Bai, P.; Mikó, E. The role of bile acids in carcinogenesis. Cell Mol. Life Sci. 2022, 79, 243. [Google Scholar] [CrossRef] [PubMed]
- Baiocchi, L.; Zhou, T.; Liangpunsakul, S.; Lenci, I.; Santopaolo, F.; Meng, F.; Kennedy, L.; Glaser, S.; Francis, H.; Alpini, G. Dual Role of Bile Acids on the Biliary Epithelium: Friend or Foe? Int. J. Mol. Sci. 2019, 20, 1869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Chen, F.; Shang, Q.; Pan, L.; Shneider, B.L.; Chiang, J.Y.; Forman, B.M.; Ananthanarayanan, M.; Tint, G.S.; Salen, G.; et al. FXR-activating ligands inhibit rabbit ASBT expression via FXR-SHP-FTF cascade. Am. J. Physiol. Gastrointest. Liver Physiol. 2005, 288, G60–G66. [Google Scholar] [CrossRef] [Green Version]
- Fu, T.; Kim, Y.C.; Byun, S.; Kim, D.H.; Seok, S.; Suino-Powell, K.; Xu, H.E.; Kemper, B.; Kemper, J.K. FXR Primes the Liver for Intestinal FGF15 Signaling by Transient Induction of β-Klotho. Mol. Endocrinol. 2016, 30, 92–103. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Chiang, J.Y. Mechanism of rifampicin and pregnane X receptor inhibition of human cholesterol 7 alpha-hydroxylase gene transcription. Am. J. Physiol. Gastrointest. Liver Physiol. 2005, 288, G74–G84. [Google Scholar] [CrossRef] [Green Version]
- Norman, A.W. Minireview: Vitamin D receptor: New assignments for an already busy receptor. Endocrinology 2006, 147, 5542–5548. [Google Scholar] [CrossRef] [Green Version]
- Masyuk, A.I.; Huang, B.Q.; Radtke, B.N.; Gajdos, G.B.; Splinter, P.L.; Masyuk, T.V.; Gradilone, S.A.; LaRusso, N.F. Ciliary subcellular localization of TGR5 determines the cholangiocyte functional response to bile acid signaling. Am. J. Physiol. Gastrointest. Liver Physiol. 2013, 304, G1013–G1024. [Google Scholar] [CrossRef] [Green Version]
- Reich, M.; Deutschmann, K.; Sommerfeld, A.; Klindt, C.; Kluge, S.; Kubitz, R.; Ullmer, C.; Knoefel, W.T.; Herebian, D.; Mayatepek, E.; et al. GR5 is essential for bile acid-dependent cholangiocyte proliferation in vivo and in vitro. Gut 2016, 65, 487–501. [Google Scholar] [CrossRef]
- Nagahashi, M.; Yuza, K.; Hirose, Y.; Nakajima, M.; Ramanathan, R.; Hait, N.C.; Hylemon, P.B.; Zhou, H.; Takabe, K.; Wakai, T. The roles of bile acids and sphingosine-1-phosphate signaling in the hepatobiliary diseases. J. Lipid Res. 2016, 57, 1636–1643. [Google Scholar] [CrossRef] [Green Version]
- Studer, E.; Zhou, X.; Zhao, R.; Wang, Y.; Takabe, K.; Nagahashi, M.; Pandak, W.M.; Dent, P.; Spiegel, S.; Shi, R.; et al. Conjugated bile acids activate the sphingosine-1-phosphate receptor 2 in primary rodent hepatocytes. Hepatology 2012, 55, 267–276. [Google Scholar] [CrossRef] [Green Version]
- Lozano, E.; Sanchez-Vicente, L.; Monte, M.J.; Herraez, E.; Briz, O.; Banales, J.M.; Marin, J.J.; Macias, R.I. Cocarcinogenic effects of intrahepatic bile acid accumulation in cholangiocarcinoma development. Mol. Cancer Res. 2014, 12, 91–100. [Google Scholar] [CrossRef] [Green Version]
- Hashim Abdalla, M.S.; Taylor-Robinson, S.D.; Sharif, A.W.; Williams, H.R.; Crossey, M.M.; Badra, G.A.; Thillainayagam, A.V.; Bansi, D.S.; Thomas, H.C.; Waked, I.A.; et al. Differences in phosphatidylcholine and bile acids in bile from Egyptian and UK patients with and without cholangiocarcinoma. HPB 2011, 13, 385–390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharif, A.W.; Williams, H.R.; Lampejo, T.; Khan, S.A.; Bansi, D.S.; Westaby, D.; Thillainayagam, A.V.; Thomas, H.C.; Cox, I.J.; Taylor-Robinson, S.D. Metabolic profiling of bile in cholangiocarcinoma using in vitro magnetic resonance spectroscopy. HPB 2010, 12, 396–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Yang, Z.; Shi, Z.; Zhu, Z.; Li, C.; Du, Z.; Zhang, Y.; Wang, Z.; Jiao, Z.; Tian, X.; et al. Analysis of bile acid profile in plasma to differentiate cholangiocarcinoma from benign biliary diseases and healthy controls. J. Steroid Biochem. Mol. Biol. 2021, 205, 105775. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Tian, S.L.; Jin, D.; Liu, B.; Wang, W.; Chang, H.; Chen, C.; Yu, Z.; Wang, Y.; Li, Y. The role of bile acid subtypes in the diagnosis of cholangiocarcinoma. Asia Pac. J. Clin. Oncol. 2022, 18, e163–e172. [Google Scholar] [CrossRef] [PubMed]
- Farhat, Z.; Freedman, N.D.; Sampson, J.N.; Falk, R.T.; Koshiol, J.; Weinstein, S.J.; Albanes, D.; Sinha, R.; Loftfield, E. A prospective investigation of serum bile acids with risk of liver cancer, fatal liver disease, and biliary tract cancer. Hepatol. Commun. 2022, 6, 2391–2399. [Google Scholar] [CrossRef]
- Di Ciaula, A.; Wang, D.Q.; Molina-Molina, E.; Lunardi Baccetto, R.; Calamita, G.; Palmieri, V.O.; Portincasa, P. Bile Acids and Cancer: Direct and Environmental-Dependent Effects. Ann. Hepatol. 2017, 16, s87–s105. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Chiang, J.Y. Regulation of bile acid and cholesterol metabolism by PPARs. PPAR Res. 2009, 2009, 501739. [Google Scholar] [CrossRef] [Green Version]
- Fujino, T.; Takeuchi, A.; Maruko-Ohtake, A.; Ohtake, Y.; Satoh, J.; Kobayashi, T.; Tanaka, T.; Ito, H.; Sakamaki, R.; Kashimura, R.; et al. Critical role of farnesoid X receptor for hepatocellular carcinoma cell proliferation. J. Biochem. 2012, 152, 577–586. [Google Scholar] [CrossRef]
- Liu, R.; Zhao, R.; Zhou, X.; Liang, X.; Campbell, D.J.; Zhang, X.; Zhang, L.; Shi, R.; Wang, G.; Pandak, W.M.; et al. Conjugated bile acids promote cholangiocarcinoma cell invasive growth through activation of sphingosine 1-phosphate receptor 2. Hepatology 2014, 60, 908–918. [Google Scholar] [CrossRef]
- Erice, O.; Labiano, I.; Arbelaiz, A.; Santos-Laso, A.; Munoz-Garrido, P.; Jimenez-Agüero, R.; Olaizola, P.; Caro-Maldonado, A.; Martín-Martín, N.; Carracedo, A.; et al. Differential effects of FXR or TGR5 activation in cholangiocarcinoma progression. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864 Pt B, 1335–1344. [Google Scholar] [CrossRef]
- Trauner, M. The nuclear bile acid receptor FXR as a novel therapeutic target in cholestatic liver diseases: Hype or hope? Hepatology 2004, 40, 260–263. [Google Scholar] [CrossRef]
- Vaquero, J.; Monte, M.J.; Dominguez, M.; Muntané, J.; Marin, J.J. Differential activation of the human farnesoid X receptor depends on the pattern of expressed isoforms and the bile acid pool composition. Biochem. Pharmacol. 2013, 86, 926–939. [Google Scholar] [CrossRef]
- Huang, X.F.; Zhao, W.Y.; Huang, W.D. FXR and liver carcinogenesis. Acta Pharmacol. Sin. 2015, 36, 37–43. [Google Scholar] [CrossRef] [Green Version]
- Dai, J.; Wang, H.; Shi, Y.; Dong, Y.; Zhang, Y.; Wang, J. Impact of bile acids on the growth of human cholangiocarcinoma via FXR. J. Hematol. Oncol. 2011, 4, 41. [Google Scholar] [CrossRef] [Green Version]
- Kim, I.; Ahn, S.H.; Inagaki, T.; Choi, M.; Ito, S.; Guo, G.L.; Kliewer, S.A.; Gonzalez, F.J. Differential regulation of bile acid homeostasis by the farnesoid X receptor in liver and intestine. J. Lipid Res. 2007, 48, 2664–2672. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.D.; Chen, W.D.; Wang, M.; Yu, D.; Forman, B.M.; Huang, W. Farnesoid X receptor antagonizes nuclear factor kappaB in hepatic inflammatory response. Hepatology 2008, 48, 1632–1643. [Google Scholar] [CrossRef] [Green Version]
- Bernstein, H.; Bernstein, C.; Payne, C.M.; Dvorak, K. Bile acids as endogenous etiologic agents in gastrointestinal cancer. World J. Gastroenterol. 2009, 15, 3329–3340. [Google Scholar] [CrossRef]
- Dai, J.; Wang, H.; Dong, Y.; Zhang, Y.; Wang, J. Bile acids affect the growth of human cholangiocarcinoma via NF-kB pathway. Cancer Investig. 2013, 31, 111–120. [Google Scholar] [CrossRef]
- García-Rodríguez, J.L.; Barbier-Torres, L.; Fernández-Álvarez, S.; Gutiérrez-de Juan, V.; Monte, M.J.; Halilbasic, E.; Herranz, D.; Álvarez, L.; Aspichueta, P.; Marín, J.J.; et al. SIRT1 controls liver regeneration by regulating bile acid metabolism through farnesoid X receptor and mammalian target of rapamycin signaling. Hepatology 2014, 59, 1972–1983. [Google Scholar] [CrossRef] [Green Version]
- Yang, F.; Hu, Y.; Liu, H.X.; Wan, Y.J. MiR-22-silenced cyclin A expression in colon and liver cancer cells is regulated by bile acid receptor. J. Biol. Chem. 2015, 290, 6507–6515. [Google Scholar] [CrossRef] [Green Version]
- Wu, W.; Zhu, B.; Peng, X.; Zhou, M.; Jia, D.; Gu, J. Activation of farnesoid X receptor attenuates hepatic injury in a murine model of alcoholic liver disease. Biochem. Biophys. Res. Commun. 2014, 443, 68–73. [Google Scholar] [CrossRef]
- Xie, G.; Zhong, W.; Li, H.; Li, Q.; Qiu, Y.; Zheng, X.; Chen, H.; Zhao, X.; Zhang, S.; Zhou, Z.; et al. Alteration of bile acid metabolism in the rat induced by chronic ethanol consumption. FASEB J. 2013, 27, 3583–3593. [Google Scholar] [CrossRef] [Green Version]
- Osawa, Y.; Hannun, Y.A.; Proia, R.L.; Brenner, D.A. Roles of AKT and sphingosine kinase in the antiapoptotic effects of bile duct ligation in mouse liver. Hepatology 2005, 42, 1320–1328. [Google Scholar] [CrossRef]
- Dumur, C.I.; Campbell, D.J.; DeWitt, J.L.; Oyesanya, R.A.; Sirica, A.E. Differential gene expression profiling of cultured neu-transformed versus spontaneously-transformed rat cholangiocytes and of corresponding cholangiocarcinomas. Exp. Mol. Pathol. 2010, 89, 227–235. [Google Scholar] [CrossRef] [Green Version]
- Spiegel, S.; Milstien, S. Sphingosine-1-phosphate: An enigmatic signalling lipid. Nat. Rev. Mol. Cell Biol. 2003, 4, 397–407. [Google Scholar] [CrossRef]
- Raggi, C.; Taddei, M.L.; Rae, C.; Braconi, C.; Marra, F. Metabolic reprogramming in cholangiocarcinoma. J. Hepatol. 2022, 77, 849–864. [Google Scholar] [CrossRef]
- Song, W.S.; Park, H.M.; Ha, J.M.; Shin, S.G.; Park, H.G.; Kim, J.; Zhang, T.; Ahn, D.-H.; Kim, S.-M.; Yang, Y.-H.; et al. Discovery of glycocholic acid and taurochenodeoxycholic acid as phenotypic biomarkers in cholangiocarcinoma. Sci. Rep. 2018, 8, 11088. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Aoki, H.; Yang, J.; Peng, K.; Liu, R.; Li, X.; Qiang, X.; Sun, L.; Gurley, E.C.; Lai, G.; et al. The role of sphingosine 1-phosphate receptor 2 in bile-acid-induced cholangiocyte proliferation and cholestasis-induced liver injury in mice. Hepatology 2017, 65, 2005–2018. [Google Scholar] [CrossRef] [Green Version]
- Keitel, V.; Reich, M.; Häussinger, D. TGR5: Pathogenetic role and/or therapeutic target in fibrosing cholangitis? Clin. Rev. Allergy Immunol. 2015, 48, 218–225. [Google Scholar] [CrossRef]
- Beuers, U.; Hohenester, S.; de Buy Wenniger, L.J.; Kremer, A.E.; Jansen, P.L.; Elferink, R.P. The biliary HCO3− umbrella: A unifying hypothesis on pathogenetic and therapeutic aspects of fibrosing cholangiopathies. Hepatology 2010, 52, 1489–1496. [Google Scholar] [CrossRef]
- Di Tommaso, N.; Gasbarrini, A.; Ponziani, F.R. Intestinal Barrier in Human Health and Disease. Int. J. Environ. Res. Public Health 2021, 18, 12836. [Google Scholar] [CrossRef]
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [Green Version]
- Schroeder, B.O.; Bäckhed, F. Signals from the gut microbiota to distant organs in physiology and disease. Nat. Med. 2016, 22, 1079–1089. [Google Scholar] [CrossRef]
- Gopalakrishnan, V.; Helmink, B.A.; Spencer, C.N.; Reuben, A.; Wargo, J.A. The Influence of the Gut Microbiome on Cancer, Immunity, and Cancer Immunotherapy. Cancer Cell 2018, 33, 570–580. [Google Scholar] [CrossRef] [Green Version]
- Fu, Z.D.; Cui, J.Y. Remote Sensing between Liver and Intestine: Importance of Microbial Metabolites. Curr. Pharmacol. Rep. 2017, 3, 101–113. [Google Scholar] [CrossRef]
- Jia, W.; Xie, G.; Jia, W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 111–128. [Google Scholar] [CrossRef] [Green Version]
- Ma, C.; Han, M.; Heinrich, B.; Fu, Q.; Zhang, Q.; Sandhu, M.; Agdashian, D.; Terabe, M.; Berzofsky, J.A.; Fako, V.; et al. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science 2018, 360, eaan5931. [Google Scholar] [CrossRef] [Green Version]
- Guzior, D.V.; Quinn, R.A. Review: Microbial transformations of human bile acids. Microbiome 2021, 9, 140. [Google Scholar] [CrossRef]
- Islam, K.B.; Fukiya, S.; Hagio, M.; Fujii, N.; Ishizuka, S.; Ooka, T.; Ogura, Y.; Hayashi, T.; Yokota, A. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology 2011, 141, 1773–1781. [Google Scholar] [CrossRef]
- Collins, S.L.; Stine, J.G.; Bisanz, J.E.; Okafor, C.D.; Patterson, A.D. Bile acids and the gut microbiota: Metabolic interactions and impacts on disease. Nat. Rev. Microbiol. 2022, 21, 236–247. [Google Scholar] [CrossRef]
- Jia, B.; Jeon, C.O. Promotion and induction of liver cancer by gut microbiome mediated modulation of bile acids. PLoS Pathog. 2019, 15, e1007954. [Google Scholar] [CrossRef]
- Loo, T.M.; Kamachi, F.; Watanabe, Y.; Yoshimoto, S.; Kanda, H.; Arai, Y.; Nakajima-Takagi, Y.; Iwama, A.; Koga, T.; Sugimoto, Y.; et al. Gut Microbiota Promotes Obesity-Associated Liver Cancer through PGE2-Mediated Suppression of Antitumor Immunity. Cancer Discov. 2017, 7, 522–538. [Google Scholar] [CrossRef] [Green Version]
- Ito, Z.; Koido, S.; Kato, K.; Odamaki, T.; Horiuchi, S.; Akasu, T.; Saruta, M.; Hata, T.; Kumagai, Y.; Fujioka, S.; et al. Dysbiosis of the Fecal and Biliary Microbiota in Biliary Tract Cancer. Cancers 2022, 14, 5379. [Google Scholar] [CrossRef]
- Jia, X.; Lu, S.; Zeng, Z.; Liu, Q.; Dong, Z.; Chen, Y.; Zhu, Z.; Hong, Z.; Zhang, T.; Du, G.; et al. Characterization of Gut Microbiota, Bile Acid Metabolism, and Cytokines in Intrahepatic Cholangiocarcinoma. Hepatology 2020, 71, 893–906. [Google Scholar] [CrossRef]
- Scott, A.J. Bacteria and disease of the biliary tract. Gut 1971, 12, 487–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nielsen, M.L.; Justesen, T. Anaerobic and aerobic bacteriological studies in biliary tract disease. Scand. J. Gastroenterol. 1976, 11, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Molinero, N.; Ruiz, L.; Milani, C.; Gutierrez-Diaz, I.; Sanchez, B.; Mangifesta, M.; Segura, J.; Cambero, I.; Campelo, A.B.; Garcia-Bernardo, C.M.; et al. The human gallbladder microbiome is related to the physiological state and the biliary metabolic profile. Microbiome 2019, 7, 100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mattos, V.C.; Nascimento, F.S.D.; Suzuki, M.O.; Taba, J.V.; Pipek, L.Z.; Moraes, W.A.F.; Cortez, V.S.; Kubrusly, M.S.; Torsani, M.B.; Iuamoto, L.; et al. MICRObiota on BILIOpancreatic malignant diseases [MICROBILIO]: A systematic review. Clinics 2022, 77, 100101. [Google Scholar] [CrossRef]
- Saab, M.; Mestivier, D.; Sohrabi, M.; Rodriguez, C.; Khonsari, M.R.; Faraji, A.; Sobhani, I. Characterization of biliary microbiota dysbiosis in extrahepatic cholangiocarcinoma. PLoS ONE 2021, 16, e0247798. [Google Scholar] [CrossRef]
- Miyabe, K.; Chandrasekhara, V.; Wongjarupong, N.; Chen, J.; Yang, L.; Johnson, S.; Chia, N.; Walther-Antonio, M.; Yao, J.Z.; Harrington, S.C.; et al. Potential Role of Inflammation-Promoting Biliary Microbiome in Primary Sclerosing Cholangitis and Cholangiocarcinoma. Cancers 2022, 14, 2120. [Google Scholar] [CrossRef]
- Okuda, S.; Hirose, Y.; Takihara, H.; Okuda, A.; Ling, Y.; Tajima, Y.; Shimada, Y.; Ichikawa, H.; Takizawa, K.; Sakata, J.; et al. Unveiling microbiome profiles in human inner body fluids and tumor tissues with pancreatic or biliary tract cancer. Sci. Rep. 2022, 12, 8766. [Google Scholar] [CrossRef]
- Avilés-Jiménez, F.; Guitron, A.; Segura-López, F.; Méndez-Tenorio, A.; Iwai, S.; Hernández-Guerrero, A.; Torres, J. Microbiota studies in the bile duct strongly suggest a role for Helicobacter pylori in extrahepatic cholangiocarcinoma. Clin. Microbiol. Infect. 2016, 22, 178.e11–178.e22. [Google Scholar] [CrossRef] [Green Version]
- Zhou, D.; Wang, J.D.; Weng, M.Z.; Zhang, Y.; Wang, X.F.; Gong, W.; Quan, Z.W. Infections of Helicobacter spp. in the biliary system are associated with biliary tract cancer: A meta-analysis. Eur. J. Gastroenterol. Hepatol. 2013, 25, 447–454. [Google Scholar] [CrossRef]
- Di Carlo, P.; Serra, N.; D’Arpa, F.; Agrusa, A.; Gulotta, G.; Fasciana, T.; Rodolico, V.; Giammanco, A.; Sergi, C. The microbiota of the bilio-pancreatic system: A cohort, STROBE-compliant study. Infect. Drug Resist. 2019, 12, 1513–1527. [Google Scholar] [CrossRef] [Green Version]
| Author, Year | Biological Specimens | Sampling Methods | Conclusions |
|---|---|---|---|
| Healthy | |||
| Molinero N, 2019 [73] | Bile and gallbladder tissue | Sterile sampling during liver transplants from liver donors | Bile samples and gallbladder tissues of individuals without gallstones are rich in Actinobacteria, Firmicutes, and Bacteroidetes as well as abundant in the Propionibacteriaceae family. |
| Cancer | |||
| Saab M, 2019 [75] | Bile | ERCP | At phyla level, Firmicutes, Fusobacteria, and Cianobacteria levels were significantly lower in CCA than in the controls. |
| Ito Z, 2022 [69] | Bile and feces | ERCP or surgery | Bile of patients with cancer was enriched in Enterobacteriaceae compared to that in patients with benign biliary disease. |
| Miyabe K, 2022 [76] | Bile and feces | ERCP or surgery | The microbiota of the bile and stool samples from the same subjects were correlated. CCA bile was characterized by increased abundance of Firmicutes, Fusobacteria, and Actinobacteria compared to control bile samples. Fusobacteria and Firmicutes were also increased with increasing PSC duration in bile from patients with PSC. |
| Okuda S, 2022 [77] | Saliva, gastric and pancreatic juice, bile, feces, tumor and nontumor tissue | Surgery | Akkermansia was the most abundant, and it was present only in bile samples. Akkermansia was detected in 7/11 patients with biliary tract cancer and in none of the 4 patients with pancreatic cancer. |
| Aviles-Jimenez F, 2016 [78] | Epithelial cells from the biliary duct | Brushing during ERCP | In extrahepatic cholangiocarcinoma, the microbiota showed significant changes in microbial composition, with a predominant presence of Phylum Proteobacteria in all samples. Fusobacterium, Prevotella, Actinomyces, Novosphingobium, and H. pylori increased in patients with CCAs. |
| Zhou D, 2013 [79] | Serum and bile | ERCP | The infection rate of Helicobacter spp was significantly higher in CCA compared to in the control and benign biliary pathology groups. |
| Di Carlo P, 2019 [80] | Bile | ERCP | E. coli, Pseudomonas aeruginosa, and Klebisiella pneumonas were identified in the cancer and were associated with reduced survival time. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cossiga, V.; Guarino, M.; Capasso, M.; Morisco, F. Relevance of Bile Acids in Cholangiocarcinoma Pathogenesis: Critical Revision and Future Directions. Cells 2023, 12, 1576. https://doi.org/10.3390/cells12121576
Cossiga V, Guarino M, Capasso M, Morisco F. Relevance of Bile Acids in Cholangiocarcinoma Pathogenesis: Critical Revision and Future Directions. Cells. 2023; 12(12):1576. https://doi.org/10.3390/cells12121576
Chicago/Turabian StyleCossiga, Valentina, Maria Guarino, Mario Capasso, and Filomena Morisco. 2023. "Relevance of Bile Acids in Cholangiocarcinoma Pathogenesis: Critical Revision and Future Directions" Cells 12, no. 12: 1576. https://doi.org/10.3390/cells12121576
APA StyleCossiga, V., Guarino, M., Capasso, M., & Morisco, F. (2023). Relevance of Bile Acids in Cholangiocarcinoma Pathogenesis: Critical Revision and Future Directions. Cells, 12(12), 1576. https://doi.org/10.3390/cells12121576








