Eupatilin Improves Cilia Defects in Human CEP290 Ciliopathy Models
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. qPCR
2.3. Western Blot
2.4. Cell Culture and Eupatilin Treatment
2.5. Immunocytochemistry
2.6. iPSC Differentiation to Retinal Organoids
2.7. Cryopreservation and Immunohistochemistry of Retinal Organoids
2.8. Cilia Quantification and Statistical Analysis
2.9. RNAseq Pre-Processing and Analysis
3. Results
3.1. Eupatilin Increases Cilia Incidence and Length in CEP290 LCA10 Patient Fibroblasts
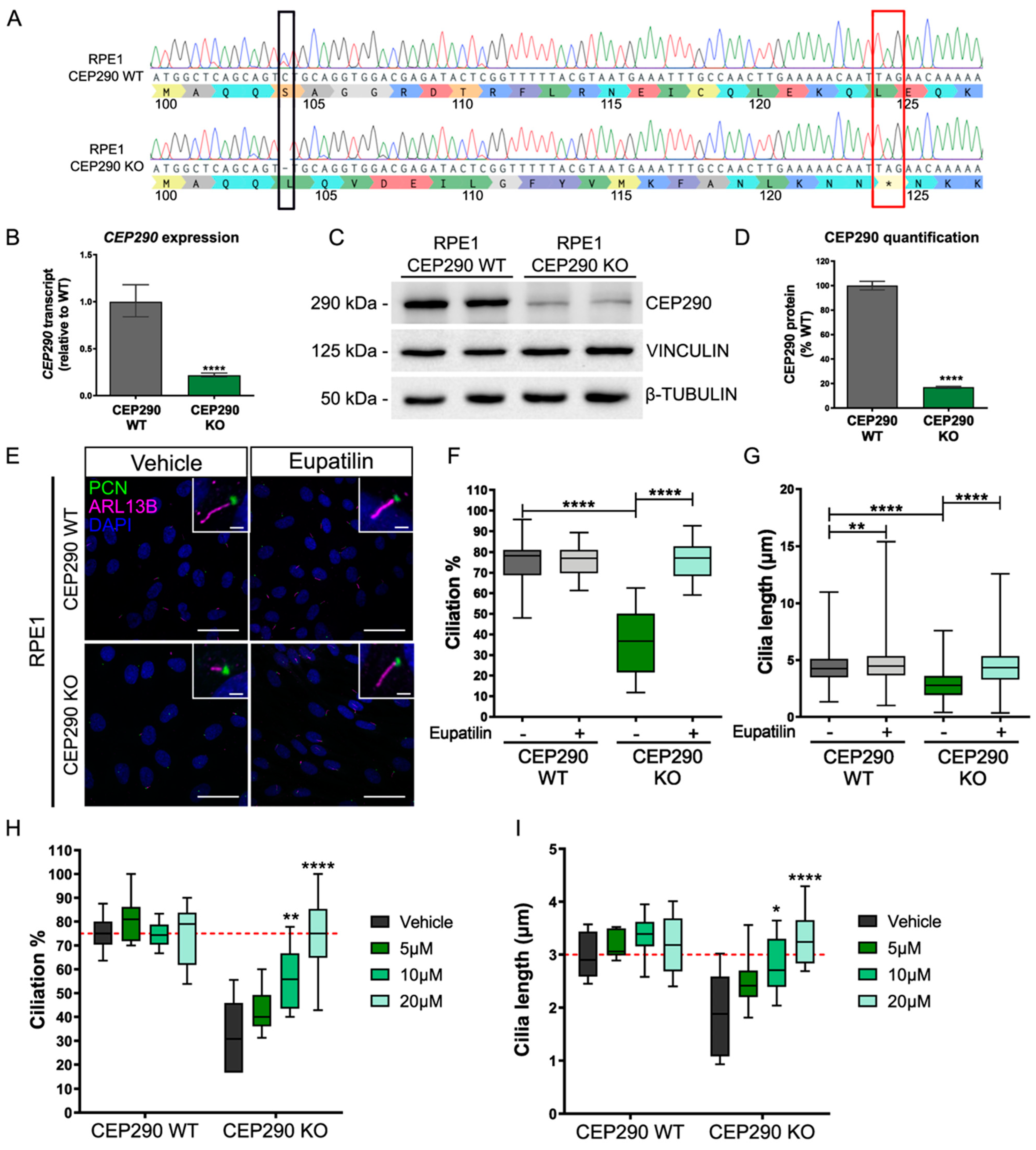
3.2. Eupatilin Restores Cilia Incidence and Length in CEP290 Knockout RPE1 Cells
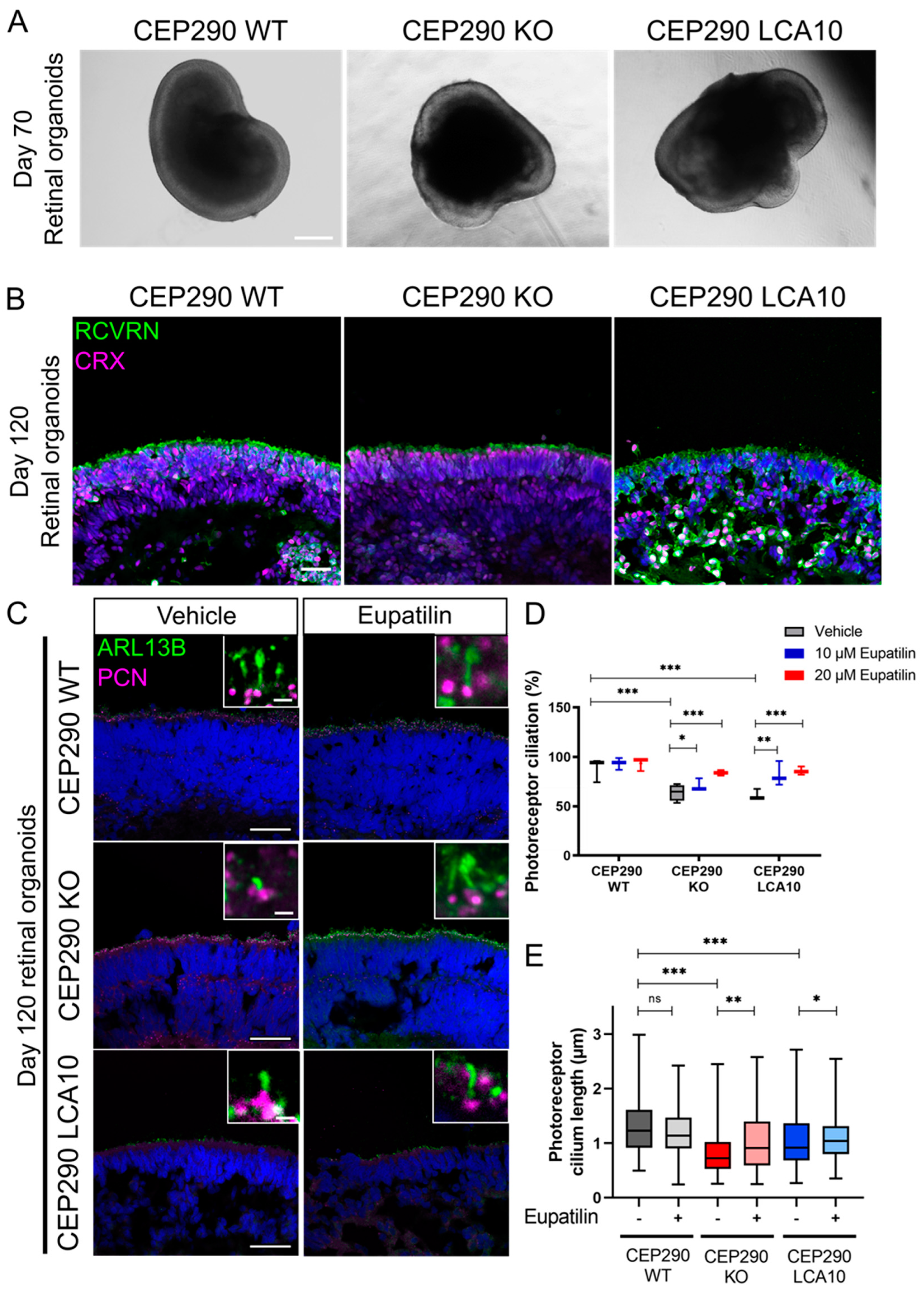
3.3. Eupatilin Treatment Restores Ciliation and Cilia Length in iPSC-Derived CEP290 LCA10 and CEP290 KO Retinal Organoids
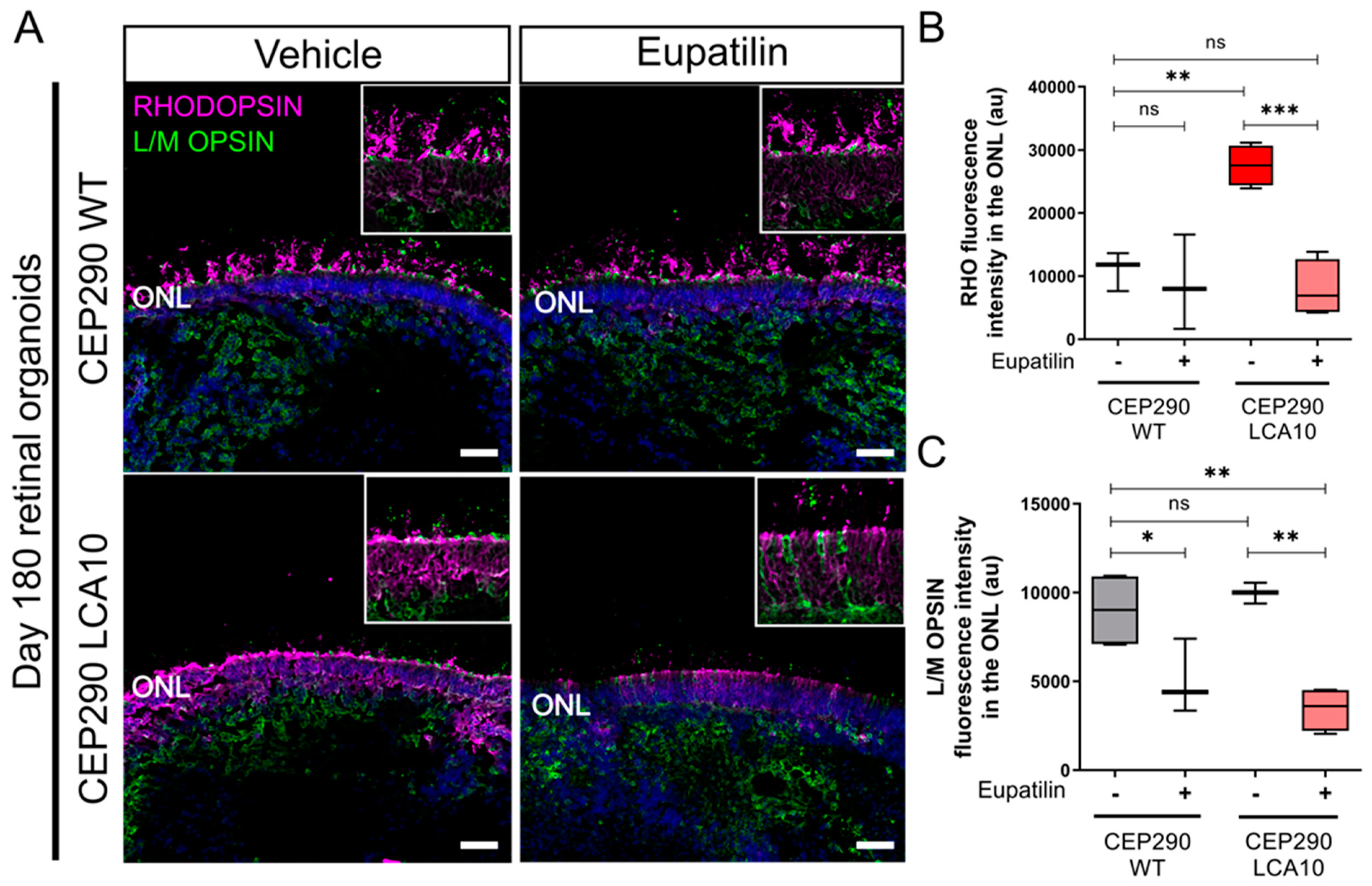
3.4. Opsin Accumulates in the Outer Nuclear Layer of CEP290 LCA10 Retinal Organoids and Is Rescued by Eupatilin
3.5. Eupatilin Treatment Reduces the Expression of Rhodopsin
3.6. Eupatilin Modulates the Expression of Ciliary and Synaptic Pathways
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sayer, J.A.; Otto, E.A.; O’Toole, J.F.; Nurnberg, G.; Kennedy, M.A.; Becker, C.; Hennies, H.C.; Helou, J.; Attanasio, M.; Fausett, B.V.; et al. The Centrosomal Protein Nephrocystin-6 Is Mutated in Joubert Syndrome and Activates Transcription Factor ATF4. Nat. Genet. 2006, 38, 674–681. [Google Scholar] [CrossRef] [PubMed]
- Valente, E.M.; Silhavy, J.L.; Brancati, F.; Barrano, G.; Krishnaswami, S.R.; Castori, M.; Lancaster, M.A.; Boltshauser, E.; Boccone, L.; Al-Gazali, L.; et al. Mutations in CEP290, Which Encodes a Centrosomal Protein, Cause Pleiotropic Forms of Joubert Syndrome. Nat. Genet. 2006, 38, 623–625. [Google Scholar] [CrossRef]
- Rachel, R.A.; Li, T.; Swaroop, A. Photoreceptor Sensory Cilia and Ciliopathies: Focus on CEP290, RPGR and Their Interacting Proteins. Cilia 2012, 1, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- den Hollander, A.I.; Koenekoop, R.K.; Yzer, S.; Lopez, I.; Arends, M.L.; Voesenek, K.E.J.; Zonneveld, M.N.; Strom, T.M.; Meitinger, T.; Brunner, H.G.; et al. Mutations in the CEP290 (NPHP6) Gene Are a Frequent Cause of Leber Congenital Amaurosis. Am. J. Hum. Genet. 2006, 79, 556–561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coppieters, F.; Lefever, S.; Leroy, B.P.; de Baere, E. CEP290, a Gene with Many Faces: Mutation Overview and Presentation of CEP290base. Hum. Mutat. 2010, 31, 1097–1108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garanto, A.; Chung, D.C.; Duijkers, L.; Corral-Serrano, J.C.; Messchaert, M.; Xiao, R.; Bennett, J.; Vandenberghe, L.H.; Collin, R.W.J. In Vitro and in Vivo Rescue of Aberrant Splicing in CEP290-Associated LCA by Antisense Oligonucleotide Delivery. Hum. Mol. Genet. 2016, 25, 2552–2563. [Google Scholar] [CrossRef] [Green Version]
- Maeder, M.L.; Stefanidakis, M.; Wilson, C.J.; Baral, R.; Barrera, L.A.; Bounoutas, G.S.; Bumcrot, D.; Chao, H.; Ciulla, D.M.; DaSilva, J.A.; et al. Development of a Gene-Editing Approach to Restore Vision Loss in Leber Congenital Amaurosis Type 10. Nat. Med. 2019, 25, 229–233. [Google Scholar] [CrossRef]
- Dulla, K.; Aguila, M.; Lane, A.; Jovanovic, K.; Parfitt, D.A.; Schulkens, I.; Chan, H.L.; Schmidt, I.; Beumer, W.; Vorthoren, L.; et al. Splice-Modulating Oligonucleotide QR-110 Restores CEP290 MRNA and Function in Human c.2991+1655A>G LCA10 Models. Mol. Ther. Nucleic Acids 2018, 12, 730–740. [Google Scholar] [CrossRef] [Green Version]
- Russell, S.R.; Drack, A.V.; Cideciyan, A.V.; Jacobson, S.G.; Leroy, B.P.; van Cauwenbergh, C.; Ho, A.C.; Dumitrescu, A.V.; Han, I.C.; Martin, M.; et al. Intravitreal Antisense Oligonucleotide Sepofarsen in Leber Congenital Amaurosis Type 10: A Phase 1b/2 Trial. Nat. Med. 2022, 28, 1014–1021. [Google Scholar] [CrossRef]
- Cideciyan, A.V.; Jacobson, S.G.; Ho, A.C.; Garafalo, A.V.; Roman, A.J.; Sumaroka, A.; Krishnan, A.K.; Swider, M.; Schwartz, M.R.; Girach, A. Durable Vision Improvement after a Single Treatment with Antisense Oligonucleotide Sepofarsen: A Case Report. Nat. Med. 2021, 27, 785–789. [Google Scholar] [CrossRef]
- Cideciyan, A.V.; Jacobson, S.G.; Drack, A.V.; Ho, A.C.; Charng, J.; Garafalo, A.V.; Roman, A.J.; Sumaroka, A.; Han, I.C.; Hochstedler, M.D.; et al. Effect of an Intravitreal Antisense Oligonucleotide on Vision in Leber Congenital Amaurosis Due to a Photoreceptor Cilium Defect. Nat. Med. 2019, 25, 225–228. [Google Scholar] [CrossRef] [PubMed]
- Cideciyan, A.V.; Jacobson, S.G.; Ho, A.C.; Krishnan, A.K.; Roman, A.J.; Garafalo, A.V.; Wu, V.; Swider, M.; Sumaroka, A.; Van Cauwenbergh, C.; et al. Restoration of Cone Sensitivity to Individuals with Congenital Photoreceptor Blindness within the Phase 1/2 Sepofarsen Trial. Ophthalmol. Sci. 2022, 2, 100133. [Google Scholar] [CrossRef]
- ProQR Therapeutics, N.V. ProQR Announces Additional Sepofarsen Illuminate Trial Analyses and Provides Update on Company Strategy; ProQR: Leiden, The Netherlands, 2022. [Google Scholar]
- Kennedy, D.O. Polyphenols and the Human Brain: Plant “Secondary Metabolite” Ecologic Roles and Endogenous Signaling Functions Drive Benefits. Adv. Nutr. 2014, 5, 515–533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rendeiro, C.; Foley, A.; Lau, V.C.; Ring, R.; Rodriguez-Mateos, A.; Vauzour, D.; Williams, C.M.; Regan, C.; Spencer, J.P.E. A Role for Hippocampal PSA-NCAM and NMDA-NR2B Receptor Function in Flavonoid-Induced Spatial Memory Improvements in Young Rats. Neuropharmacology 2014, 79, 335–344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, C.M.; el Mohsen, M.A.; Vauzour, D.; Rendeiro, C.; Butler, L.T.; Ellis, J.A.; Whiteman, M.; Spencer, J.P.E. Blueberry-Induced Changes in Spatial Working Memory Correlate with Changes in Hippocampal CREB Phosphorylation and Brain-Derived Neurotrophic Factor (BDNF) Levels. Free Radic. Biol. Med. 2008, 45, 295–305. [Google Scholar] [CrossRef]
- Kim, Y.J.; Kim, S.; Jung, Y.; Jung, E.; Kwon, H.J.; Kim, J. Eupatilin Rescues Ciliary Transition Zone Defects to Ameliorate Ciliopathy-Related Phenotypes. J. Clin. Investig. 2018, 128, 3642–3648. [Google Scholar] [CrossRef] [Green Version]
- Wiegering, A.; Dildrop, R.; Vesque, C.; Khanna, H.; Schneider-Maunoury, S.; Gerhardt, C. Rpgrip1l Controls Ciliary Gating by Ensuring the Proper Amount of CEP290 at the Vertebrate Transition Zone. Mol. Biol. Cell. 2021, 32, 675–689. [Google Scholar] [CrossRef]
- Garcia-Gonzalo, F.R.; Corbit, K.C.; Sirerol-Piquer, M.S.; Ramaswami, G.; Otto, E.A.; Noriega, T.R.; Seol, A.D.; Robinson, J.F.; Bennett, C.L.; Josifova, D.J.; et al. A Transition Zone Complex Regulates Mammalian Ciliogenesis and Ciliary Membrane Composition. Nat. Genet. 2011, 43, 776–784. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.; Pang, N.; Zhang, Y.; Chen, H.; Peng, Y.; Fu, J.; Wei, Q. CEP290 Is Essential for the Initiation of Ciliary Transition Zone Assembly. PLoS Biol. 2020, 18, e3001034. [Google Scholar] [CrossRef]
- Jensen, V.L.; Li, C.; Bowie, R.V.; Clarke, L.; Mohan, S.; Blacque, O.E.; Leroux, M.R. Formation of the Transition Zone by Mks5/Rpgrip1L Establishes a Ciliary Zone of Exclusion (CIZE) That Compartmentalises Ciliary Signalling Proteins and Controls PIP2 Ciliary Abundance. EMBO J. 2015, 34, 2537–2556. [Google Scholar] [CrossRef] [Green Version]
- Ryoo, S.B.; Oh, H.K.; Yu, S.A.; Moon, S.H.; Choe, E.K.; Oh, T.Y.; Park, K.J. The Effects of Eupatilin (Stillen®) on Motility of Human Lower Gastrointestinal Tracts. Korean J. Physiol. Pharmacol. 2014, 18, 383–390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parfitt, D.A.; Lane, A.; Ramsden, C.M.; Carr, A.J.F.; Munro, P.M.; Jovanovic, K.; Schwarz, N.; Kanuga, N.; Muthiah, M.N.; Hull, S.; et al. Identification and Correction of Mechanisms Underlying Inherited Blindness in Human iPSC-Derived Optic Cups. Cell Stem Cell 2016, 18, 769–781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwarz, N.; Carr, A.J.; Lane, A.; Moeller, F.; Chen, L.L.; Aguilà, M.; Nommiste, B.; Muthiah, M.N.; Kanuga, N.; Wolfrum, U.; et al. Translational Read-through of the RP2 Arg120stop Mutation in Patient iPSC-Derived Retinal Pigment Epithelium Cells. Hum. Mol. Genet. 2015, 24, 972–986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howden, S.E.; Thomson, J.A.; Little, M.H. Simultaneous Reprogramming and Gene Editing of Human Fibroblasts. Nat. Protoc. 2018, 13, 875–898. [Google Scholar] [CrossRef]
- Sladen, P.E.; Jovanovic, K.; Guarascio, R.; Ottaviani, D.; Salsbury, G.; Novoselova, T.; Chapple, J.P.; Yu-Wai-Man, P.; Cheetham, M.E. Modelling Autosomal Dominant Optic Atrophy Associated with OPA1 Variants in iPSC-Derived Retinal Ganglion Cells. Hum. Mol. Genet. 2022, 31, 3478–3493. [Google Scholar] [CrossRef] [PubMed]
- Okita, K.; Matsumura, Y.; Sato, Y.; Okada, A.; Morizane, A.; Okamoto, S.; Hong, H.; Nakagawa, M.; Tanabe, K.; Tezuka, K.I.; et al. A More Efficient Method to Generate Integration-Free Human IPS Cells. Nat. Methods 2011, 8, 409–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corral-Serrano, J.C.; Lamers, I.J.C.; Van Reeuwijk, J.; Duijkers, L.; Hoogendoorn, A.D.M.; Yildirim, A.; Argyrou, N.; Ruigrok, R.A.A.; Letteboer, S.J.F.; Butcher, R.; et al. PCARE and WASF3 Regulate Ciliary F-Actin Assembly That Is Required for the Initiation of Photoreceptor Outer Segment Disk Formation. Proc. Natl. Acad. Sci. USA 2020, 117, 9922–9931. [Google Scholar] [CrossRef] [Green Version]
- Hansen, J.N.; Rassmann, S.; Stüven, B.; Jurisch-Yaksi, N.; Wachten, D. CiliaQ: A Simple, Open-Source Software for Automated Quantification of Ciliary Morphology and Fluorescence in 2D, 3D, and 4D Images. Eur. Phys. J. E Soft Matter 2021, 8, 44. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Zhu, A.; Ibrahim, J.G.; Love, M.I. Heavy-Tailed Prior Distributions for Sequence Count Data: Removing the Noise and Preserving Large Differences. Bioinformatics 2019, 35, 2084–2092. [Google Scholar] [CrossRef] [Green Version]
- Shimada, H.; Lu, Q.; Insinna-Kettenhofen, C.; Nagashima, K.; English, M.A.; Semler, E.M.; Mahgerefteh, J.; Cideciyan, A.V.; Li, T.; Brooks, B.P.; et al. In Vitro Modeling Using Ciliopathy-Patient-Derived Cells Reveals Distinct Cilia Dysfunctions Caused by CEP290 Mutations. Cell. Rep. 2017, 20, 384–396. [Google Scholar] [CrossRef] [Green Version]
- Hishimoto, A.; Pletnikova, O.; Lang, D.L.; Troncoso, J.C.; Egan, J.M.; Liu, Q.R. Neurexin 3 Transmembrane and Soluble Isoform Expression and Splicing Haplotype Are Associated with Neuron Inflammasome and Alzheimer’s Disease. Alzheimers Res. Ther. 2019, 11, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.-K.; Zhang, K.; Church-Kopish, J.; Huang, W.; Zhang, H.; Chen, Y.-J.; Frederick, J.M.; Baehr, W. Characterization of human GRK7 as a potential cone opsin kinase. Mol. Vis. 2001, 7, 305–313. Available online: http://www.molvis.org/molvis/v7/a43/ (accessed on 2 September 2022). [PubMed]
- Silva, J.-P.; Lelianova, V.G.; Ermolyuk, Y.S.; Vysokov, N.; Hitchen, P.G.; Berninghausen, O.; Rahman, M.A.; Zangrandi, A.; Fidalgo, S.; Tonevitsky, A.G.; et al. Latrophilin 1 and Its Endogenous Ligand Lasso/ Teneurin-2 Form a High-Affinity Transsynaptic Receptor Pair with Signaling Capabilities. Proc. Natl. Acad. Sci. USA 2011, 108, 12113–12118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramsbottom, S.A.; Molinari, E.; Srivastava, S.; Silberman, F.; Henry, C.; Alkanderi, S.; Devlin, L.A.; White, K.; Steel, D.H.; Saunier, S.; et al. Targeted Exon Skipping of a CEP290 Mutation Rescues Joubert Syndrome Phenotypes in Vitro and in a Murine Model. Proc. Natl. Acad. Sci. USA 2018, 115, 12489–12494. [Google Scholar] [CrossRef] [Green Version]
- Editas Medicine, Inc. Editas Medicine Announces Clinical Data Demonstrating Proof of Concept of EDIT-101 from Phase 1/2 BRILLIANCE Trial; Editas Medicine, Inc.: Cambridge, MA, USA, 2022. [Google Scholar]
- Davinelli, S.; Ali, S.; Scapagnini, G.; Costagliola, C. Effects of Flavonoid Supplementation on Common Eye Disorders: A Systematic Review and Meta-Analysis of Clinical Trials. Front. Nutr. 2021, 8, 651441. [Google Scholar] [CrossRef]
- Li, H.L.; Ashpole, N.E.; Navarro, I.D.; Lam, T.C.; Chan, H.L.H.; To, C.H.; Stamer, W.D.; Do, C. Baicalein Lowers Intraocular Pressure and Increases Outflow Facility in Mouse Eye. Invest. Ophthalmol. Vis. Sci. 2015, 56, 4853. [Google Scholar]
- Zhao, N.; Shi, J.; Xu, H.; Luo, Q.; Li, Q.; Liu, M. Baicalin Suppresses Glaucoma Pathogenesis by Regulating the PI3K/AKT Signaling in Vitro and in Vivo. Bioengineered 2021, 12, 10187–10198. [Google Scholar] [CrossRef] [PubMed]
- Ola, M.S.; Ahmed, M.M.; Shams, S.; Al-Rejaie, S.S. Neuroprotective Effects of Quercetin in Diabetic Rat Retina. Saudi J. Biol. Sci. 2017, 24, 1186–1194. [Google Scholar] [CrossRef]
- Ortega, J.T.; Parmar, T.; Golczak, M.; Jastrzebska, B. Protective Effects of Flavonoids in Acute Models of Light-Induced Retinal Degeneration. Mol. Pharmacol. 2021, 99, 60–77. [Google Scholar] [CrossRef]
- Jegal, K.H.; Ko, H.L.; Park, S.M.; Byun, S.H.; Kang, K.W.; Cho, I.J.; Kim, S.C. Eupatilin Induces Sestrin2-Dependent Autophagy to Prevent Oxidative Stress. Apoptosis 2016, 21, 642–656. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Yang, C.; Song, G.; Lim, W. Eupatilin Impacts on the Progression of Colon Cancer by Mitochondria Dysfunction and Oxidative Stress. Antioxidants 2021, 10, 957. [Google Scholar] [CrossRef] [PubMed]
- Huh, K.; Kwon, T.H.; Shin, U.S.; Kim, W.B.; Ahn, B.O.; Oh, T.Y.; Kim, J.A. Inhibitory Effects of DA-9601 on Ethanol-Induced Gastrohemorrhagic Lesions and Gastric Xanthine Oxidase Activity in Rats. J. Ethnopharmacol. 2003, 88, 269–273. [Google Scholar] [CrossRef]
- Kim, M.J.; Kim, D.H.; Na, H.K.; Oh, T.Y.; Shin, C.Y.; Surh, Y.J. Eupatilin, a Pharmacologically Active Flavone Derived from Artemisia Plants, Induces Apoptosis in Human Gastric Cancer (AGS) Cells. J. Environ. Pathol. Toxicol. Oncol. 2005, 24, 261–269. [Google Scholar] [CrossRef]
- Wang, Y.; Hou, H.; Li, M.; Yang, Y.; Sun, L. Anticancer Effect of Eupatilin on Glioma Cells through Inhibition of the Notch-1 Signaling Pathway. Mol. Med. Rep. 2016, 13, 1141–1146. [Google Scholar] [CrossRef] [Green Version]
- Giangaspero, A.; Ponti, C.; Pollastro, F.; Del Favero, G.; Della Loggia, R.; Tubaro, A.; Appendino, G.; Sosa, S. Topical Anti-Inflammatory Activity of Eupatilin, A Lipophilic Flavonoid from Mountain Wormwood (Artemisia umbelliformis Lam.). J. Agric. Food Chem. 2009, 57, 7726–7730. [Google Scholar] [CrossRef] [PubMed]
- Choi, E.J.; Lee, S.; Chae, J.R.; Lee, H.S.; Jun, C.D.; Kim, S.H. Eupatilin Inhibits Lipopolysaccharide-Induced Expression of Inflammatory Mediators in Macrophages. Life Sci. 2011, 88, 1121–1126. [Google Scholar] [CrossRef] [PubMed]
- Nageen, B.; Sarfraz, I.; Rasul, A.; Hussain, G.; Rukhsar, F.; Irshad, S.; Riaz, A.; Selamoglu, Z.; Ali, M. Eupatilin: A Natural Pharmacologically Active Flavone Compound with Its Wide Range Applications. J. Asian Nat. Prod. Res. 2018, 22, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Seol, S.-Y.; Kim, M.-H.; Rew, J.-S.; Choi, M.-G. A Phase III Clinical Trial of Stillen™ for Erosive Gastritis. Korean J. Gastrointest. Endosc. 2004, 28, 230–236. [Google Scholar]
- May-Simera, H.L.; Wan, Q.; Jha, B.S.; Hartford, J.; Khristov, V.; Dejene, R.; Chang, J.; Patnaik, S.; Lu, Q.; Banerjee, P.; et al. Primary Cilium-Mediated Retinal Pigment Epithelium Maturation Is Disrupted in Ciliopathy Patient Cells. Cell. Rep. 2018, 22, 189–205. [Google Scholar] [CrossRef] [Green Version]
- Ra, R.; Yamamoto, E.A.; Dewanjee, M.; Munasinghe, J.; May-Simera, H.L.; Dong, L.; Swaroop, A. CEP290 Is Required for Photoreceptor Ciliogenesis and Other Cilia Related Functions. Cilia 2012, 1, P98. [Google Scholar] [CrossRef] [Green Version]
- Potter, V.L.; Moye, A.R.; Robichaux, M.A.; Wensel, T.G. Super-Resolution Microscopy Reveals Photoreceptor-Specific Subciliary Location and Function of Ciliopathy-Associated Protein CEP290. JCI Insight 2021, 6, e145256. [Google Scholar] [CrossRef] [PubMed]
- Mercey, O.; Kostic, C.; Bertiaux, E.; Giroud, A.; Sadian, Y.; Gaboriau, D.C.A.; Morrison, C.G.; Chang, N.; Arsenijevic, Y.; Guichard, P.; et al. The Connecting Cilium Inner Scaffold Provides a Structural Foundation That Protects against Retinal Degeneration. PLoS Biol. 2022, 20, e3001649. [Google Scholar] [CrossRef] [PubMed]
- Drivas, T.G.; Wojno, A.P.; Tucker, B.A.; Stone, E.M.; Bennett, J. Basal Exon Skipping and Genetic Pleiotropy: A Predictive Model of Disease Pathogenesis. Sci. Transl. Med. 2015, 7, 291ra97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garanto, A.; van Beersum, S.E.C.; Peters, T.A.; Roepman, R.; Cremers, F.P.M.; Collin, R.W.J. Unexpected CEP290 MRNA Splicing in a Humanized Knock-in Mouse Model for Leber Congenital Amaurosis. PLoS ONE 2013, 8, e79369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kruczek, K.; Qu, Z.; Welby, E.; Shimada, H.; Hiriyanna, S.; English, M.A.; Zein, W.M.; Brooks, B.P.; Swaroop, A. In Vitro Modeling and Rescue of Ciliopathy Associated with IQCB1/NPHP5 Mutations Using Patient-Derived Cells. Stem Cell. Rep. 2022, 17, 2172–2186. [Google Scholar] [CrossRef]
- Kim, J.; Krishnaswami, S.R.; Gleeson, J.G. CEP290 Interacts with the Centriolar Satellite Component PCM-1 and Is Required for Rab8 Localization to the Primary Cilium. Hum. Mol. Genet. 2008, 17, 3796–3805. [Google Scholar] [CrossRef] [Green Version]
- Prosser, S.L.; Morrison, C.G. Centrin2 Regulates CP110 Removal in Primary Cilium Formation. J. Cell. Biol. 2015, 208, 693–701. [Google Scholar] [CrossRef] [Green Version]
- Spektor, A.; Tsang, W.Y.; Khoo, D.; Dynlacht, B.D. Cep97 and CP110 Suppress a Cilia Assembly Program. Cell 2007, 130, 678–690. [Google Scholar] [CrossRef] [Green Version]
- Tsang, W.Y.; Bossard, C.; Khanna, H.; Peränen, J.; Swaroop, A.; Malhotra, V.; Dynlacht, B.D. CP110 Suppresses Primary Cilia Formation through Its Interaction with CEP290, a Protein Deficient in Human Ciliary Disease. Dev. Cell 2008, 15, 187–197. [Google Scholar] [CrossRef] [Green Version]
- Tsang, W.Y.; Spektor, A.; Luciano, D.J.; Indjeian, V.B.; Chen, Z.; Salisbury, J.L.; Sánchez, I.; Dynlacht, B.D. CP110 Cooperates with Two Calcium-Binding Proteins to Regulate Cytokinesis and Genome Stability. Mol. Biol. Cell 2006, 17, 3423–3434. [Google Scholar] [CrossRef] [Green Version]
- Arnon, A.; Cook, B.; Montell, C.; Selinger, Z.; Minke, B. Calmodulin Regulation of Calcium Stores in Phototransduction of Drosophila. Science 1997, 275, 1119–1121. [Google Scholar] [CrossRef]
- Ikura, M.; Osawa, M.; Ames, J.B. The Role of Calcium-Binding Proteins in the Control of Transcription: Structure to Function. BioEssays 2002, 24, 625–636. [Google Scholar] [CrossRef] [PubMed]
- Carrión, A.M.; Link, W.A.; Ledo, F.; Mellström, B.; Naranjo, J.R. DREAM Is a Ca2+-Regulated Transcriptional Repressor. Nature 1999, 398, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Naranjo, R.; González, P.; Lopez-Hurtado, A.; Dopazo, X.M.; Mellström, B.; Naranjo, J.R. Inhibition of the Neuronal Calcium Sensor DREAM Modulates Presenilin-2 Endoproteolysis. Front. Mol. Neurosci. 2018, 11, 449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Oliveira, D.R.; Zamberlam, C.R.; Rêgo, G.M.; Cavalheiro, A.; Cerutti, J.M.; Cerutti, S.M. Effects of a Flavonoid-Rich Fraction on the Acquisition and Extinction of Fear Memory: Pharmacological and Molecular Approaches. Front. Behav. Neurosci. 2016, 9, 345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cichon, N.; Saluk-Bijak, J.; Gorniak, L.; Przyslo, L.; Bijak, M. Flavonoids as a Natural Enhancer of Neuroplasticity-An Overview of the Mechanism of Neurorestorative Action. Antioxidants 2020, 9, 1035. [Google Scholar] [CrossRef]
- Carbonel, A.A.F.; Cecyn, M.N.; Girão, J.H.R.C.; da Silva Sasso, G.R.; de Mello Ponteciano, B.; Pereira Vellozo, E.; Santos Simões, R.; Simões, M.d.J.; Girão, M.J.B.C.; Rodrigues de Oliveira, D. Flavonoids as Modulators of Synaptic Plasticity: Implications for the Development of Novel Therapeutic Strategies for Healthy Lifestyle. In Flavonoids—A Coloring Model for Cheering Up Life; IntechOpen: London, UK, 2019. [Google Scholar] [CrossRef] [Green Version]
- Kayama, M.; Nakazawa, T.; Thanos, A.; Morizane, Y.; Murakami, Y.; Theodoropoulou, S.; Abe, T.; Vavvas, D.; Miller, J.W. Heat Shock Protein 70 (HSP70) Is Critical for the Photoreceptor Stress Response after Retinal Detachment via Modulating Anti-Apoptotic Akt Kinase. Am. J. Pathol. 2011, 178, 1080–1091. [Google Scholar] [CrossRef]
- Jiang, K.; Fairless, E.; Kanda, A.; Gotoh, N.; Cogliati, T.; Li, T.; Swaroop, A. Divergent Effects of HSP70 Overexpression in Photoreceptors During Inherited Retinal Degeneration. Investig. Ophthalmol. Vis. Sci. 2020, 61, 25. [Google Scholar] [CrossRef]
- Deane, C.A.S.; Brown, I.R. Knockdown of Heat Shock Proteins HSPA6 (Hsp70B’) and HSPA1A (Hsp70-1) Sensitizes Differentiated Human Neuronal Cells to Cellular Stress. Neurochem. Res. 2018, 43, 340–350. [Google Scholar] [CrossRef]
- Shorbagi, S.; Brown, I.R. Dynamics of the Association of Heat Shock Protein HSPA6 (Hsp70B’) and HSPA1A (Hsp70-1) with Stress-Sensitive Cytoplasmic and Nuclear Structures in Differentiated Human Neuronal Cells. Cell. Stress. Chaperones 2016, 21, 993–1003. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Corral-Serrano, J.C.; Sladen, P.E.; Ottaviani, D.; Rezek, O.F.; Athanasiou, D.; Jovanovic, K.; van der Spuy, J.; Mansfield, B.C.; Cheetham, M.E. Eupatilin Improves Cilia Defects in Human CEP290 Ciliopathy Models. Cells 2023, 12, 1575. https://doi.org/10.3390/cells12121575
Corral-Serrano JC, Sladen PE, Ottaviani D, Rezek OF, Athanasiou D, Jovanovic K, van der Spuy J, Mansfield BC, Cheetham ME. Eupatilin Improves Cilia Defects in Human CEP290 Ciliopathy Models. Cells. 2023; 12(12):1575. https://doi.org/10.3390/cells12121575
Chicago/Turabian StyleCorral-Serrano, Julio C., Paul E. Sladen, Daniele Ottaviani, Olivia F. Rezek, Dimitra Athanasiou, Katarina Jovanovic, Jacqueline van der Spuy, Brian C. Mansfield, and Michael E. Cheetham. 2023. "Eupatilin Improves Cilia Defects in Human CEP290 Ciliopathy Models" Cells 12, no. 12: 1575. https://doi.org/10.3390/cells12121575
APA StyleCorral-Serrano, J. C., Sladen, P. E., Ottaviani, D., Rezek, O. F., Athanasiou, D., Jovanovic, K., van der Spuy, J., Mansfield, B. C., & Cheetham, M. E. (2023). Eupatilin Improves Cilia Defects in Human CEP290 Ciliopathy Models. Cells, 12(12), 1575. https://doi.org/10.3390/cells12121575






