Insights into Gene Regulation under Temozolomide-Promoted Cellular Dormancy and Its Connection to Stemness in Human Glioblastoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Human Specimens
2.2. Human Glioblastoma (GBM) Cell Lines, Primary Culture Cells, and Stem-like Cells
2.3. Stimulation of Glioblastoma (GBM) Cells
2.4. Reverse Transcription and Quantitative Real-Time PCR (qRT–PCR)
2.5. Immunofluorescence Staining
2.6. Gene Set Enrichment Analysis
2.7. Cytotoxicity Assay and Determination of Proliferation
2.8. Self-Renewal Capacity and Extreme Limiting Dilution Assay
2.9. Statistical and Correlation Analysis
3. Results
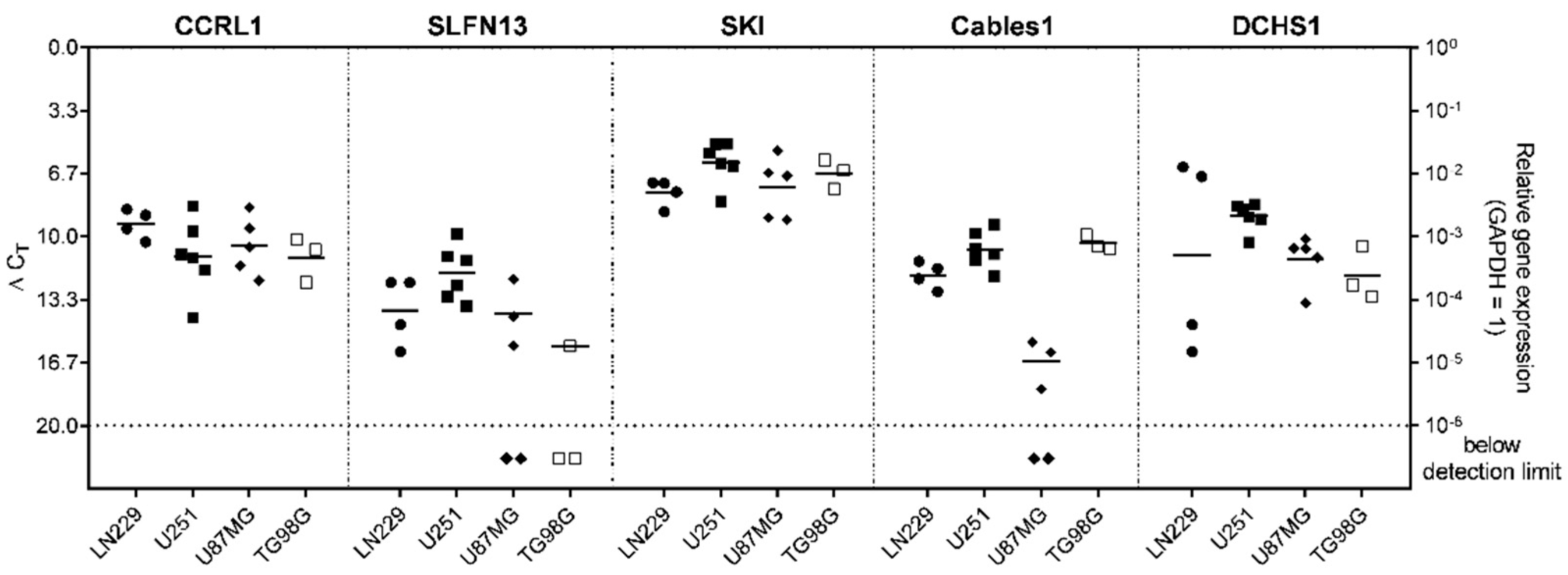
3.1. Expression and Regulation of Selected Genes under Temozolomide (TMZ)-Promoted Cellular Dormancy in Glioblastoma (GBM) Cell Lines and Patient-Derived Primary Cultures
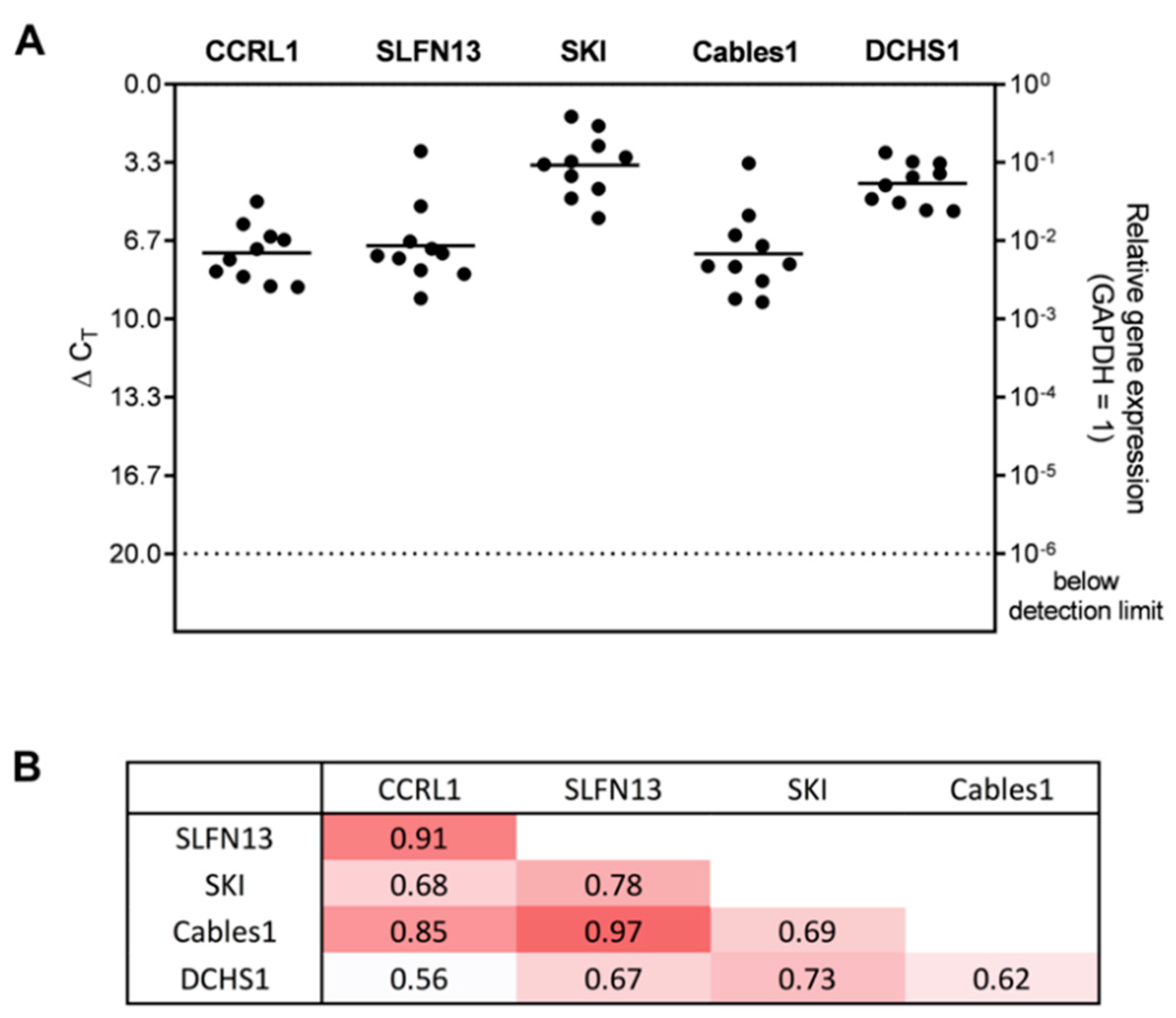
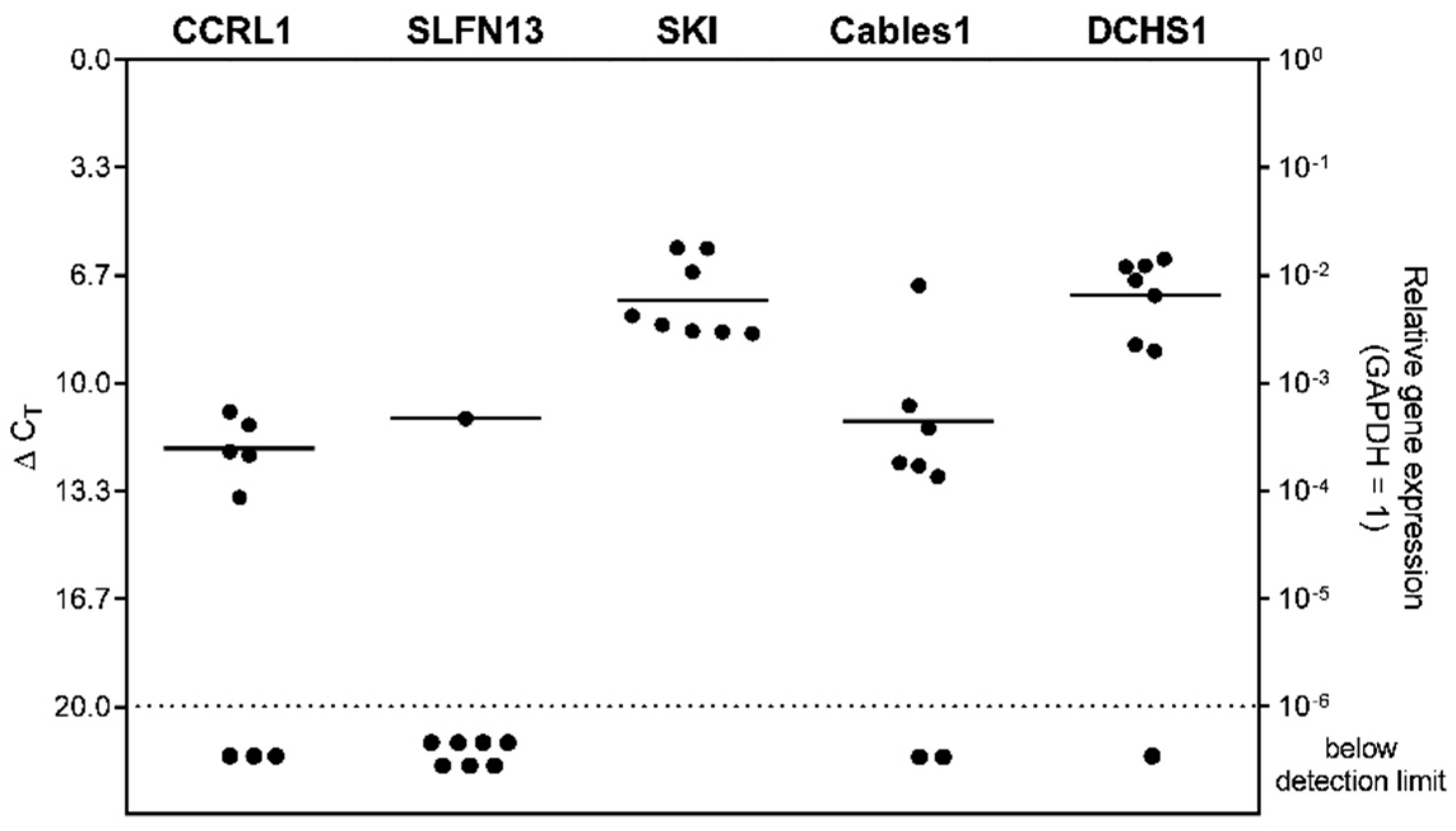
3.2. Expression and Correlation of Selected Genes with Each Other in Patient-Derived Glioblastoma (GBM) Ex Vivo Samples
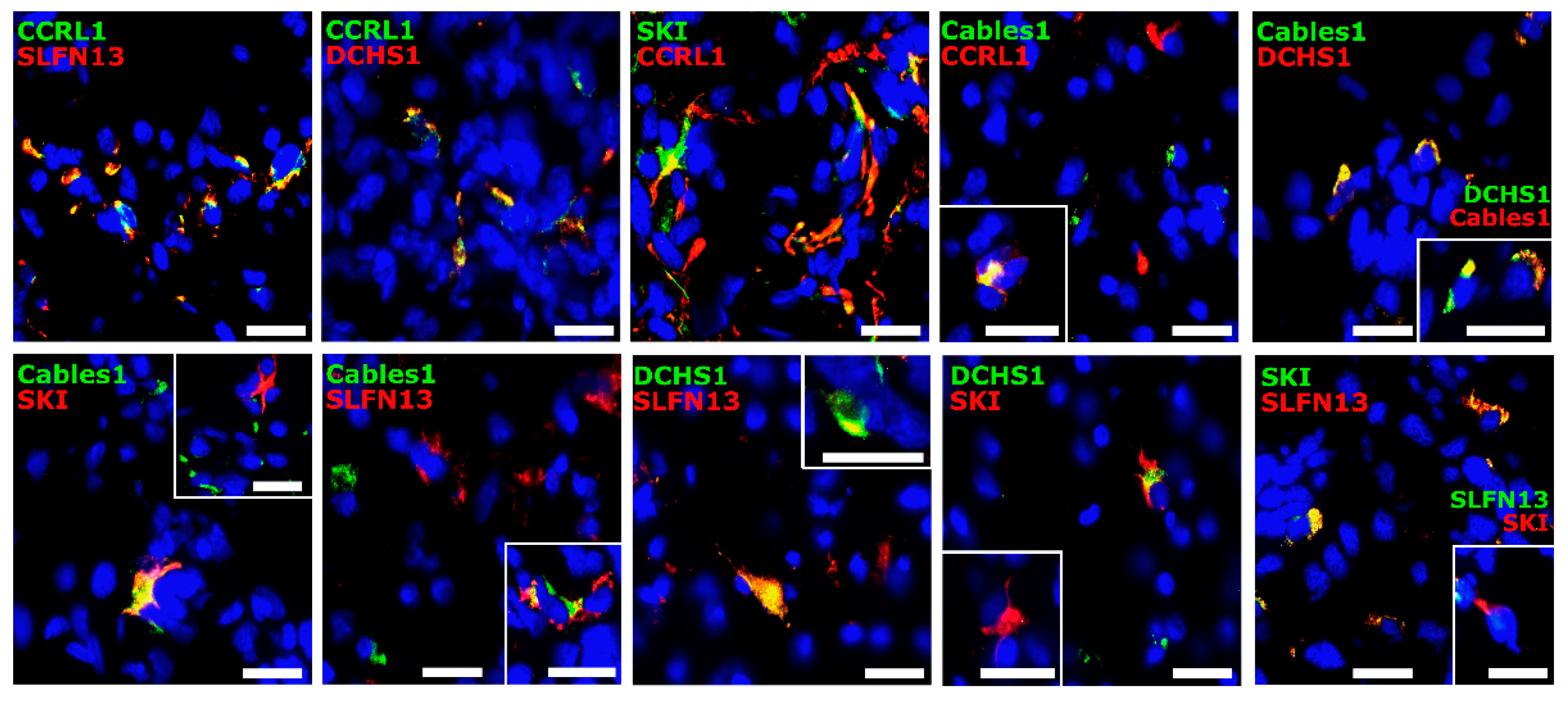
3.3. Co-Staining Patterns of Selected Molecules with Each Other in Patient-Derived Glioblastoma (GBM) Ex Vivo Samples
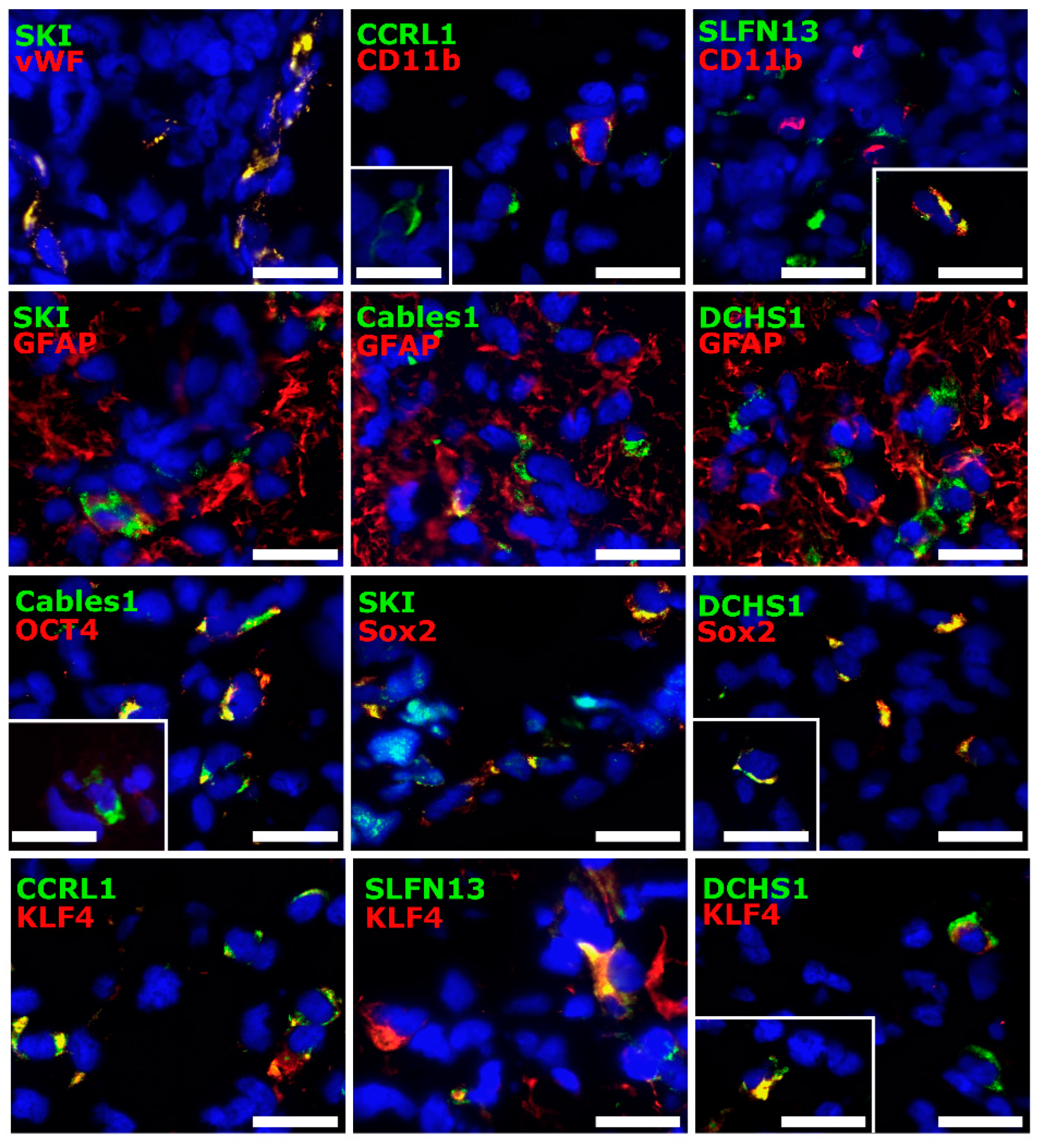
3.4. Cellular Sources of Selected Molecules in Patient-Derived Glioblastoma (GBM) Ex Vivo Samples
3.5. Correlation Analysis of Dormancy-Associated Genes and Stemness Markers in Patient-Derived Glioblastoma (GBM) Ex Vivo Samples
3.6. Expression of Selected Genes in Stem-like Cells Generated from Glioblastoma (GBM) Cell Lines or Patient-Derived Primary Cultures
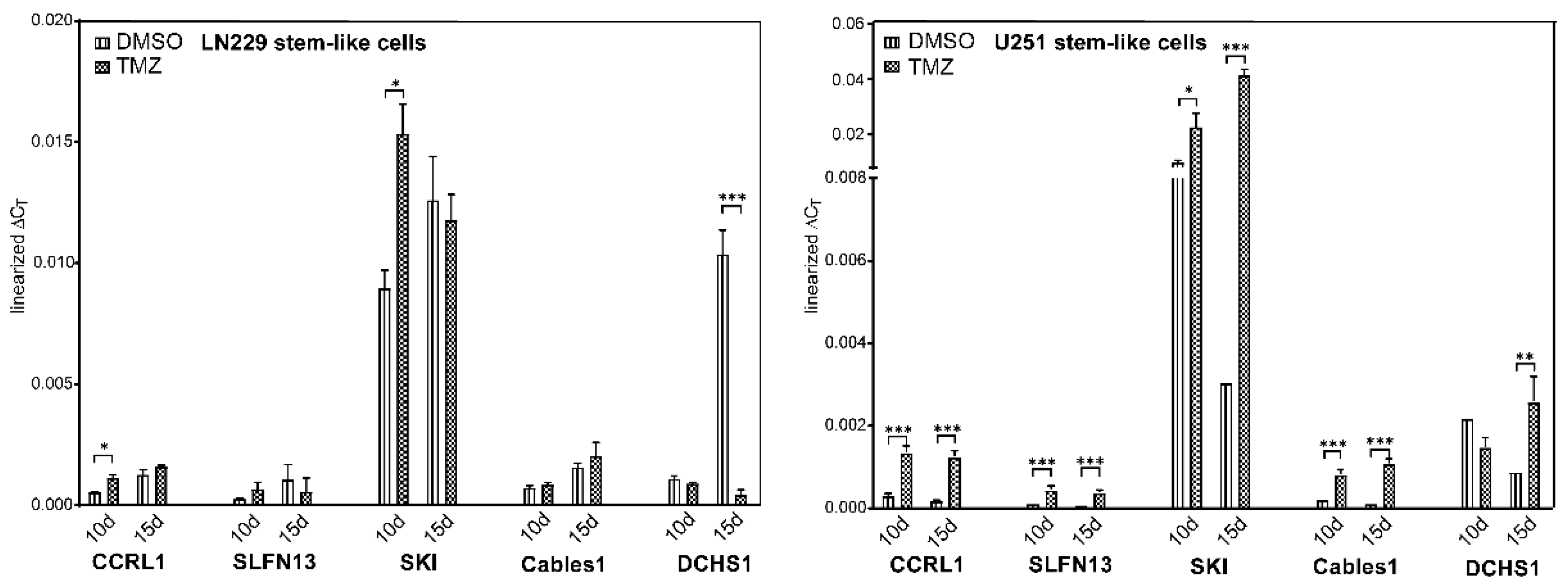
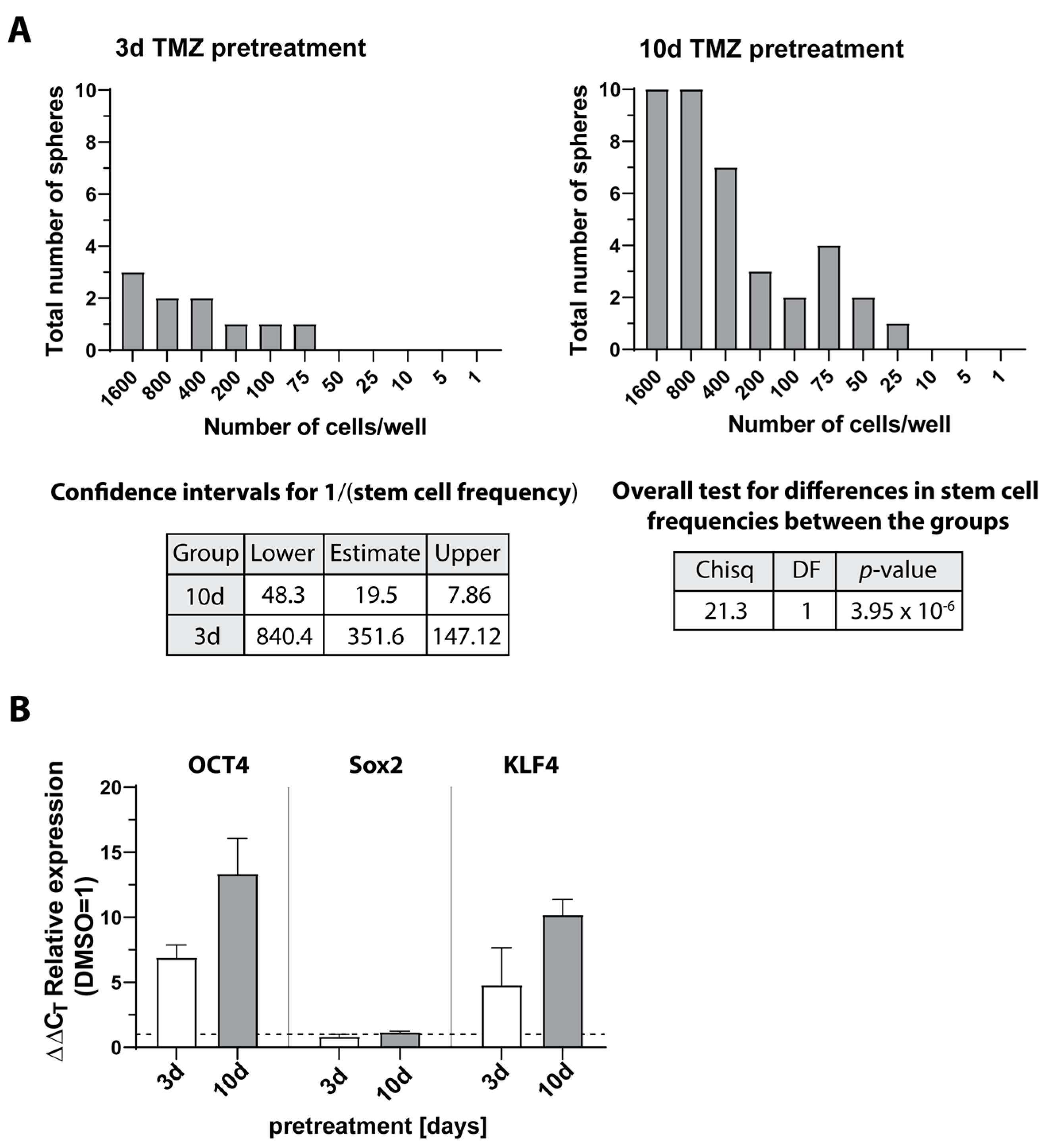
3.7. Expression and Regulation of Selected Genes under Temozolomide-Promoted Cellular Dormancy in Stem-like Cells and Neurosphere Formation Assay
3.8. Gene Set Enrichment Analysis and Inhibition of Sloan-Kettering Institute (SKI)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GBM | Glioblastoma |
| TMZ | Temozolomide |
| CCRL1 | Chemokine (C-C Motif) Receptor-Like 1 |
| SLFN13 | Schlafen 13 |
| SKI | Sloan-Kettering Institute |
| Cables1 | Like1Cdk5 and Abl Enzyme Substrate 1 |
| DCHS1 | Dachsous Cadherin-Related 1 |
| qRT-PCR | Reverse transcription and quantitative real-time PCR |
| GSC | Glioma stem-like cells |
| EMT | Epithelial-to-mesenchymal transition |
| CDH19 | Cadherin 19 |
| WHO | World Health Organization |
| UKE | University Medical Center Hamburg-Eppendorf |
| ANOVA | Analysis of variance |
| PCa/b | Primary culture a/b |
| vWF | Von Willebrand factor |
| CD11b | Cluster of differentiation molecule 11b |
| GFAP | Glial fibrillary acidic protein |
| OCT4 | Octamer binding transcription factor 4 |
| Sox2 | Sex determining region Y-box 2 |
| KLF4 | Krüppel-like factor 4 |
| DMSO | Dimethyl sulfoxide |
| MGMT | O6-methylguanine-DNA methyltransferase |
| TGF-ß | Transforming growth factor |
| Shh | Sonic hedgehog |
References
- Amman, J.; Tamimi, A.F.; Juweid, M. Epidemiology and Outcome of Glioblastoma. In Glioblastoma; De Vleeschouwer, S., Ed.; Codon Publications: Brisbane, Australia, 2017; pp. 143–153. [Google Scholar] [CrossRef]
- Verhaak, R.G.W.; Hoadley, K.A.; Purdom, E.; Wang, V.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Jill, P.; Alexe, G.; et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Adamski, V.; Hempelmann, A.; Flüh, C.; Lucius, R.; Synowitz, M.; Hattermann, K.; Held-Feindt, J. Dormant glioblastoma cells acquire stem cell characteristics and are differentially affected by Temozolomide and AT101 treatment. Oncotarget 2017, 8, 108064–108078. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Nishimura, M.C.; Bumbaca, S.M.; Kharbanda, S.; Forrest, W.F.; Kasman, I.M.; Greve, J.M.; Soriano, R.H.; Gilmour, L.L.; Rivers, C.S.; et al. A Hierarchy of Self-Renewing Tumor-Initiating Cell Types in Glioblastoma. Cancer Cell 2010, 17, 362–375. [Google Scholar] [CrossRef]
- Tong, L.; Yi, L.; Liu, P.; Abeysekera, I.R.; Hai, L.; Li, T.; Tao, Z.; Ma, H.; Xie, Y.; Huang, Y.; et al. Tumour cell dormancy as a contributor to the reduced survival of GBM patients who received standard therapy. Oncol. Rep. 2018, 40, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Adamski, V.; Hattermann, K.; Kubelt, C.; Cohrs, G.; Lucius, R.; Synowitz, M.; Sebens, S.; Held-Feindt, J. Entry and exit of chemotherapeutically-promoted cellular dormancy in glioblastoma cells is differentially affected by the chemokines CXCL12, CXCL16, and CX3CL1. Oncogene 2020, 39, 4421–4435. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.-Y.; Yang, L.-X.; Wang, Z.-C.; Wang, L.-Y.; Zhou, J.; Wang, X.-Y.; Shi, G.-M.; Ding, Z.-B.; Ke, A.-W.; Dai, Z.; et al. CC chemokine receptor-like 1 functions as a tumour suppressor by impairing CCR7-related chemotaxis in hepatocellular carcinoma. J. Pathol. 2015, 235, 546–558. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.Y.; Ou, Z.L.; Wu, F.Y.; Shen, Z.Z.; Shao, Z.M. Involvement of a Novel Chemokine Decoy Receptor CCX-CKR in Breast Cancer Growth, Metastasis and Patient Survival. Clin. Cancer Res. 2009, 15, 2962–2970. [Google Scholar] [CrossRef] [PubMed]
- Ju, Y.; Sun, C.; Wang, X. Loss of atypical chemokine receptor 4 facilitates C-C motif chemokine ligand 21-mediated tumor growth and invasion in nasopharyngeal carcinoma. Exp. Ther. Med. 2019, 17, 613–620. [Google Scholar] [CrossRef]
- Harata-Lee, Y.; Turvey, M.E.; Brazzatti, J.A.; Gregor, C.E.; Brown, M.P.; Smyth, M.J.; Comerford, I.; McColl, S.R. The atypical chemokine receptor CCX-CKR regulates metastasis of mammary carcinoma via an effect on EMT. Immunol. Cell Biol. 2014, 92, 815–824. [Google Scholar] [CrossRef]
- Al-Marsoummi, S.; Vomhof-DeKrey, E.E.; Basson, M.D. Schlafens: Emerging Proteins in Cancer Cell Biology. Cells 2021, 10, 2238. [Google Scholar] [CrossRef]
- Chen, W.; Lam, S.S.; Srinath, H.; Schiffer, C.A.; Royer, W.E.; Lin, K. Competition between Ski and CREB-binding protein for binding to Smad proteins in transforming growth factor-beta signaling. J. Biol. Chem. 2007, 282, 11365–11376. [Google Scholar] [CrossRef]
- Buess, M.; Terracciano, L.; Reuter, J.; Ballabeni, P.; Boulay, J.-L.; Laffer, U.; Metzger, U.; Herrmann, R.; Rochlitz, C.F. Amplification of SKI Is a Prognostic Marker in Early Colorectal Cancer. Neoplasia 2004, 6, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Fukuchi, M.; Nakajima, M.; Fukai, Y.; Miyazaki, T.; Masuda, N.; Sohda, M.; Manda, R.; Tsukada, K.; Kato, H.; Kuwano, H. Increased expression of c-Ski as a co-repressor in transforming growth factor-? signaling correlates with progression of esophageal squamous cell carcinoma. Int. J. Cancer 2004, 108, 818–824. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Wu, X.; Zhang, J.; Zhang, J.; Li, X. Ski regulates Smads and TAZ signaling to suppress lung cancer progression. Mol. Carcinog. 2017, 56, 2178–2189. [Google Scholar] [CrossRef]
- Kirley, S.D.; Rueda, B.R.; Chung, D.C.; Zukerberg, L.R. Increased growth rate, delayed senescense, and decreased serum dependence characterize CABLES-deficient cells. Cancer Biol. Ther. 2005, 4, 654–658. [Google Scholar] [CrossRef] [PubMed]
- Groeneweg, J.W.; White, Y.A.; Kokel, D.; Peterson, R.T.; Zukerberg, L.R.; Berin, I.; Rueda, B.R.; Wood, A.W. CABLES1 is required for embryonic neural development: Molecular, cellular, and behavioral evidence from the zebrafish. Mol. Reprod. Dev. 2011, 78, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Dong, Q.; Kirley, S.; Rueda, B.; Zhao, C.; Zukerberg, L.; Oliva, E. Loss of CABLES, a novel gene on chromosome 18q, in ovarian cancer. Mod. Pathol. 2003, 16, 863–868. [Google Scholar] [CrossRef]
- Wu, C.L.; Kirley, S.D.; Xiao, H.; Chuang, Y.; Chung, D.C.; Zukerberg, L.R. CABLES enhances cdk2 tyrosine 15 phosphorylation by Wee1, inhibits cell growth, and is lost in many human colon and squamous cancers. Cancer Res. 2001, 61, 7325–7332. [Google Scholar]
- Cappello, S.; Gray, M.J.; Badouel, C.; Lange, S.; Einsiedler, M.; Srour, M.; Chitayat, D.; Hamdan, F.F.; Jenkins, Z.A.; Morgan, T.R.; et al. Mutations in genes encoding the cadherin receptor-ligand pair DCHS1 and FAT4 disrupt cerebral cortical development. Nat. Genet. 2013, 45, 1300–1308. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, Z.; Zhu, J. In silico screening using bulk and single-cell RNA-seq data identifies RIMS2 as a prognostic marker in basal-like breast cancer: A retrospective study. Medicine 2021, 100, e25414. [Google Scholar] [CrossRef]
- de Mello, J.B.H.; Cirilo, P.D.R.; Michelin, O.C.; Domingues, M.A.C.; Rudge, M.V.C.; Rogatto, S.R.; Maestá, I. Genomic profile in gestational and non-gestational choriocarcinomas. Placenta 2017, 50, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Bujko, M.; Kober, P.; Mikula, M.; Ligaj, M.; Ostrowski, J.; Siedlecki, J.A. Expression changes of cell-cell adhesion-related genes in colorectal tumors. Oncol. Lett. 2015, 9, 2463–2470. [Google Scholar] [CrossRef]
- Hattermann, K.; Gebhardt, H.; Krossa, S.; Ludwig, A.; Lucius, R.; Held-Feindt, J.; Mentlein, R. Transmembrane chemokines act as receptors in a novel mechanism termed inverse signaling. eLife 2016, 5, e10820. [Google Scholar] [CrossRef]
- Flüh, C.; Chitadze, G.; Adamski, V.; Hattermann, K.; Synowitz, M.; Kabelitz, D.; Held-Feindt, J. NKG2D ligands in glioma stem-like cells: Expression in situ and in vitro. Histochem Cell Biol. 2018, 149, 219–233. [Google Scholar] [CrossRef] [PubMed]
- Hattermann, K.; Held-Feindt, J.; Lucius, R.; Müerköster, S.S.; Penfold, M.E.; Schall, T.J.; Mentlein, R. The chemokine receptor CXCR7 is highly expressed in human glioma cells and mediates antiapoptotic effects. Cancer Res. 2010, 70, 3299–3308. [Google Scholar] [CrossRef] [PubMed]
- Caylioglu, D.; Meyer, R.J.; Hellmold, D.; Kubelt, C.; Synowitz, M.; Held-Feindt, J. Effects of the Anti-Tumorigenic Agent AT101 on Human Glioblastoma Cells in the Microenvironmental Glioma Stem Cell Niche. Int. J. Mol. Sci. 2021, 22, 3606. [Google Scholar] [CrossRef] [PubMed]
- Kubelt, C.; Peters, S.; Ahmeti, H.; Huhndorf, M.; Huber, L.; Cohrs, G.; Hövener, J.-B.; Jansen, O.; Synowitz, M.; Held-Feindt, J. Intratumoral Distribution of Lactate and the Monocarboxylate Transporters 1 and 4 in Human Glioblastoma Multiforme and Their Relationships to Tumor Progression-Associated Markers. Int. J. Mol. Sci. 2020, 21, 6254. [Google Scholar] [CrossRef]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic. Acids Res. 2019, 7, W191–W198. [Google Scholar] [CrossRef]
- Hu, Y.; Smyth, G.K. ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods 2009, 347, 70–78. [Google Scholar] [CrossRef]
- Nikitin, P.V.; Musina, G.R.; Pekov, S.I.; Kuzin, A.A.; Popov, I.A.; Belyaev, A.Y.; Kobyakov, G.L.; Usachev, D.Y.; Nikolaev, V.N.; Mikhailov, V.P. Cell-Population Dynamics in Diffuse Gliomas during Gliomagenesis and Its Impact on Patient Survival. Cancers 2023, 15, 145. [Google Scholar] [CrossRef]
- Gallo-Oller, G.; Vollmann-Zwerenz, A.; Meléndez, B.; Rey, J.A.; Hau, P.; Dotor, J.; Castresana, J.S. P144, a Transforming Growth Factor beta inhibitor peptide, generates antitumoral effects and modifies SMAD7 and SKI levels in human glioblastoma cell lines. Cancer Lett. 2016, 381, 67–75. [Google Scholar] [CrossRef]
- Hegi, M.E.; Diserens, A.-C.; Gorlia, T.; Hamou, M.-F.; De Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Houillier, C.; Wang, X.; Kaloshi, G.; Mokhtari, K.; Guillevin, R.; Laffaire, J.; Paris, S.; Boisselier, B.; Idbaih, A.; Laigle-Donadey, F.; et al. IDH1 or IDH2 mutations predict longer survival and response to temozolomide in low-grade gliomas. Neurology 2010, 75, 1560–1566. [Google Scholar] [CrossRef]
- Han, B.; Meng, X.; Wu, P.; Li, Z.; Li, S.; Zhang, Y.; Zha, C.; Ye, Q.; Jiang, C.; Cai, J.; et al. ATRX/EZH2 complex epigenetically regulates FADD/PARP1 axis, contributing to TMZ resistance in glioma. Theranostics 2020, 10, 3351–3365. [Google Scholar] [CrossRef] [PubMed]
- Amen, A.M.; Fellmann, C.; Soczek, K.M.; Ren, S.M.; Lew, R.J.; Knott, G.J.; Park, J.E.; McKinney, A.M.; Mancini, A.; Doudna, J.A.; et al. Cancer-specific loss of TERT activation sensitizes glioblastoma to DNA damage. Proc. Natl. Acad. Sci. USA 2021, 118, e2008772118. [Google Scholar] [CrossRef]
- Isci, D.; D’uonnolo, G.; Wantz, M.; Rogister, B.; Lombard, A.; Chevigné, A.; Szpakowska, M.; Neirinckx, V. Patient-Oriented Perspective on Chemokine Receptor Expression and Function in Glioma. Cancers 2021, 14, 130. [Google Scholar] [CrossRef]
- Arslan, A.D.; Sassano, A.; Saleiro, D.; Lisowski, P.; Kosciuczuk, E.M.; Fischietti, M.; Eckerdt, F.; Fish, E.N.; Platanias, L.C. Human SLFN5 is a transcriptional co-repressor of STAT1-mediated interferon responses and promotes the malignant phenotype in glioblastoma. Oncogene 2017, 36, 6006–6019. [Google Scholar] [CrossRef]
- Morein, D.; Erlichman, N.; Ben-Baruch, A. Beyond Cell Motility: The Expanding Roles of Chemokines and Their Receptors in Malignancy. Front. Immunol. 2020, 11, 952. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Xu, W.; Bales, E.; Colmenares, C.; Conacci-Sorrell, M.; Ishii, S.; Stavnezer, E.; Campisi, J.; E Fisher, D.; Ben-Ze’Ev, A.; et al. SKI activates Wnt/beta-catenin signaling in human melanoma. Cancer Res. 2003, 63, 6626–6634. [Google Scholar]
- Zhao, X.; Fang, Y.; Wang, X.; Yang, Z.; Li, D.; Tian, M.; Kang, P. Knockdown of Ski decreases osteosarcoma cell proliferation and migration by suppressing the PI3K/Akt signaling pathway. Int. J. Oncol. 2020, 56, 206–218. [Google Scholar] [CrossRef]
- Liu, P.; Wang, Q.-S.; Zhai, Y.; Xiong, R.-P.; Chen, X.; Peng, Y.; Zhao, Y.; Ning, Y.-L.; Yang, N.; Zhou, Y.-G. Ski mediates TGF-β1-induced fibrosarcoma cell proliferation and promotes tumor growth. J. Cancer 2020, 11, 5929–5940. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Jin, C.; Liu, J.; Hua, D.; Zhou, F.; Lou, X.; Zhao, N.; Lan, Q.; Huang, Q.; Yoon, J.-G.; et al. Next generation sequencing analysis of miRNAs: MiR-127-3p inhibits glioblastoma proliferation and activates TGF-β signaling by targeting SKI. Omics J. Integr. Biol. 2014, 18, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Maier, H.J.; Wirth, T.; Beug, H. Epithelial-Mesenchymal Transition in Pancreatic Carcinoma. Cancers 2010, 2, 2058–2083. [Google Scholar] [CrossRef] [PubMed]
- Arnason, T.; Pino, M.S.; Yilmaz, O.; Kirley, S.D.; Rueda, B.R.; Chung, D.C.; Zukerberg, L.R. CABLES1 is a tumor suppressor gene that regulates intestinal tumor progression in Apc(Min) mice. Cancer Biol. Ther. 2013, 14, 672–678. [Google Scholar] [CrossRef]
- Safran, M.; Rosen, N.; Twik, M.; BarShir, R.; Stein, T.I.; Dahary, D.; Fishilevich, S.; Lancet, D. The GeneCards Suite. In Practical Guide to Life Science Databases; Abugessaisa, I., Kasukawa, T., Eds.; Springer: Singapore, 2021; pp. 27–56. [Google Scholar] [CrossRef]
- Sjöberg, E.; Meyrath, M.; Chevigné, A.; Östman, A.; Augsten, M.; Szpakowska, M. The diverse and complex roles of atypical chemokine receptors in cancer: From molecular biology to clinical relevance and therapy. Adv. Cancer Res. 2020, 145, 99–138. [Google Scholar] [CrossRef]
- Lucas, B.; White, A.J.; Ulvmar, M.H.; Nibbs, R.J.B.; Sitnik, K.M.; Agace, W.W.; Jenkinson, W.E.; Anderson, G.; Rot, A. CCRL1/ACKR4 is expressed in key thymic microenvironments but is dispensable for T lymphopoiesis at steady state in adult mice. Eur. J. Immunol. 2015, 45, 574–583. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, M.; Zhang, C.; Méar, L.; Zhong, W.; Digre, A.; Katona, B.; Sjöstedt, E.; Butler, L.; Odeberg, J.; Dusart, P.; et al. A single–cell type transcriptomics map of human tissues. Sci. Adv. 2021, 7, eabh2169. [Google Scholar] [CrossRef]
- Teresa, M.; Tan, M.; Tarango, M.; Fink, L.; Mihm, M.; Ma, Y.; Waner, M. Differential expression of SKI oncogene protein in hemangiomas. Otolaryngol. Head Neck Surg. 2009, 141, 213–218. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Beghi, F.; Baral, V.; Dépond, M.; Zhang, Y.; Joulin, V.; Rueda, B.R.; Gonin, P.; Foudi, A.; Wittner, M.; et al. CABLES1 Deficiency Impairs Quiescence and Stress Responses of Hematopoietic Stem Cells in Intrinsic and Extrinsic Manners. Stem Cell Rep. 2019, 13, 274–290. [Google Scholar] [CrossRef] [PubMed]
- Spittau, B.; Wullkopf, L.; Zhou, X.; Rilka, J.; Pfeifer, D.; Krieglstein, K. Endogenous transforming growth factor-beta promotes quiescence of primary microglia in vitro. Glia 2013, 61, 287–300. [Google Scholar] [CrossRef]
- Zhai, Y.; Ye, S.-Y.; Wang, Q.-S.; Xiong, R.-P.; Fu, S.-Y.; Du, H.; Xu, Y.-W.; Peng, Y.; Huang, Z.-Z.; Yang, N.; et al. Overexpressed ski efficiently promotes neurorestoration, increases neuronal regeneration, and reduces astrogliosis after traumatic brain injury. Gene Ther. 2022, 30, 75–87. [Google Scholar] [CrossRef]
- Abud, E.M.; Ramirez, R.N.; Martinez, E.S.; Healy, L.M.; Nguyen, C.H.H.; Newman, S.A.; Yeromin, A.V.; Scarfone, V.M.; Marsh, S.E.; Fimbres, C.; et al. iPSC-Derived Human Microglia-like Cells to Study Neurological Diseases. Neuron 2017, 94, 278–293e9. [Google Scholar] [CrossRef]
- Dubbelaar, M.; Misrielal, C.; Bajramovic, J.; Burm, S.; Zuiderwijk-Sick, E.; Brouwer, N.; Grit, C.; Kooistra, S.; Shinjo, S.; Marie, S.; et al. Transcriptional profiling of macaque microglia reveals an evolutionary preserved gene expression program. Brain Behav. Immun. Health 2021, 15, 100265. [Google Scholar] [CrossRef]
- Galatro, T.F.; Holtman, I.R.; Lerario, A.M.; Vainchtein, I.D.; Brouwer, N.; Sola, P.R.; Veras, M.M.; Pereira, T.F.; Leite, R.E.P.; Möller, T.; et al. Transcriptomic analysis of purified human cortical microglia reveals age-associated changes. Nat. Neurosci. 2017, 20, 1162–1171. [Google Scholar] [CrossRef]
- Whyte, C.E.; Osman, M.; Kara, E.E.; Abbott, C.; Foeng, J.; McKenzie, D.R.; Fenix, K.A.; Harata-Lee, Y.; Foyle, K.; Boyle, S.T.; et al. ACKR4 restrains antitumor immunity by regulating CCL21. J. Exp. Med. 2020, 217, e20190634. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhou, D.; Wu, F.; Liang, Q.; He, Q.; Peng, M.; Yao, T.; Hu, Y.; Qian, B.; Tang, J.; et al. Immune Microenvironment Signatures as Biomarkers to Predict Early Recurrence of Stage Ia-b Lung Cancer. Front. Oncol. 2021, 11, 680287. [Google Scholar] [CrossRef] [PubMed]
- Zorniak, M.; Clark, P.A.; Kuo, J.S. Myelin-forming cell-specific cadherin-19 is a marker for minimally infiltrative glioblastoma stem-like cells. J. Neurosurg. 2015, 122, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Chen, X.; Gao, S.; Zhang, C.; Qu, C.; Wang, P.; Liu, L. Ski modulate the characteristics of pancreatic cancer stem cells via regulating sonic hedgehog signaling pathway. Tumor Biol. 2016, 37, 16115–16125. [Google Scholar] [CrossRef] [PubMed]
- Kaminska, B.; Kocyk, M.; Kijewska, M. TGF beta signaling and its role in glioma pathogenesis. Adv. Exp. Med. Biol. 2013, 986, 171–187. [Google Scholar] [CrossRef]
- Taghizadeh, S.; Heiner, M.; Vazquez-Armendariz, A.I.; Wilhelm, J.; Herold, S.; Chen, C.; Zhang, J.S.; Bellusci, S. Characterization in Mice of the Resident Mesenchymal Niche Maintaining At2 Stem Cell Proliferation in Homeostasis and Disease. Stem Cells 2021, 39, 1382–1394. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kubelt, C.; Hellmold, D.; Esser, D.; Ahmeti, H.; Synowitz, M.; Held-Feindt, J. Insights into Gene Regulation under Temozolomide-Promoted Cellular Dormancy and Its Connection to Stemness in Human Glioblastoma. Cells 2023, 12, 1491. https://doi.org/10.3390/cells12111491
Kubelt C, Hellmold D, Esser D, Ahmeti H, Synowitz M, Held-Feindt J. Insights into Gene Regulation under Temozolomide-Promoted Cellular Dormancy and Its Connection to Stemness in Human Glioblastoma. Cells. 2023; 12(11):1491. https://doi.org/10.3390/cells12111491
Chicago/Turabian StyleKubelt, Carolin, Dana Hellmold, Daniela Esser, Hajrullah Ahmeti, Michael Synowitz, and Janka Held-Feindt. 2023. "Insights into Gene Regulation under Temozolomide-Promoted Cellular Dormancy and Its Connection to Stemness in Human Glioblastoma" Cells 12, no. 11: 1491. https://doi.org/10.3390/cells12111491
APA StyleKubelt, C., Hellmold, D., Esser, D., Ahmeti, H., Synowitz, M., & Held-Feindt, J. (2023). Insights into Gene Regulation under Temozolomide-Promoted Cellular Dormancy and Its Connection to Stemness in Human Glioblastoma. Cells, 12(11), 1491. https://doi.org/10.3390/cells12111491




