PNPLA3(I148M) Inhibits Lipolysis by Perilipin-5-Dependent Competition with ATGL
Abstract
1. Introduction
2. Materials and Methods
2.1. Tissues and Cell Culture
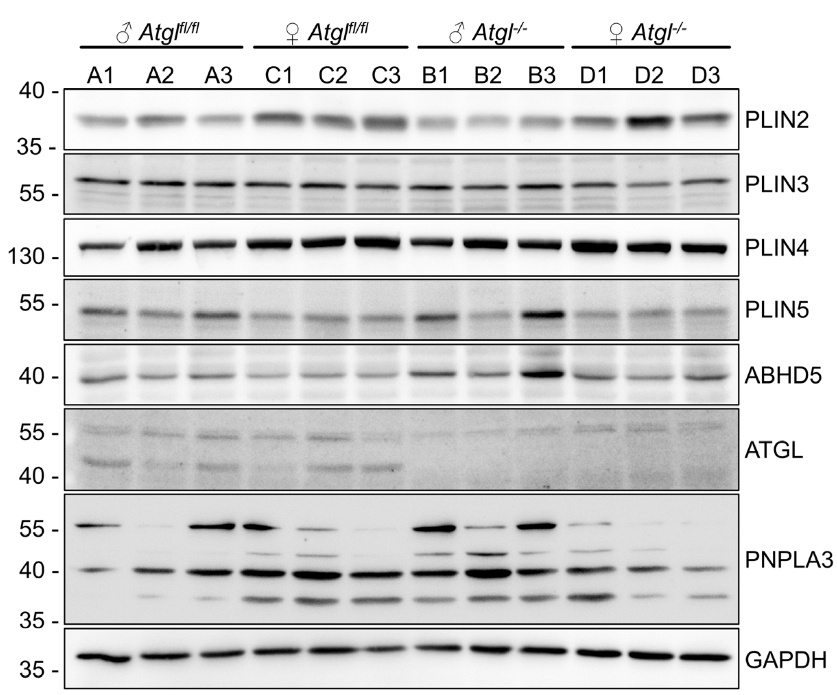
2.2. Mouse Models
2.3. Protein Isolation
2.4. Antibodies
2.5. Transfection
2.6. Co-Immunoprecipitation
2.7. Immunofluorescence Microscopy
2.8. Image Analysis
2.9. Immunohistochemistry
2.10. Transmission Electron Microscopy
2.11. CRISPR/Cas
2.12. Lipolysis/Steatogenesis
2.13. Quantitative PCR
2.14. Statistical Analysis
3. Results
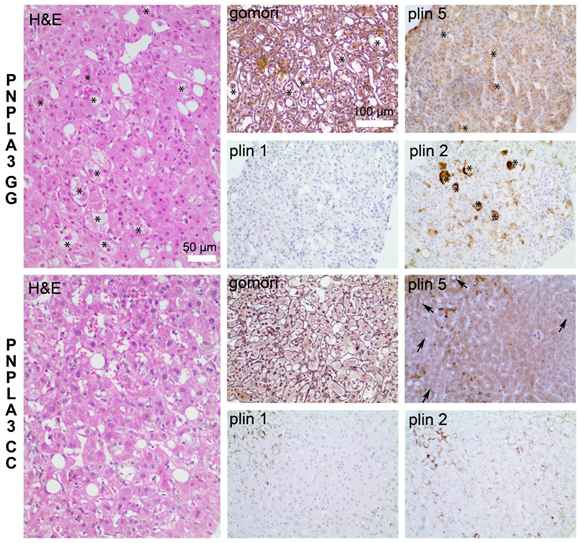
3.1. Increased Inflammation and Fibrosis in Patients with PNPLA3(I148M)
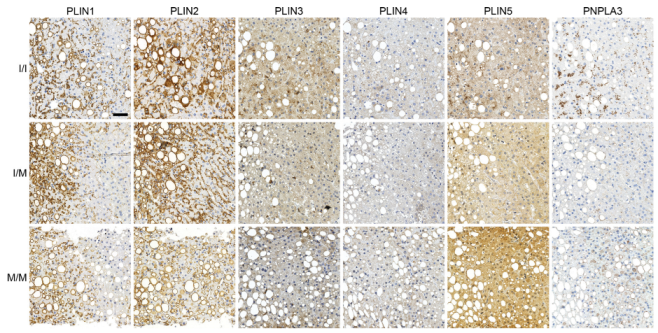
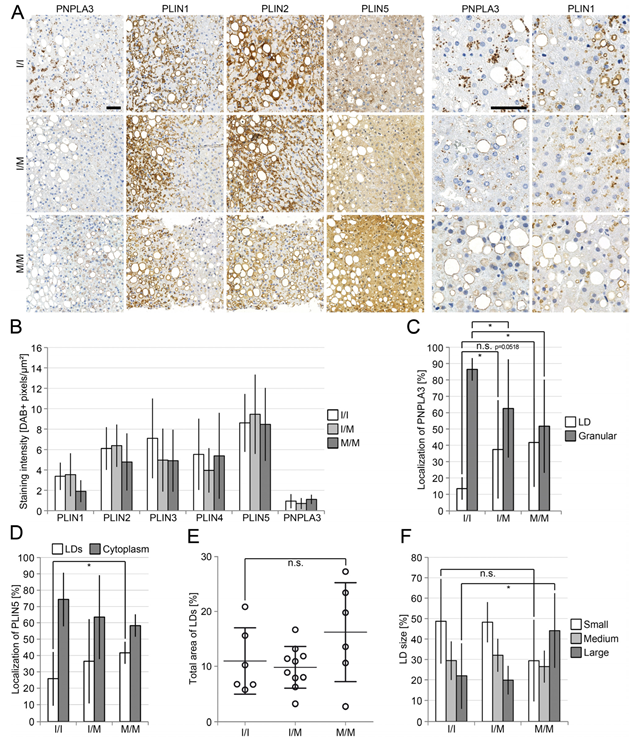
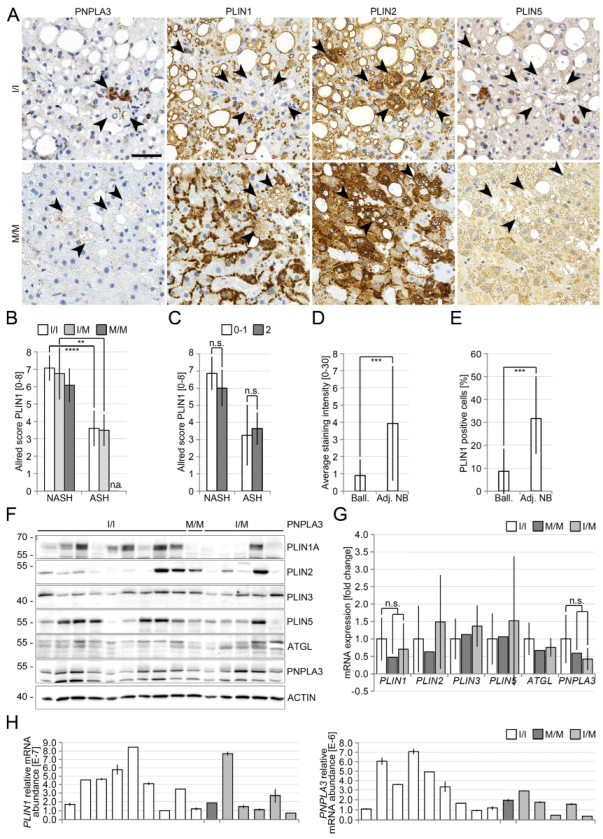
3.2. Reduced Perilipin 1 Expression in PNPLA3(I148M)-Hepatocytes In Situ
3.3. Opposing Effects of Perilipin 1 and 5 on the Recruitment of PNPLA3 to LDs
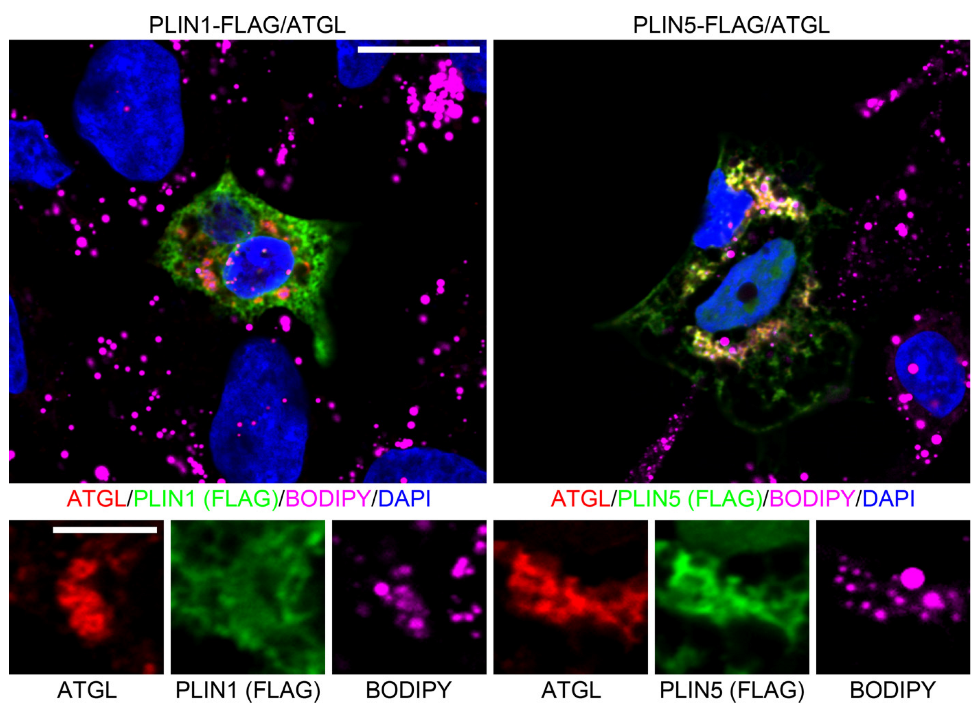
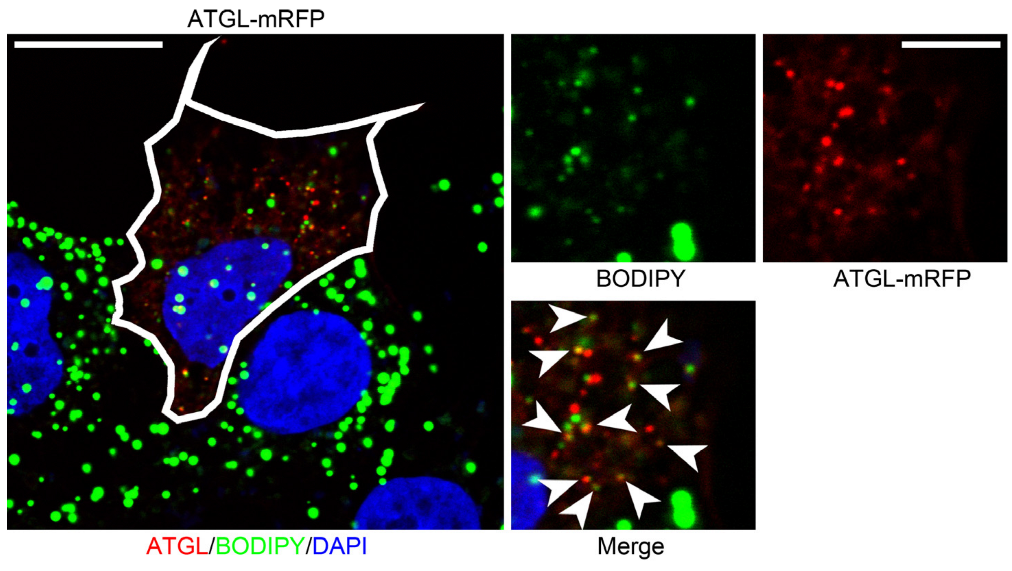
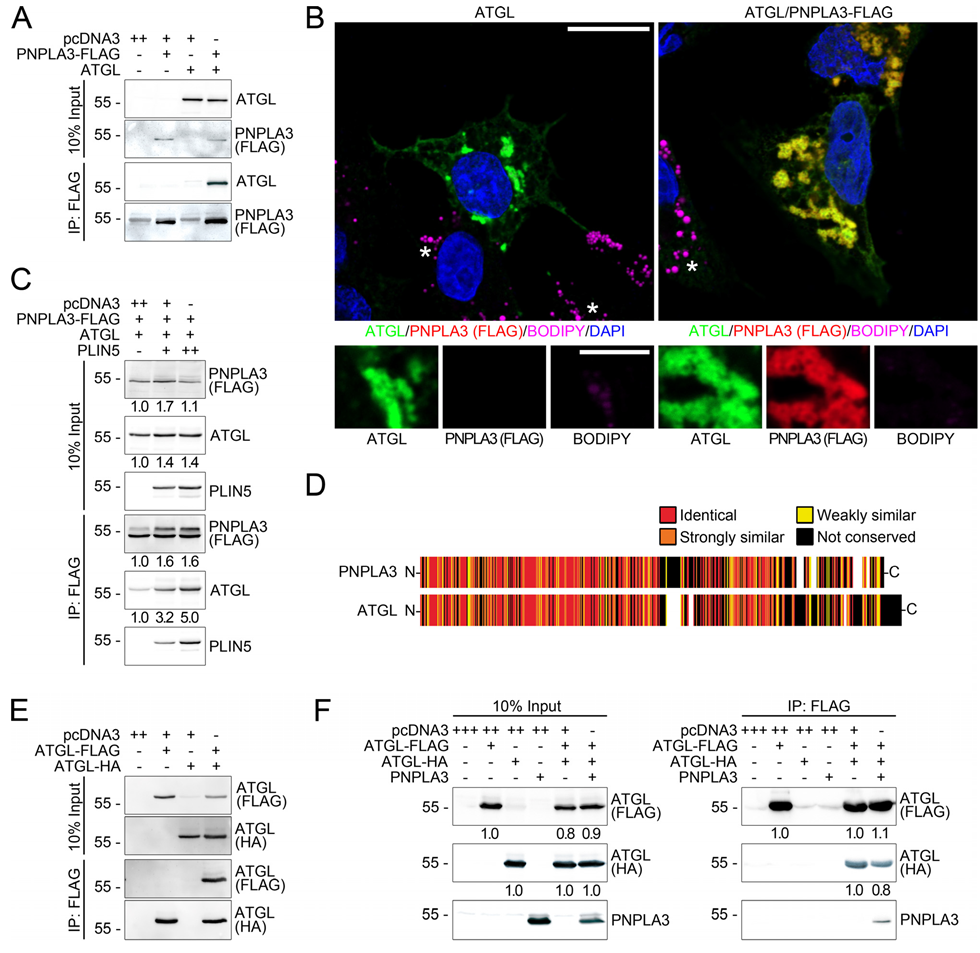
3.4. PNPLA3 Interacts with ATGL, the Rate-Limiting Enzyme in Lipolysis, in a Perilipin 5-Dependent Manner
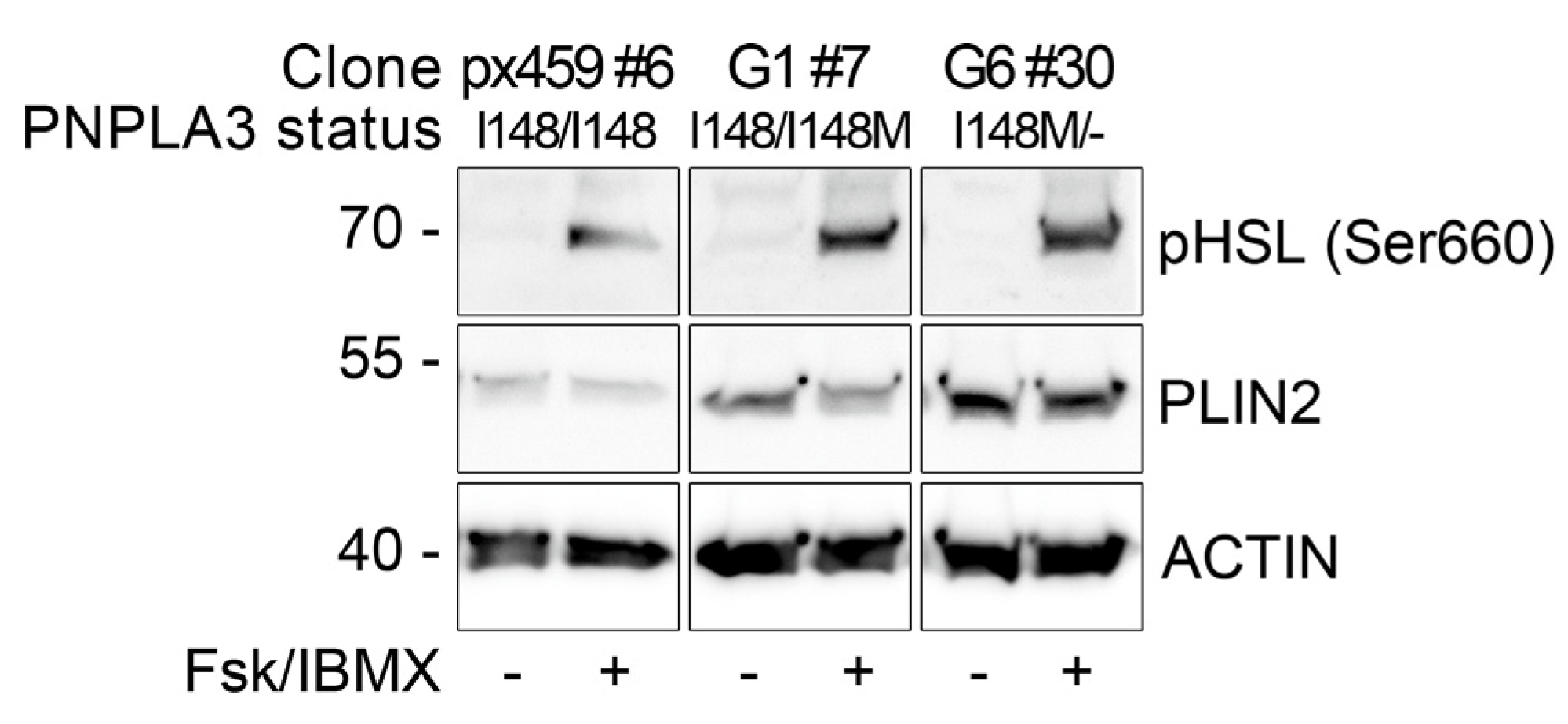
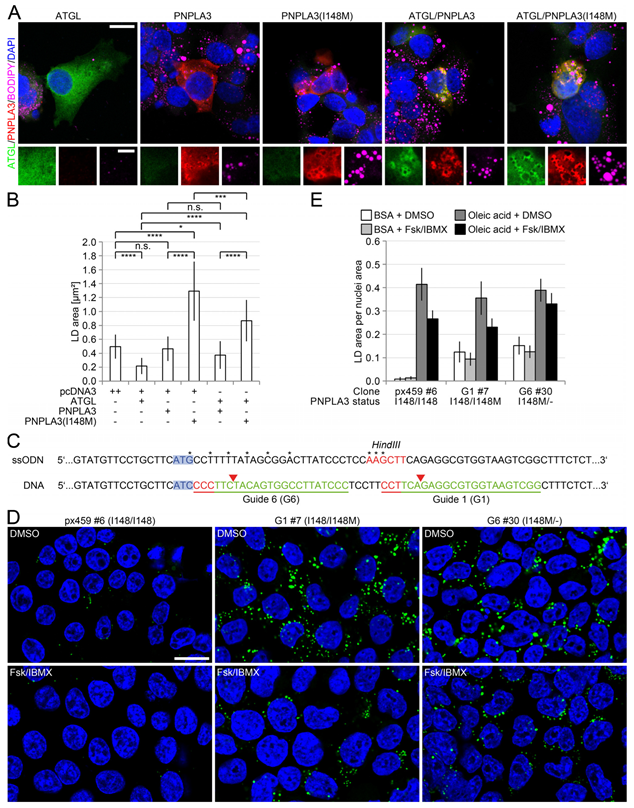
3.5. PNPLA3(I148M) Negatively Regulates ATGL Activity
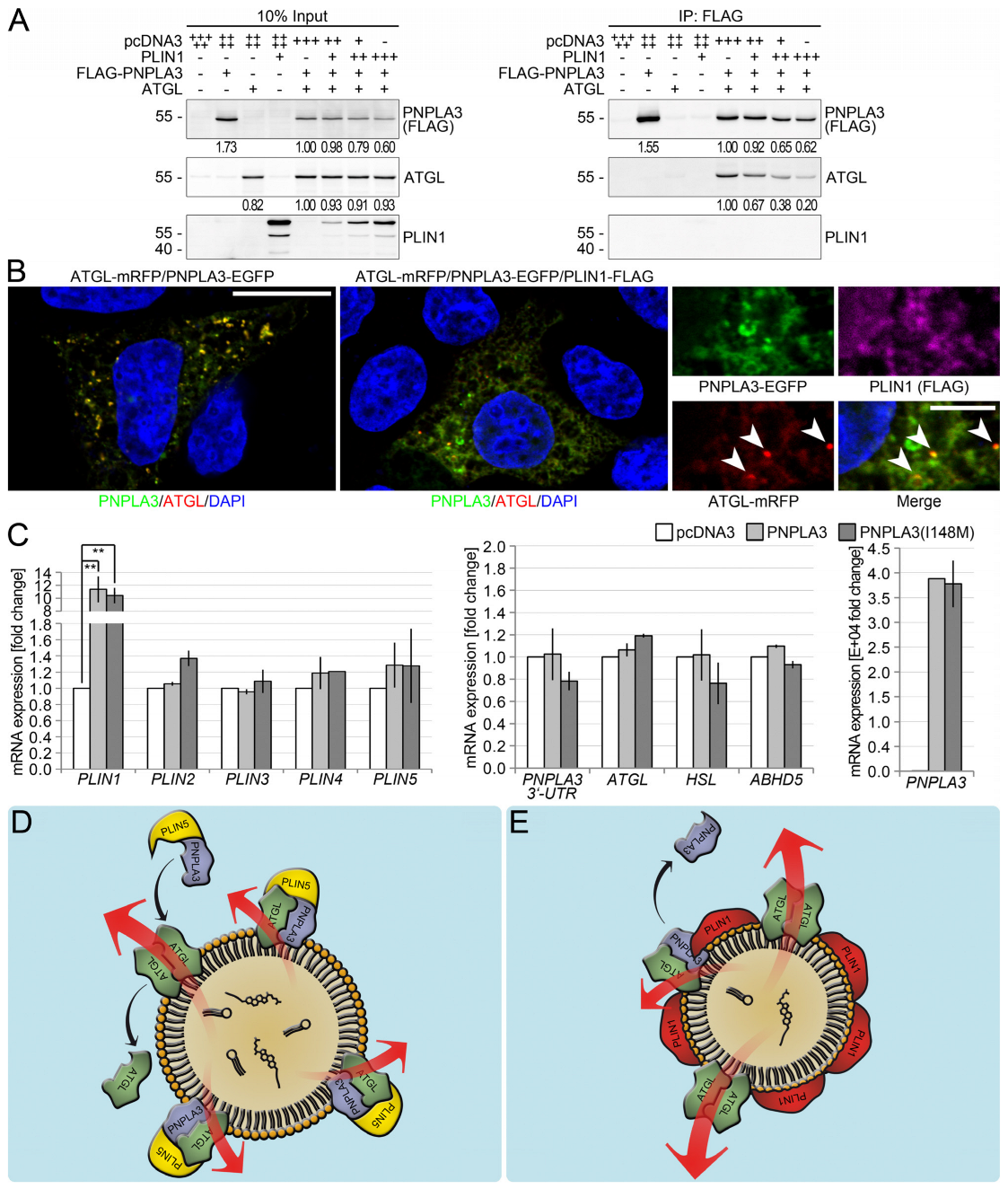
3.6. Perilipin 1 Displaces PNPLA3 from the ATGL Complex and Drives ATGL-Mediated Lipolysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A



References
- Gluchowski, N.L.; Becuwe, M.; Walther, T.C.; Farese, R.V., Jr. Lipid droplets and liver disease: From basic biology to clinical implications. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 343–355. [Google Scholar] [CrossRef] [PubMed]
- Schattenberg, J.M.; Schuppan, D. Nonalcoholic steatohepatitis: The therapeutic challenge of a global epidemic. Curr. Opin. Lipidol. 2011, 22, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Hardy, T.; Oakley, F.; Anstee, Q.M.; Day, C.P. Nonalcoholic Fatty Liver Disease: Pathogenesis and Disease Spectrum. Annu. Rev. Pathol. 2016, 11, 451–496. [Google Scholar] [CrossRef]
- Kleiner, D.E.; Brunt, E.M.; Van Natta, M.; Behling, C.; Contos, M.J.; Cummings, O.W.; Ferrell, L.D.; Liu, Y.C.; Torbenson, M.S.; Unalp-Arida, A.; et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 2005, 41, 1313–1321. [Google Scholar] [CrossRef] [PubMed]
- Bedossa, P.; Poynard, T. An algorithm for the grading of activity in chronic hepatitis C. The METAVIR Cooperative Study Group. Hepatology 1996, 24, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Romeo, S.; Kozlitina, J.; Xing, C.; Pertsemlidis, A.; Cox, D.; Pennacchio, L.A.; Boerwinkle, E.; Cohen, J.C.; Hobbs, H.H. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 2008, 40, 1461–1465. [Google Scholar] [CrossRef]
- Sookoian, S.; Castano, G.O.; Burgueno, A.L.; Gianotti, T.F.; Rosselli, M.S.; Pirola, C.J. A nonsynonymous gene variant in the adiponutrin gene is associated with nonalcoholic fatty liver disease severity. J. Lipid Res. 2009, 50, 2111–2116. [Google Scholar] [CrossRef]
- Valenti, L.; Dongiovanni, P.; Ginanni Corradini, S.; Burza, M.A.; Romeo, S. PNPLA3 I148M variant and hepatocellular carcinoma: A common genetic variant for a rare disease. Dig. Liver Dis. 2013, 45, 619–624. [Google Scholar] [CrossRef]
- BasuRay, S.; Wang, Y.; Smagris, E.; Cohen, J.C.; Hobbs, H.H. Accumulation of PNPLA3 on lipid droplets is the basis of associated hepatic steatosis. Proc. Natl. Acad. Sci. USA 2019, 116, 9521–9526. [Google Scholar] [CrossRef]
- Huang, Y.; Cohen, J.C.; Hobbs, H.H. Expression and characterization of a PNPLA3 protein isoform (I148M) associated with nonalcoholic fatty liver disease. J. Biol. Chem. 2011, 286, 37085–37093. [Google Scholar] [CrossRef]
- Chen, W.; Chang, B.; Li, L.; Chan, L. Patatin-like phospholipase domain-containing 3/adiponutrin deficiency in mice is not associated with fatty liver disease. Hepatology 2010, 52, 1134–1142. [Google Scholar] [CrossRef] [PubMed]
- Basantani, M.K.; Sitnick, M.T.; Cai, L.; Brenner, D.S.; Gardner, N.P.; Li, J.Z.; Schoiswohl, G.; Yang, K.; Kumari, M.; Gross, R.W.; et al. Pnpla3/Adiponutrin deficiency in mice does not contribute to fatty liver disease or metabolic syndrome. J. Lipid Res. 2011, 52, 318–329. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Z.; Huang, Y.; Karaman, R.; Ivanova, P.T.; Brown, H.A.; Roddy, T.; Castro-Perez, J.; Cohen, J.C.; Hobbs, H.H. Chronic overexpression of PNPLA3I148M in mouse liver causes hepatic steatosis. J. Clin. Investig. 2012, 122, 4130–4144. [Google Scholar] [CrossRef] [PubMed]
- Smagris, E.; BasuRay, S.; Li, J.; Huang, Y.; Lai, K.M.; Gromada, J.; Cohen, J.C.; Hobbs, H.H. Pnpla3I148M knockin mice accumulate PNPLA3 on lipid droplets and develop hepatic steatosis. Hepatology 2015, 61, 108–118. [Google Scholar] [CrossRef] [PubMed]
- BasuRay, S.; Smagris, E.; Cohen, J.C.; Hobbs, H.H. The PNPLA3 variant associated with fatty liver disease (I148M) accumulates on lipid droplets by evading ubiquitylation. Hepatology 2017, 66, 1111–1124. [Google Scholar] [CrossRef] [PubMed]
- Kumari, M.; Schoiswohl, G.; Chitraju, C.; Paar, M.; Cornaciu, I.; Rangrez, A.Y.; Wongsiriroj, N.; Nagy, H.M.; Ivanova, P.T.; Scott, S.A.; et al. Adiponutrin functions as a nutritionally regulated lysophosphatidic acid acyltransferase. Cell Metab. 2012, 15, 691–702. [Google Scholar] [CrossRef]
- Pirazzi, C.; Adiels, M.; Burza, M.A.; Mancina, R.M.; Levin, M.; Stahlman, M.; Taskinen, M.R.; Orho-Melander, M.; Perman, J.; Pujia, A.; et al. Patatin-like phospholipase domain-containing 3 (PNPLA3) I148M (rs738409) affects hepatic VLDL secretion in humans and in vitro. J. Hepatol. 2012, 57, 1276–1282. [Google Scholar] [CrossRef]
- Wang, Y.; Kory, N.; BasuRay, S.; Cohen, J.C.; Hobbs, H.H. PNPLA3, CGI-58, and Inhibition of Hepatic Triglyceride Hydrolysis in Mice. Hepatology 2019, 69, 2427–2441. [Google Scholar] [CrossRef]
- Haemmerle, G.; Lass, A.; Zimmermann, R.; Gorkiewicz, G.; Meyer, C.; Rozman, J.; Heldmaier, G.; Maier, R.; Theussl, C.; Eder, S.; et al. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science 2006, 312, 734–737. [Google Scholar] [CrossRef]
- Sztalryd, C.; Brasaemle, D.L. The perilipin family of lipid droplet proteins: Gatekeepers of intracellular lipolysis. Biochim. Biophys. Acta Mol. Cell. Biol. Lipids 2017, 1862, 1221–1232. [Google Scholar] [CrossRef]
- Heid, H.; Rickelt, S.; Zimbelmann, R.; Winter, S.; Schumacher, H.; Dorflinger, Y. Lipid droplets, perilipins and cytokeratins–unravelled liaisons in epithelium-derived cells. PLoS ONE 2013, 8, e63061. [Google Scholar] [CrossRef] [PubMed]
- MacPherson, R.E.; Ramos, S.V.; Vandenboom, R.; Roy, B.D.; Peters, S.J. Skeletal muscle PLIN proteins, ATGL and CGI-58, interactions at rest and following stimulated contraction. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2013, 304, R644–R650. [Google Scholar] [CrossRef]
- Tansey, J.T.; Sztalryd, C.; Gruia-Gray, J.; Roush, D.L.; Zee, J.V.; Gavrilova, O.; Reitman, M.L.; Deng, C.X.; Li, C.; Kimmel, A.R.; et al. Perilipin ablation results in a lean mouse with aberrant adipocyte lipolysis, enhanced leptin production, and resistance to diet-induced obesity. Proc. Natl. Acad. Sci. USA 2001, 98, 6494–6499. [Google Scholar] [CrossRef]
- Martinez-Botas, J.; Anderson, J.B.; Tessier, D.; Lapillonne, A.; Chang, B.H.; Quast, M.J.; Gorenstein, D.; Chen, K.H.; Chan, L. Absence of perilipin results in leanness and reverses obesity in Lepr(db/db) mice. Nat. Genet. 2000, 26, 474–479. [Google Scholar] [CrossRef] [PubMed]
- Chang, B.H.; Li, L.; Paul, A.; Taniguchi, S.; Nannegari, V.; Heird, W.C.; Chan, L. Protection against fatty liver but normal adipogenesis in mice lacking adipose differentiation-related protein. Mol. Cell. Biol. 2006, 26, 1063–1076. [Google Scholar] [CrossRef]
- Kuramoto, K.; Okamura, T.; Yamaguchi, T.; Nakamura, T.Y.; Wakabayashi, S.; Morinaga, H.; Nomura, M.; Yanase, T.; Otsu, K.; Usuda, N.; et al. Perilipin 5, a lipid droplet-binding protein, protects heart from oxidative burden by sequestering fatty acid from excessive oxidation. J. Biol. Chem. 2012, 287, 23852–23863. [Google Scholar] [CrossRef] [PubMed]
- Gandotra, S.; Le Dour, C.; Bottomley, W.; Cervera, P.; Giral, P.; Reznik, Y.; Charpentier, G.; Auclair, M.; Delepine, M.; Barroso, I.; et al. Perilipin deficiency and autosomal dominant partial lipodystrophy. N. Engl. J. Med. 2011, 364, 740–748. [Google Scholar] [CrossRef]
- Straub, B.K.; Stoeffel, P.; Heid, H.; Zimbelmann, R.; Schirmacher, P. Differential pattern of lipid droplet-associated proteins and de novo perilipin expression in hepatocyte steatogenesis. Hepatology 2008, 47, 1936–1946. [Google Scholar] [CrossRef]
- Straub, B.K.; Herpel, E.; Singer, S.; Zimbelmann, R.; Breuhahn, K.; Macher-Goeppinger, S.; Warth, A.; Lehmann-Koch, J.; Longerich, T.; Heid, H.; et al. Lipid droplet-associated PAT-proteins show frequent and differential expression in neoplastic steatogenesis. Mod. Pathol. 2010, 23, 480–492. [Google Scholar] [CrossRef] [PubMed]
- Pawella, L.M.; Hashani, M.; Eiteneuer, E.; Renner, M.; Bartenschlager, R.; Schirmacher, P.; Straub, B.K. Perilipin discerns chronic from acute hepatocellular steatosis. J. Hepatol. 2014, 60, 633–642. [Google Scholar] [CrossRef]
- Hashani, M.; Witzel, H.R.; Pawella, L.M.; Lehmann-Koch, J.; Schumacher, J.; Mechtersheimer, G.; Schnolzer, M.; Schirmacher, P.; Roth, W.; Straub, B.K. Widespread expression of perilipin 5 in normal human tissues and in diseases is restricted to distinct lipid droplet subpopulations. Cell Tissue Res. 2018, 374, 121–136. [Google Scholar] [CrossRef] [PubMed]
- Rausch, V.; Mueller, S. Suppressed Fat Mobilization Due to PNPLA3 rs738409 -Associated Liver Damage in Heavy Drinkers: The Liver Damage Feedback Hypothesis. Adv. Exp. Med. Biol. 2018, 1032, 153–172. [Google Scholar] [CrossRef] [PubMed]
- Witzel, H.R.; Jungblut, B.; Choe, C.P.; Crump, J.G.; Braun, T.; Dobreva, G. The LIM protein Ajuba restricts the second heart field progenitor pool by regulating Isl1 activity. Dev. Cell 2012, 23, 58–70. [Google Scholar] [CrossRef] [PubMed]
- Bankhead, P.; Loughrey, M.B.; Fernandez, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef]
- Bedossa, P.; Poitou, C.; Veyrie, N.; Bouillot, J.L.; Basdevant, A.; Paradis, V.; Tordjman, J.; Clement, K. Histopathological algorithm and scoring system for evaluation of liver lesions in morbidly obese patients. Hepatology 2012, 56, 1751–1759. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef]
- Granneman, J.G.; Moore, H.P.; Mottillo, E.P.; Zhu, Z.; Zhou, L. Interactions of perilipin-5 (Plin5) with adipose triglyceride lipase. J. Biol. Chem. 2011, 286, 5126–5135. [Google Scholar] [CrossRef]
- Smirnova, E.; Goldberg, E.B.; Makarova, K.S.; Lin, L.; Brown, W.J.; Jackson, C.L. ATGL has a key role in lipid droplet/adiposome degradation in mammalian cells. EMBO Rep. 2006, 7, 106–113. [Google Scholar] [CrossRef]
- Jenkins, C.M.; Mancuso, D.J.; Yan, W.; Sims, H.F.; Gibson, B.; Gross, R.W. Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J. Biol. Chem. 2004, 279, 48968–48975. [Google Scholar] [CrossRef]
- Zimmermann, R.; Strauss, J.G.; Haemmerle, G.; Schoiswohl, G.; Birner-Gruenberger, R.; Riederer, M.; Lass, A.; Neuberger, G.; Eisenhaber, F.; Hermetter, A.; et al. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science 2004, 306, 1383–1386. [Google Scholar] [CrossRef]
- Schratter, M.; Lass, A.; Radner, F.P.W. ABHD5-A Regulator of Lipid Metabolism Essential for Diverse Cellular Functions. Metabolites 2022, 12, 1015. [Google Scholar] [CrossRef] [PubMed]
- Brasaemle, D.L.; Rubin, B.; Harten, I.A.; Gruia-Gray, J.; Kimmel, A.R.; Londos, C. Perilipin A increases triacylglycerol storage by decreasing the rate of triacylglycerol hydrolysis. J. Biol. Chem. 2000, 275, 38486–38493. [Google Scholar] [CrossRef]
- Perttila, J.; Huaman-Samanez, C.; Caron, S.; Tanhuanpaa, K.; Staels, B.; Yki-Jarvinen, H.; Olkkonen, V.M. PNPLA3 is regulated by glucose in human hepatocytes, and its I148M mutant slows down triglyceride hydrolysis. Am. J. Physiol. Endocrinol. Metab. 2012, 302, E1063–E1069. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.; Mottillo, E.P.; Mladenovic-Lucas, L.; Zhou, L.; Granneman, J.G. Dynamic interactions of ABHD5 with PNPLA3 regulate triacylglycerol metabolism in brown adipocytes. Nat. Metab. 2019, 1, 560–569. [Google Scholar] [CrossRef] [PubMed]
- Oliner, J.D.; Kinzler, K.W.; Vogelstein, B. In vivo cloning of PCR products in E. coli. Nucleic Acids Res. 1993, 21, 5192–5197. [Google Scholar] [CrossRef] [PubMed]

| PNPLA3 Status | Number of Cases | Sex (% Male) | Age | Brunt-Score (1–3) | Ballooning (0–2) | NAS-Score (0–7) | Inflammation (0–3) | Fibrosis (% Cirrhosis) | |
|---|---|---|---|---|---|---|---|---|---|
| NASH | C/C | 7 | 42.9 | 54 ± 14 | 2.00 ± 0.58 | 1.14 ± 0.38 | 4.83 ± 1.47 | 1.00 ± 0.58 | 14.3 |
| C/G | 10 | 50 | 48 ± 11 | 2.00 ± 0.00 | 1.40 ± 0.52 | 4.78 ± 1.30 | 1.50 ± 0.71 | 20.0 | |
| G/G | 6 | 50 | 43 ± 15 | 2.67 ± 0.52 | 1.00 ± 0.00 | 4.43 ± 1.51 | 1.00 ± 0.63 | 0.0 | |
| C/C | 6 | 66.7 | 54 ± 10 | 1.50 ± 0.84 | 1.83 ± 0.41 | 6.00 ± 1.26 | 2.67 ± 0.52 | 83.3 | |
| ASH | C/G | 8 | 75 | 46 ± 12 | 2.50 ± 0.76 | 1.88 ± 0.35 | 6.50 ± 1.20 | 2.25 ± 0.46 | 88.9 |
| G/G | 1 | 100 | 70 | 2.00 | 2.00 | 7.00 | 3.00 | 100 | |
| C/C | 2 | 0 | 64 ± 13 | 2.00 ± 1.41 | 1.50 ± 0.71 | 5.00 ± 2.83 | 1.50 ± 0.71 | 50.0 | |
| BASH | C/G | 1 | 0 | 59 | 1.00 | 2.00 | 5.00 | 2.00 | 100.0 |
| G/G | 1 | 100 | 31 | 3.00 | 2.00 | 6.00 | 1.00 | 0.0 | |
| C/C | 3 | 33.3 | 52 ± 7 | 1.00 ± 0.00 | 0.67 ± 0.58 | 2.67 ± 0.58 | 1.00 ± 0.00 | 33.0 | |
| HCV | C/G | 3 | 0 | 66 ± 9 | 1.00 ± 0.00 | 1.00 ± 0.00 | 3.00 ± 0.00 | 1.00 ± 0.00 | 66.0 |
| G/G | 3 | 33.3 | 53 ± 6 | 1.33 ± 0.58 | 0.67 ± 0.58 | 3.00 ± 2.00 | 1.00 ± 1.00 | 100 | |
| C/C | 1 | 100 | n.a. | 1.00 | 0.00 | 1.00 | 0.00 | 0.0 | |
| Control | C/G | 1 | n.a. | n.a. | 1.00 | 0.00 | 0.00 | 0.00 | 0.0 |
| G/G | 1 | n.a. | 14 | 2.00 | 0.00 | 4.00 | 1.00 | 0.0 | |
| Unclear | C/C | 2 | 100 | 46 ± 0 | 2.50 ± 0.71 | 1.00 ± 0.00 | 5.00 ± 1.41 | 1.50 ± 0.71 | 50.0 |
| C/G | 2 | 50 | 19 ± 2 | 2.50 ± 0.71 | 1.50 ± 0.71 | 5.00 ± 0.00 | 1.00 ± 0.00 | 0.0 | |
| n.a. | 1 | 100 | 34 | 3.00 ± 0.00 | 1.00 ± 0.00 | 5.00 | 1.00 ± 0.00 | 0.0 |
| PNPLA3 Status | Steatosis (%) | Perilipin 1 (0–3) | Perilipin 2 (0–3) | Perilipin 3 (0–3) | Perilipin 4 (0–3) | Perilipin 5 (0–3) | |
|---|---|---|---|---|---|---|---|
| NASH | C/C | 43.6 ± 20.6 | 2.50 ± 0.29 | 2.71 ± 0.27 | 0.79 ± 0.27 | 0.79 ± 0.27 | 1.07 ± 0.19 |
| C/G | 48.5 ± 11.3 | 2.55 ± 0.64 | 2.70 ± 0.26 | 0.95 ± 0.16 | 0.70 ± 0.26 | 1.30 ± 0.63 | |
| G/G | 65.0 ± 21.5 | 2.08 ± 0.38 | 2.58 ± 0.38 | 0.92 ± 0.20 | 0.67 ± 0.41 | 1.17 ± 0.41 | |
| C/C | 29.2 ± 27.1 | 0.83 ± 0.61 | 2.50 ± 0.45 | 2.17 ± 0.41 | 2.00 ± 0.00 | 2.00 ± 0.00 | |
| ASH | C/G | 56.3 ± 22.5 | 1.19 ± 0.46 | 2.72 ± 0.53 | 2.00 ± 0.38 | 2.00 ± 0.00 | 2.00 ± 0.00 |
| G/G | 35.0 | 1.00 | 2.50 | 1.50 | 2.00 | 2.00 | |
| C/C | 52.5 ± 38.9 | 0.75 ± 0.35 | 2.75 ± 0.35 | 1.25 ± 0.35 | 2.00 ± 0.00 | 2.00 ± 0.00 | |
| BASH | C/G | 25.0 | 2.00 | 2.00 | 1.50 | 2.00 | 2.00 |
| G/G | 80.0 | 0.50 | 3.00 | 2.50 | 2.00 | 2.00 | |
| C/C | 18.3 ± 7.6 | 1.16 ± 0.29 | 2.00 ± 0.00 | 1.50 ± 0.00 | 2.00 ± 0.00 | 2.00 ± 0.00 | |
| HCV | C/G | 16.7 ± 12.6 | 1.26 ± 0.76 | 2.33 ± 0.29 | 1.50 ± 0.00 | 2.00 ± 0.00 | 2.00 ± 0.00 |
| G/G | 23.3 ± 12.6 | 1.16 ± 0.29 | 2.33 ± 0.29 | 1.50 ± 0.00 | 2.00 ± 0.00 | 2.00 ± 0.00 | |
| C/C | 15.0 | 2.00 | 3.00 | 2.00 | 2.00 | 2.00 | |
| Control | C/G | 5.0 | 1.50 | 1.50 | 1.50 | 2.00 | 2.00 |
| G/G | 60.0 | 2.00 | 2.50 | 1.00 | 2.00 | 2.00 | |
| Unclear | C/C | 40.0 ± 21.0 | 1.50 ± 0.71 | 2.75 ± 0.35 | 1.75 ± 0.35 | 2.00 ± 0.00 | 2.00 ± 0.00 |
| C/G | 42.5 ± 38.9 | 1.50 ± 0.71 | 3.00 ± 0.00 | 1.50 ± 0.00 | 2.00 ± 0.00 | 2.00 ± 0.00 | |
| n.a. | 55.0 | 2.00 | 3.00 | 2.50 | 2.00 | 2.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Witzel, H.R.; Schwittai, I.M.G.; Hartmann, N.; Mueller, S.; Schattenberg, J.M.; Gong, X.-M.; Backs, J.; Schirmacher, P.; Schuppan, D.; Roth, W.; et al. PNPLA3(I148M) Inhibits Lipolysis by Perilipin-5-Dependent Competition with ATGL. Cells 2023, 12, 73. https://doi.org/10.3390/cells12010073
Witzel HR, Schwittai IMG, Hartmann N, Mueller S, Schattenberg JM, Gong X-M, Backs J, Schirmacher P, Schuppan D, Roth W, et al. PNPLA3(I148M) Inhibits Lipolysis by Perilipin-5-Dependent Competition with ATGL. Cells. 2023; 12(1):73. https://doi.org/10.3390/cells12010073
Chicago/Turabian StyleWitzel, Hagen Roland, Inga Maria Gertrud Schwittai, Nils Hartmann, Sebastian Mueller, Jörn M. Schattenberg, Xue-Min Gong, Johannes Backs, Peter Schirmacher, Detlef Schuppan, Wilfried Roth, and et al. 2023. "PNPLA3(I148M) Inhibits Lipolysis by Perilipin-5-Dependent Competition with ATGL" Cells 12, no. 1: 73. https://doi.org/10.3390/cells12010073
APA StyleWitzel, H. R., Schwittai, I. M. G., Hartmann, N., Mueller, S., Schattenberg, J. M., Gong, X.-M., Backs, J., Schirmacher, P., Schuppan, D., Roth, W., & Straub, B. K. (2023). PNPLA3(I148M) Inhibits Lipolysis by Perilipin-5-Dependent Competition with ATGL. Cells, 12(1), 73. https://doi.org/10.3390/cells12010073









