Changes in miR-124-1, miR-212, miR-132, miR-134, and miR-155 Expression Patterns after 7,12-Dimethylbenz(a)anthracene Treatment in CBA/Ca Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Treatment
2.2. Isolation of Total RNA
2.3. Reverse Transcription Polymerase Chain Reaction (RT-PCR)
2.4. Calculations and Statistical Analysis
3. Results
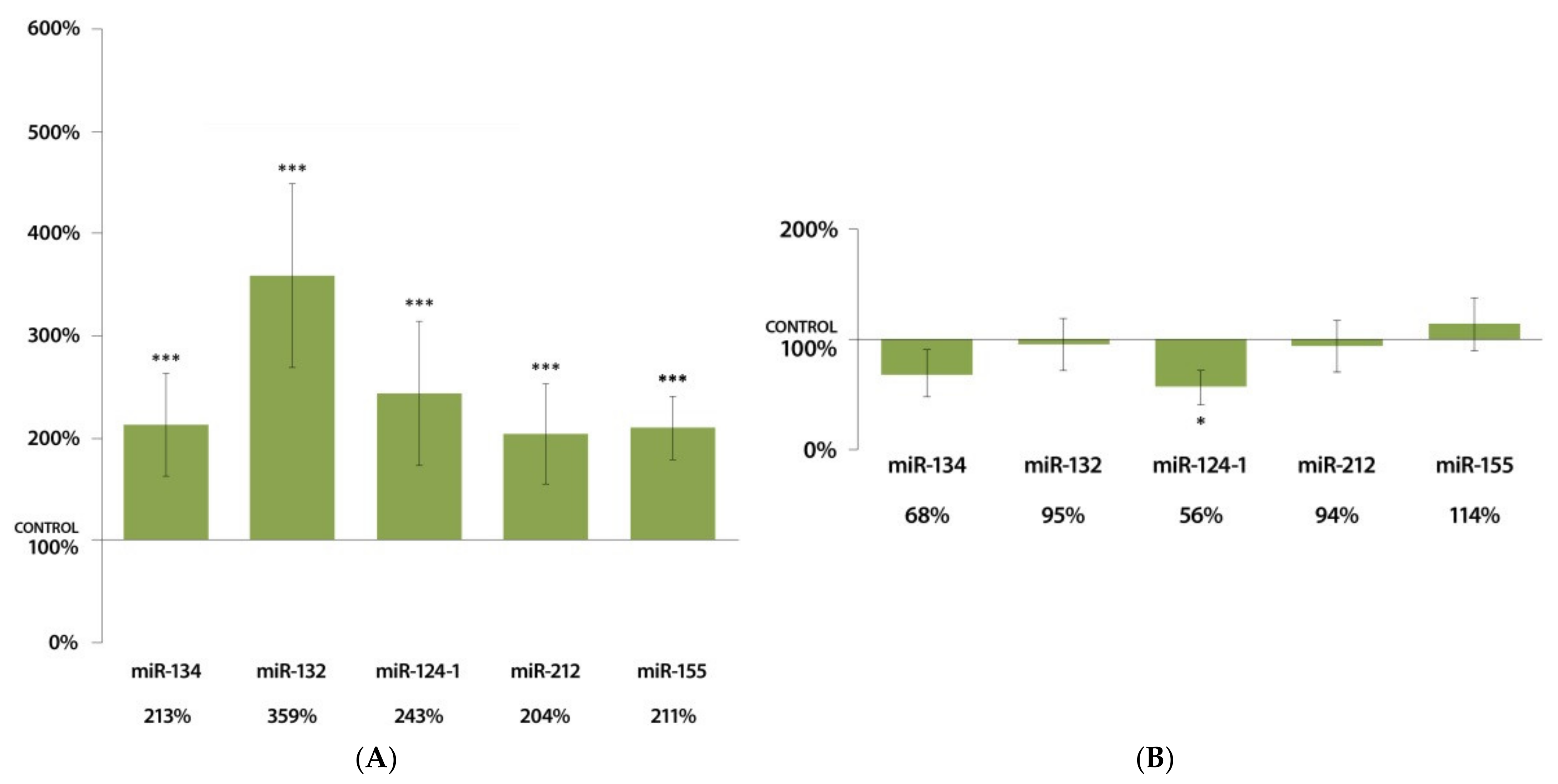
3.1. Changes in miRNA Expression in the Liver 24 h after DMBA Treatment
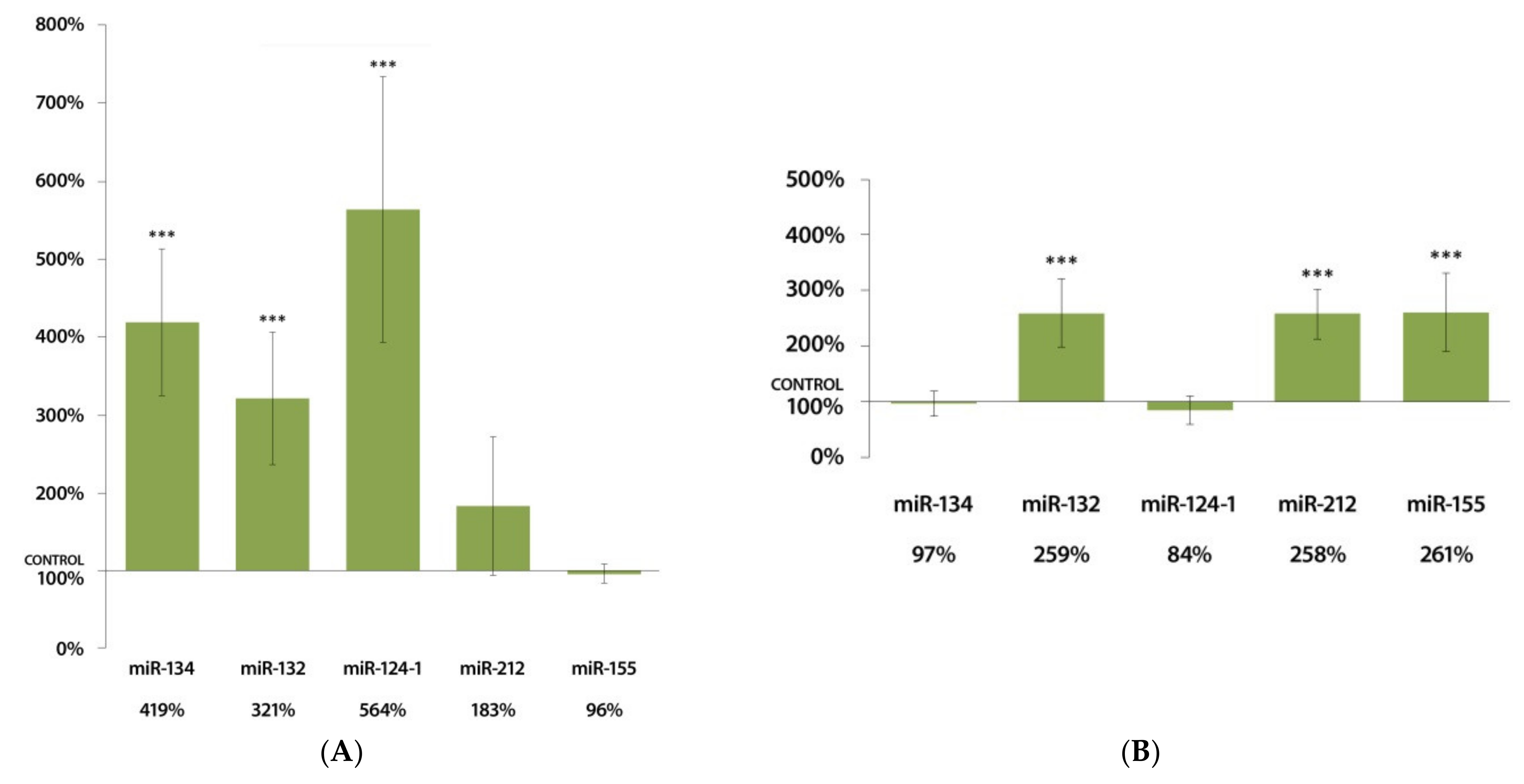
3.2. Changes in miRNA Expression in the Spleen 24 h after DMBA Treatment
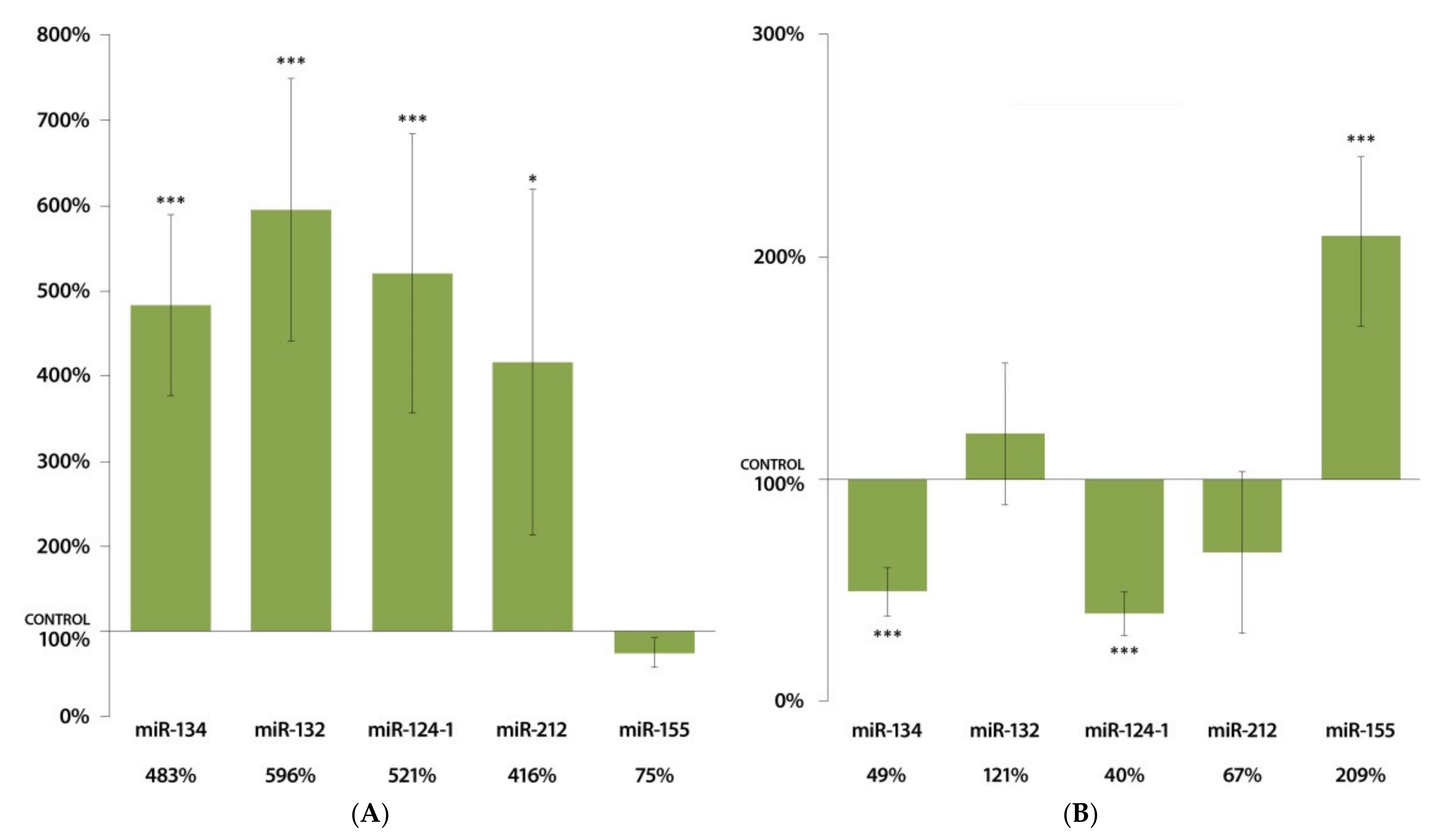
3.3. Changes in miRNA Expression in the Kidneys 24 h after DMBA Treatment
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Danaei, G.; Vander Hoorn, S.; Lopez, A.D.; Murray, C.J.; Ezzati, M. Comparative Risk Assessment Collaborating Group (Cancers). Causes of cancer in the world: Comparative risk assessment of nine behavioural and environmental risk factors. Lancet 2005, 366, 1784–1793. [Google Scholar] [CrossRef]
- Frenk, S.; Houseley, J. Gene expression hallmarks of cellular ageing. Biogerontology 2018, 19, 547–566. [Google Scholar] [CrossRef]
- Kwa, M.; Makris, A.; Esteva, F.J. Clinical utility of gene-expression signatures in early stage breast cancer. Nat. Rev. Clin. Oncol. 2017, 14, 595–610. [Google Scholar] [CrossRef]
- Yang, Z.-Y.; Liu, X.-Y.; Shu, J.; Zhang, H.; Ren, Y.-Q.; Xu, Z.-B.; Liang, Y. Multi-view based integrative analysis of gene expression data for identifying biomarkers. Sci. Rep. 2019, 9, 1775–1781. [Google Scholar] [CrossRef]
- Budán, F.; Varjas, T.; Nowrasteh, G.; Varga, Z.; Boncz, I.; Cseh, J.; Prantner, I.; Antal, T.; Pázsit, E.; Gobel, G.; et al. Early modification of c-myc, Ha-ras and p53 expressions by N-methyl-N-nitrosourea. Vivo 2008, 22, 793–797. [Google Scholar]
- Tomesz, A.; Szabo, L.; Molnar, R.; Deutsch, A.; Darago, R.; Mathe, D.; Budan, F.; Ghodratollah, N.; Varjas, T.; Nemeth, B.; et al. Effect of 7,12-Dimethylbenz(α)anthracene on the Expression of miR-330, miR-29a, miR-9-1, miR-9-3 and the mTORC1 Gene in CBA/Ca Mice. Vivo 2020, 34, 2337–2343. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Rajewsky, N. microRNA target predictions in animals. Nat. Genet. 2006, 38, S8–S13. [Google Scholar] [CrossRef]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef]
- Calin, G.A.; Sevignani, C.; Dumitru, C.D.; Hyslop, T.; Noch, E.; Yendamuri, S.; Shimizu, M.; Rattan, S.; Bullrich, F.; Negrini, M.; et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA 2004, 101, 2999–3004. [Google Scholar] [CrossRef] [PubMed]
- Visone, R.; Croce, C.M. MiRNAs and Cancer. Am. J. Pathol. 2009, 174, 1131–1138. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Huo, X.; Davuljigari, C.B.; Dai, Q.; Xu, X. MicroRNAs and their role in environmental chemical carcinogenesis. Environ. Geochem. Health 2019, 41, 225–247. [Google Scholar] [CrossRef] [PubMed]
- Ji, Q.; Hao, X.; Meng, Y.; Zhang, M.; DeSano, J.; Fan, D.; Xu, L. Restoration of tumor suppressor miR-34 inhibits human p53-mutant gastric cancer tumorspheres. BMC Cancer 2008, 8, 266. [Google Scholar] [CrossRef]
- Sun, Y.-M.; Lin, K.-Y.; Chen, Y.-Q. Diverse functions of miR-125 family in different cell contexts. J. Hematol. Oncol. 2013, 6, 314. [Google Scholar] [CrossRef]
- Rajabi, H.; Jin, C.; Ahmad, R.; McClary, A.C.; Joshi, M.D.; Kufe, D. Mucin 1 Oncoprotein Expression Is Suppressed by the miR-125b Oncomir. Genes Cancer 2010, 1, 62–68. [Google Scholar] [CrossRef]
- Bi, Q.; Tang, S.; Xia, L.; Du, R.; Fan, R.; Gao, L.; Jin, J.; Liang, S.; Chen, Z.; Xu, G.; et al. Ectopic Expression of MiR-125a Inhibits the Proliferation and Metastasis of Hepatocellular Carcinoma by Targeting MMP11 and VEGF. PLoS ONE 2012, 7, e40169. [Google Scholar] [CrossRef]
- Furuta, M.; Kozaki, K.-I.; Tanaka, S.; Arii, S.; Imoto, I.; Inazawa, J. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis 2010, 31, 766–776. [Google Scholar] [CrossRef]
- Lujambio, A.; Esteller, M. CpG island hypermethylation of tumor suppressor microRNAs in human cancer. Cell Cycle 2007, 6, 1455–1459. [Google Scholar] [CrossRef]
- Alzahrani, A.; Hanieh, H. Differential modulation of Ahr and Arid5a: A promising therapeutic strategy for autoimmune encephalomyelitis. Saudi Pharm. J. 2020, 28, 1605–1615. [Google Scholar] [CrossRef]
- Chen, Y.; Zhou, Y.; Han, F.; Zhao, Y.; Tu, M.; Wang, Y.; Huang, C.; Fan, S.; Chen, P.; Yao, X.; et al. A novel miR-1291-ERRα-CPT1C axis modulates tumor cell proliferation, metabolism and tumorigenesis. Theranostics 2020, 10, 7193–7210. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Song, J.; Bian, H.; Yang, X.; Xie, X.; Zhu, Q.; Qin, C.; Qi, J. The functions and targets of miR-212 as a potential biomarker of cancer diagnosis and therapy. J. Cell. Mol. Med. 2020, 24, 2392–2401. [Google Scholar] [CrossRef] [PubMed]
- Hanieh, H.; Ahmed, E.A.; Vishnubalaji, R.; Alajez, N. SOX4: Epigenetic regulation and role in tumorigenesis. Semin. Cancer Biol. 2020, 67, 91–104. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, A.; Feng, X.; Tian, L.; Bo, W.; Wang, H.; Hu, Y. MiR-132 promotes the proliferation, invasion and migration of human pancreatic carcinoma by inhibition of the tumor suppressor gene PTEN. Prog. Biophys. Mol. Biol. 2019, 148, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Zhang, X.; Chen, Q.; Bao, Y.; Dong, C.; Wang, X. miR-132 suppresses the migration and invasion of lung cancer cells by blocking USP9X-induced epithelial-mesenchymal transition. Am. J. Transl. Res. 2018, 10, 224–234. [Google Scholar] [PubMed]
- Sun, C.-C.; Li, S.-J.; Li, D.-J. Hsa-miR-134 suppresses non-small cell lung cancer (NSCLC) development through down-regulation of CCND1. Oncotarget 2016, 7, 35960–35978. [Google Scholar] [CrossRef] [PubMed]
- Qin, Q.; Wei, F.; Zhang, J.; Wang, X.; Li, B. miR-134 inhibits non-small cell lung cancer growth by targeting the epidermal growth factor receptor. J. Cell. Mol. Med. 2016, 20, 1974–1983. [Google Scholar] [CrossRef]
- Park, S.; Eom, K.; Kim, J.; Bang, H.; Wang, H.Y.; Ahn, S.; Kim, G.; Jang, H.; Kim, S.; Lee, D.; et al. MiR 9, miR 21, and miR 155 as potential biomarkers for HPV positive and negative cervical cancer. BMC Cancer 2017, 17, 658. [Google Scholar] [CrossRef]
- Bosland, M.C.; Prinsen, M.K. Induction of dorsolateral prostate adenocarcinomas and other accessory sex gland lesions in male Wistar rats by a single administration of N methyl N nitrosourea, 7,12 dimethylbenz(a)anthracene, and 3,2′ dimethyl 4 aminobiphenyl after sequential treatment with cyproterone acetate and testosterone propionate. Cancer Res. 1990, 50, 691–699. [Google Scholar]
- El-Sohemy, A.; Archer, M.C. Inhibition of N-methyl-N-nitrosourea- and 7,12-dimethylbenz[a] anthracene-induced rat mammary tumorigenesis by dietary cholesterol is independent of Ha-ras mutations. Carcinogenesis 2000, 21, 827–831. [Google Scholar] [CrossRef][Green Version]
- Lee, L.; Lee, J.; Waldman, S.; Casper, R.; Grynpas, M. Polycyclic aromatic hydrocarbons present in cigarette smoke cause bone loss in an ovariectomized rat model. Bone 2002, 30, 917–923. [Google Scholar] [CrossRef]
- Gao, J.; Lauer, F.T.; Dunaway, S.; Burchiel, S.W. Cytochrome P450 1B1 Is Required for 7,12-Dimethylbenz(a)-anthracene (DMBA) Induced Spleen Cell Immunotoxicity. Toxicol. Sci. 2005, 86, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Lindhe, O.; Granberg, L.; Brandt, I. Target cells for cytochrome p450 catalysed irreversible binding of 7,12 dime-thylbenz[a]anthracene (DMBA) in rodent adrenal glands. Arch Toxicol. 2002, 76, 460–466. [Google Scholar] [CrossRef] [PubMed]
- Hosny, S.; Sahyon, H.; Youssef, M.; Negm, A. Oleanolic Acid Suppressed DMBA-Induced Liver Carcinogenesis through Induction of Mitochondrial-Mediated Apoptosis and Autophagy. Nutr. Cancer 2020, 73, 968–982. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Jia, Z.; Trush, M.A. Defining ROS in biology and medicine. React. Oxyg. Species 2016, 1, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Currier, N.; Solomon, S.E.; Demicco, E.G.; Chang, D.L.F.; Farago, M.; Ying, H.; Dominguez, I.; Sonenshein, G.E.; Cardiff, R.D.; Xiao, Z.-X.J.; et al. Oncogenic Signaling Pathways Activated in DMBA-Induced Mouse Mammary Tumors. Toxicol. Pathol. 2005, 33, 726–737. [Google Scholar] [CrossRef]
- Li, N.; Chen, X.; Liao, J.; Yang, G.; Wang, S.; Josephson, Y.; Han, C.; Chen, J.; Huang, M.-T.; Yang, C.S. Inhibition of 7,12-dimethylbenz[a]anthracene (DMBA)-induced oral carcinogenesis in hamsters by tea and curcumin. Carcinogenesis 2002, 23, 1307–1313. [Google Scholar] [CrossRef]
- Budán, F.; Varjas, T.; Nowrasteh, G.; Prantner, I.; Varga, Z.; Ember, A.; Cseh, J.; Gombos, K.; Pázsit, E.; Gobel, G.; et al. Early modification of c-myc, Ha-ras and p53 expressions by chemical carcinogens (DMBA, MNU). Vivo 2009, 23, 591–598. [Google Scholar]
- Juhász, K.; Gombos, K.; Szirmai, M.; Révész, P.; Magda, I.; Gocze, K.; Ember, I. DMBA induces deregulation of miRNA expression of let-7, miR-21 and miR-146a in CBA/CA mice. Vivo 2012, 26, 113–117. [Google Scholar]
- Volinia, S.; Calin, G.A.; Liu, C.G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef]
- He, H.; Jazdzewski, K.; Li, W.; Liyanarachchi, S.; Nagy, R.; Volinia, S.; Calin, G.A.; Liu, C.-G.; Franssila, K.; Suster, S.; et al. The role of microRNA genes in papillary thyroid carcinoma. Proc. Natl. Acad. Sci. USA 2005, 102, 19075–19080. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef] [PubMed]
- Shor, B.; Cavender, D.; Harris, C. A kinase dead knock in mutation in mTOR leads to early embryonic lethality and is dis-pensable for the immune system in heterozygous mice. BMC Immunol. 2009, 10, 28. [Google Scholar] [CrossRef]
- Uchida, S.; Hara, K.; Kobayashi, A.; Funato, H.; Hobara, T.; Otsuki, K.; Yamagata, H.; McEwen, B.S.; Watanabe, Y. Early Life Stress Enhances Behavioral Vulnerability to Stress through the Activation of REST4-Mediated Gene Transcription in the Medial Prefrontal Cortex of Rodents. J. Neurosci. 2010, 30, 15007–15018. [Google Scholar] [CrossRef]
- Kong, Y.-H.; Xu, S.-P. Salidroside prevents skin carcinogenesis induced by DMBA/TPA in a mouse model through suppression of inflammation and promotion of apoptosis. Oncol. Rep. 2018, 39, 2513–2526. [Google Scholar] [CrossRef]
- Wang, Y.; Han, B.; Wang, Y.; Wang, C.; Zhang, H.; Xue, J.; Wang, X.; Niu, T.; Niu, Z.; Chen, Y. Mesenchymal stem cell–secreted extracellular vesicles carrying TGF-β1 up-regulate miR-132 and promote mouse M2 macrophage polarization. J. Cell. Mol. Med. 2020, 24, 12750–12764. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhu, Q.; Lu, L.; Liu, Y. MiR-132 inhibits migration and invasion and increases chemosensitivity of cisplatin-resistant oral squamous cell carcinoma cells via targeting TGF-β1. Bioengineered 2020, 11, 91–102. [Google Scholar] [CrossRef]
- Huang, J.; Lu, D.; Xiang, T.; Wu, X.; Ge, S.; Wang, Y.; Wang, J.; Cheng, N. MicroRNA-132-3p regulates cell proliferation, apoptosis, migration and invasion of liver cancer by targeting Sox4. Oncol. Lett. 2020, 19, 3173–3180. [Google Scholar] [CrossRef]
- Foronda, M.; Martínez, P.; Schoeftner, S.; López, G.G.; Schneider, R.; Flores, J.M.; Pisano, D.; Blasco, M.A. Sox4 Links Tumor Suppression to Accelerated Aging in Mice by Modulating Stem Cell Activation. Cell Rep. 2014, 8, 487–500. [Google Scholar] [CrossRef]
- Sobinoff, A.P.; Mahony, M.; Nixon, B.; Roman, S.D.; McLaughlin, E.A. Understanding the Villain: DMBA-Induced Preantral Ovotoxicity Involves Selective Follicular Destruction and Primordial Follicle Activation through PI3K/Akt and mTOR Signaling. Toxicol. Sci. 2011, 123, 563–575. [Google Scholar] [CrossRef]
- Deng, J.-H.; Zheng, G.-Y.; Li, H.-Z.; Ji, Z.-G. MiR-212-5p inhibits the malignant behavior of clear cell renal cell carcinoma cells by targeting TBX15. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 10699–10707. [Google Scholar] [PubMed]
- Gu, C.; Wang, Z.; Jin, Z.; Li, G.; Kou, Y.; Jia, Z.; Yang, J.; Tian, F. MicroRNA-212 inhibits the proliferation, migration and invasion of renal cell carcinoma by targeting X-linked inhibitor of apoptosis protein (XIAP). Oncotarget 2017, 8, 92119–92133. [Google Scholar] [CrossRef] [PubMed]
- Dou, C.; Wang, Y.; Li, C.; Liu, Z.; Jia, Y.; Li, Q.; Yang, W.; Yao, Y.; Liu, Q.; Tu, K. MicroRNA-212 suppresses tumor growth of human hepatocellular carcinoma by targeting FOXA1. Oncotarget 2015, 6, 13216–13228. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Li, Z. Interplay of estrogen receptors and FOXA factors in the liver cancer. Mol. Cell. Endocrinol. 2015, 418 Pt 3, 334–339. [Google Scholar] [CrossRef]
- Li, Z.; Tuteja, G.; Schug, J.; Kaestner, K.H. Foxa1 and Foxa2 Are Essential for Sexual Dimorphism in Liver Cancer. Cell 2012, 148, 72–83. [Google Scholar] [CrossRef]
- Zhang, C.; Zhao, J.; Deng, H. 17β-Estradiol up-regulates miR-155 expression and reduces TP53INP1 expression in MCF-7 breast cancer cells. Mol. Cell. Biochem. 2013, 379, 201–211. [Google Scholar] [CrossRef]
- Granberg, A.L.; Brunström, B.; Brandt, I. Cytochrome P450-dependent binding of 7,12-dimethylbenz[a]anthracene (DMBA) and benzo[a]pyrene (B[a]P) in murine heart, lung, and liver endothelial cells. Arch. Toxicol. 2000, 74, 593–601. [Google Scholar] [CrossRef]
- Yuan, K.; Zhang, X.; Lv, L.; Zhang, J.; Liang, W.; Wang, P. Fine-tuning the expression of microRNA-155 controls acetaminophen-induced liver inflammation. Int. Immunopharmacol. 2016, 40, 339–346. [Google Scholar] [CrossRef]
- Kong, W.; He, L.; Richards, E.J.; Challa, S.; Xu, C.-X.; Permuthwey, J.; Lancaster, J.M.; Coppola, D.M.; Sellers, T.; Djeu, J.Y.; et al. Upregulation of miRNA-155 promotes tumour angiogenesis by targeting VHL and is associated with poor prognosis and triple-negative breast cancer. Oncogene 2013, 33, 679–689. [Google Scholar] [CrossRef]
- Gironella, M.; Seux, M.; Xie, M.-J.; Cano, C.; Tomasini, R.; Gommeaux, J.; Garcia, S.; Nowak, J.; Yeung, M.L.; Jeang, K.-T.; et al. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc. Natl. Acad. Sci. USA 2007, 104, 16170–16175. [Google Scholar] [CrossRef]
- Xu, L.; Dai, W.; Li, J.; He, L.; Wang, F.; Xia, Y.; Chen, K.; Li, S.; Liu, T.; Lu, J.; et al. Methylation-regulated miR-124-1 suppresses tumorigenesis in hepatocellular carcinoma by targeting CASC3. Oncotarget 2016, 7, 26027–26041. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Kang, J.; Sun, S.; Luo, Y.; Ji, X.; Zeng, X.; Zhao, S. iASPP, a microRNA-124 target, is aberrantly expressed in astrocytoma and regulates malignant glioma cell migration and viability. Mol. Med. Rep. 2017, 17, 1970–1978. [Google Scholar] [CrossRef] [PubMed]
- Kawano, S.; Nakamachi, Y. miR-124a as a key regulator of proliferation and MCP-1 secretion in synoviocytes from patients with rheumatoid arthritis. Ann. Rheum. Dis. 2011, 70 (Suppl. 1), i88–i91. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.-J.; Ye, D.-J.; Baek, H.-S.; Chun, Y.-J. 7,12-Dimethylbenz[α]anthracene increases cell proliferation and invasion through induction of Wnt/β-catenin signaling and EMT process. Environ. Toxicol. 2018, 33, 729–742. [Google Scholar] [CrossRef] [PubMed]
- Pang, Y.; Wu, J.; Li, X.; Wang, C.; Wang, M.; Liu, J.; Yang, G. NEAT1/miR-124/STAT3 feedback loop promotes breast cancer progression. Int. J. Oncol. 2019, 55, 745–754. [Google Scholar] [CrossRef]
- Ho, C.-H.; Lu, Y.-C.; Fan, C.-K.; Yu, H.-J.; Liu, H.-T.; Wu, C.-C.; Chen, K.-C.; Liu, S.-P.; Cheng, P.-C. Testosterone regulates the intracellular bacterial community formation of uropathogenic Escherichia coli in prostate cells via STAT3. Int. J. Med. Microbiol. 2020, 310, 151450. [Google Scholar] [CrossRef]
- Razavipour, S.F.; Harikumar, K.; Slingerland, J.M. p27 as a Transcriptional Regulator: New Roles in Development and Cancer. Cancer Res. 2020, 80, 3451–3458. [Google Scholar] [CrossRef]
- Ahn, H.; Im, E.; Lee, D.Y.; Lee, H.-J.; Jung, J.H.; Kim, S.-H. Antitumor Effect of Pyrogallol via miR-134 Mediated S Phase Arrest and Inhibition of PI3K/AKT/Skp2/cMyc Signaling in Hepatocellular Carcinoma. Int. J. Mol. Sci. 2019, 20, 3985. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, M.; Qian, J.; Bao, M.; Meng, X.; Zhang, S.; Zhang, L.; Zhao, R.; Li, S.; Cao, Q.; et al. miR-134 Functions as a Tumor Suppressor in Cell Proliferation and Epithelial-to-Mesenchymal Transition by Targeting KRAS in Renal Cell Carcinoma Cells. DNA Cell Biol. 2015, 34, 429–436. [Google Scholar] [CrossRef]
- Vitiello, P.P.; Cardone, C.; Martini, G.; Ciardiello, D.; Belli, V.; Matrone, N.; Martinelli, E. Receptor tyrosine kinase-dependent PI3K activation is an escape mechanism to vertical suppression of the EGFR/RAS/MAPK pathway in KRAS-mutated human colorectal cancer cell lines. J. Exp. Clin. Cancer Res. 2019, 38, 41. [Google Scholar] [CrossRef]
- Dolcet, X.; Llobet, D.; Pallares, J.; Matias Guiu, X. NF kB in development and progression of human cancer. Virchows Arch. 2005, 446, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Karnam, K.C.; Ellutla, M.; Bodduluru, L.N.; Kasala, E.R.; Uppulapu, S.K.; Kalyankumarraju, M.; Lahkar, M. Preventive effect of berberine against DMBA-induced breast cancer in female Sprague Dawley rats. Biomed. Pharmacother. 2017, 92, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Shuang, T.; Wang, M.; Zhou, Y.; Shi, C.; Wang, D. NF-κB1, c-Rel, and ELK1 inhibit miR-134 expression leading to TAB1 upregulation in paclitaxel-resistant human ovarian cancer. Oncotarget 2017, 8, 24853–24868. [Google Scholar] [CrossRef] [PubMed]
- Biswas, D.K.; Singh, S.; Shi, Q.; Pardee, A.B.; Iglehart, J.D. Crossroads of Estrogen Receptor and NF-κB Signaling. Sci. STKE 2005, 2005, pe27. [Google Scholar] [CrossRef]
- Xie, C.; Zhang, L.-Z.; Chen, Z.-L.; Zhong, W.-J.; Fang, J.-H.; Zhu, Y.; Xiao, M.-H.; Guo, Z.-W.; Zhao, N.; He, X.; et al. A hMTR4-PDIA3P1-miR-125/124-TRAF6 Regulatory Axis and Its Function in NF kappa B Signaling and Chemoresistance. Hepatology 2020, 71, 1660–1677. [Google Scholar] [CrossRef]
- Molnar, R.; Szabo, L.; Tomesz, A.; Deutsch, A.; Darago, R.; Ghodratollah, N.; Varjas, T.; Nemeth, B.; Budan, F.; Kiss, I. In vivo effects of olive oil and trans-fatty acids on miR-134, miR-132, miR-124-1, miR-9-3 and mTORC1 gene expression in a DMBA-treated mouse model. PLoS ONE 2021, 16, e0246022. [Google Scholar] [CrossRef]
- Szabo, L.; Molnar, R.; Tomesz, A.; Deutsch, A.; Darago, R.; Nowrasteh, G.; Varjas, T.; Nemeth, B.; Budan, F.; Kiss, I. The effects of flavonoids, green tea polyphenols and coffee on DMBA induced LINE-1 DNA hypomethylation. PLoS ONE 2021, 16, e0250157. [Google Scholar] [CrossRef]
- Varjas, T.; Nowrasteh, G.; Budán, F.; Nadasi, E.; Horváth, G.; Makai, S.; Gracza, T.; Cseh, J.; Ember, I. Chemopreventive effect ofPanax ginseng. Phytother. Res. 2009, 23, 1399–1403. [Google Scholar] [CrossRef]
- Varjas, T.; Nowrasteh, G.; Budán, F.; Horváth, G.; Cseh, J.; Gyöngyi, Z.; Makai, S.; Ember, I. The effect of fenugreek on the gene expression of arachidonic acid metabolizing enzymes. Phytother. Res. 2011, 25, 221–227. [Google Scholar] [CrossRef]
- Szabo, L.; Molnar, R.; Tomesz, A.; Deutsch, A.; Darago, R.; Varjas, T.; Ritter, Z.; Szentpeteri, J.L.; Andreidesz, K.; Mathe, D.; et al. Olive Oil Improves While Trans Fatty Acids Further Aggravate the Hypomethylation of LINE-1 Retrotransposon DNA in an Environmental Carcinogen Model. Nutrients 2022, 14, 908. [Google Scholar] [CrossRef]


| miRNA | FORWARD | REVERSE |
|---|---|---|
| miR-134 | TGTGACTGGTTGACCAGAGG | GTGACTAGGTGGCCCACAG |
| miR-132 | ACCGTGGCTTTCGATTGTTA | CGACCATGGCTGTAGACTGTT |
| miR-124-1 | TCTCTCTCCGTGTTCACAGC | ACCGCGTGCCTTAATTGTAT |
| miR-212 | GGCACCTTGGCTCTAGACTG | GCCGTGACTGGAGACTGTTA |
| miR-155 | GACTGTTAATGCTAATCGTGATAG | GTGCAGGGTCCGAGGTATTC |
| mouse U6 | CGCTTCGGCAGCACATATAC | TTCACGAATTTGCGTGTCAT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tomesz, A.; Szabo, L.; Molnar, R.; Deutsch, A.; Darago, R.; Raposa, B.L.; Ghodratollah, N.; Varjas, T.; Nemeth, B.; Orsos, Z.; et al. Changes in miR-124-1, miR-212, miR-132, miR-134, and miR-155 Expression Patterns after 7,12-Dimethylbenz(a)anthracene Treatment in CBA/Ca Mice. Cells 2022, 11, 1020. https://doi.org/10.3390/cells11061020
Tomesz A, Szabo L, Molnar R, Deutsch A, Darago R, Raposa BL, Ghodratollah N, Varjas T, Nemeth B, Orsos Z, et al. Changes in miR-124-1, miR-212, miR-132, miR-134, and miR-155 Expression Patterns after 7,12-Dimethylbenz(a)anthracene Treatment in CBA/Ca Mice. Cells. 2022; 11(6):1020. https://doi.org/10.3390/cells11061020
Chicago/Turabian StyleTomesz, Andras, Laszlo Szabo, Richard Molnar, Arpad Deutsch, Richard Darago, Bence L. Raposa, Nowrasteh Ghodratollah, Timea Varjas, Balazs Nemeth, Zsuzsanna Orsos, and et al. 2022. "Changes in miR-124-1, miR-212, miR-132, miR-134, and miR-155 Expression Patterns after 7,12-Dimethylbenz(a)anthracene Treatment in CBA/Ca Mice" Cells 11, no. 6: 1020. https://doi.org/10.3390/cells11061020
APA StyleTomesz, A., Szabo, L., Molnar, R., Deutsch, A., Darago, R., Raposa, B. L., Ghodratollah, N., Varjas, T., Nemeth, B., Orsos, Z., Pozsgai, E., Szentpeteri, J. L., Budan, F., & Kiss, I. (2022). Changes in miR-124-1, miR-212, miR-132, miR-134, and miR-155 Expression Patterns after 7,12-Dimethylbenz(a)anthracene Treatment in CBA/Ca Mice. Cells, 11(6), 1020. https://doi.org/10.3390/cells11061020








