Clinical and Prognostic Value of PPIA, SQSTM1, and CCL20 in Hepatocellular Carcinoma Patients by Single-Cell Transcriptome Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection and Preparation
2.2. scRNA-seq Data Processing
2.3. Identification of Signature
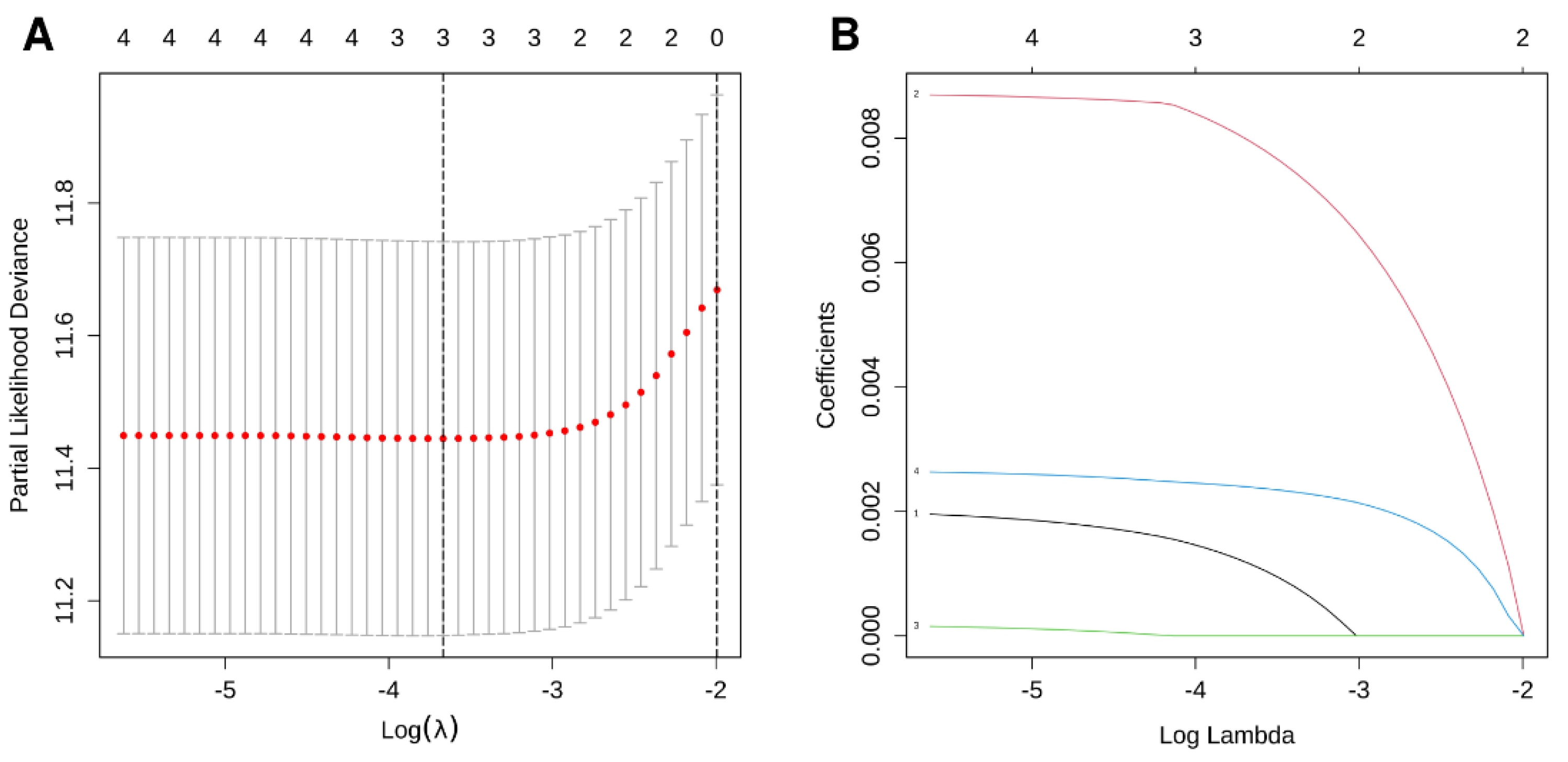
2.4. Model Construction
2.5. Histological Data Analysis of HCC
2.6. Tumor Mutational Burden (TMB)
2.7. Gene set Enrichment Analysis (GSEA)
2.8. Immune Infiltration
2.9. Statistical Analysis
3. Results
3.1. Identification of 26 Cytokine Signaling in Immune-Related Genes (CSIRG) in GSE149614
3.2. Construction of a Three-Gene Signature Based on PPIA, SQSTM1, and CCL20
3.3. Integration Nomogram Based on RS and Cancer Status
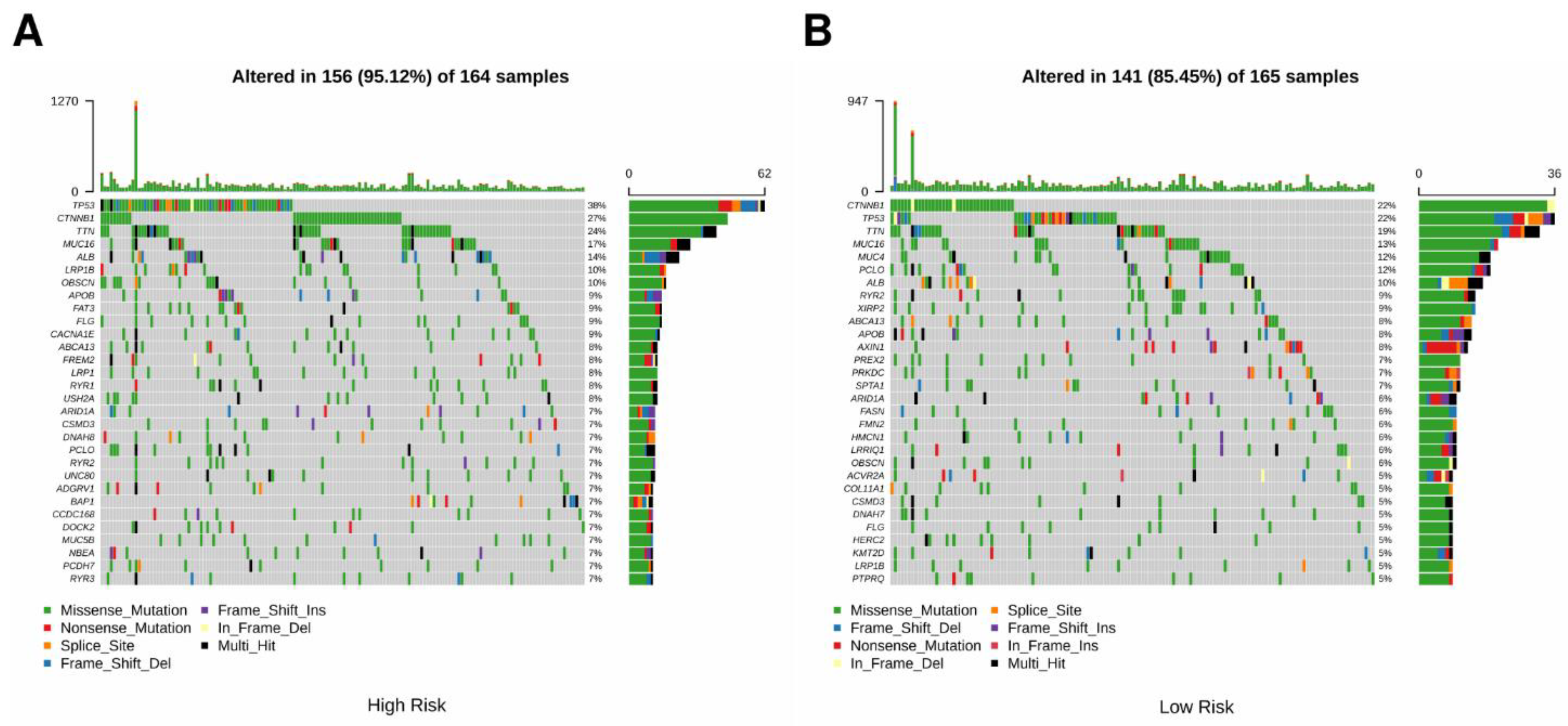
3.4. Comparison of Genomic Mutations in the Three-Gene Signature
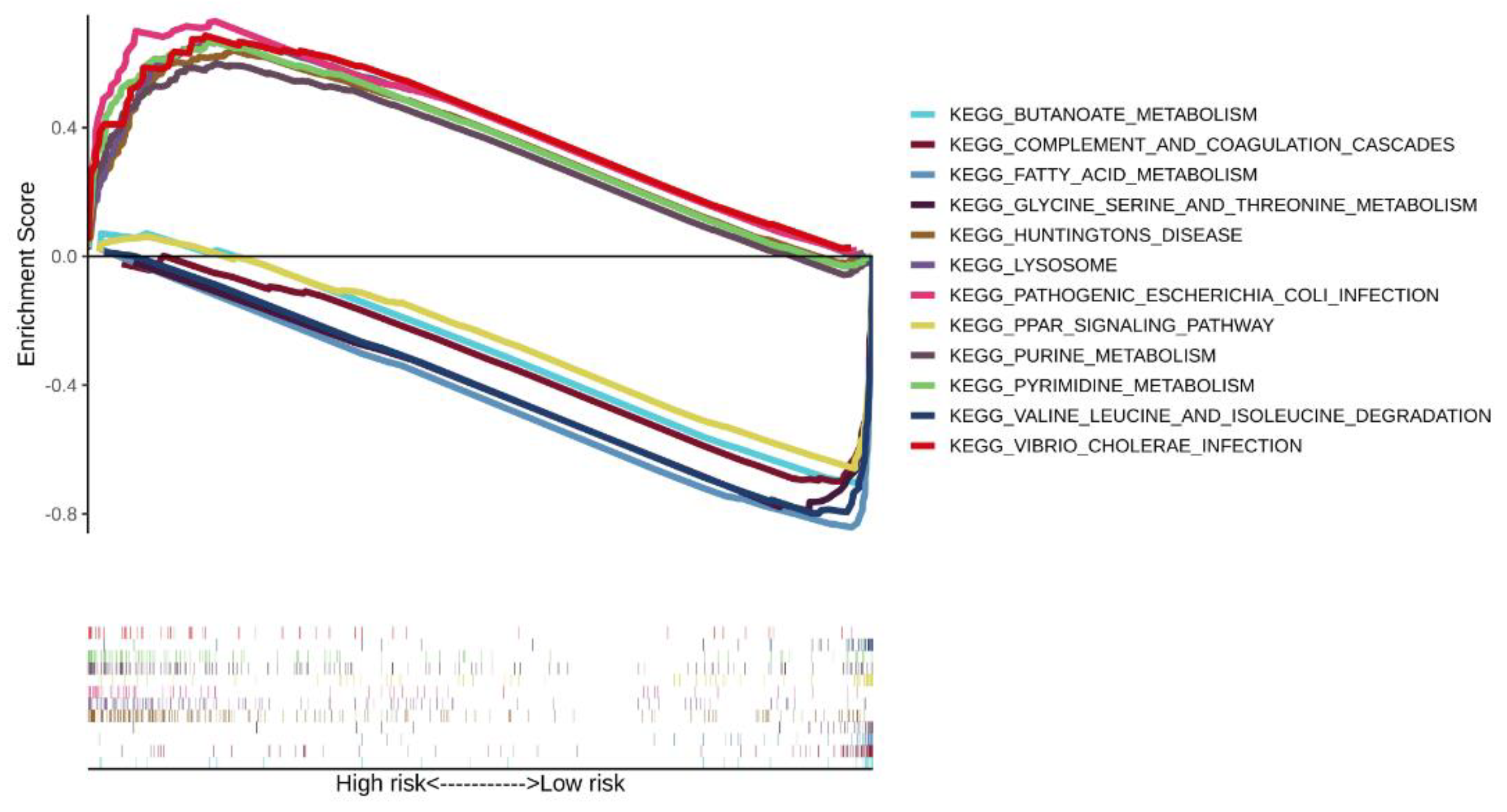
3.5. Enrichment Analysis of the Three-Gene Signature
3.6. Associations of the Three-Gene Signature with the Immune Microenvironment
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Gomaa, A.I.; Khan, S.A.; Toledano, M.B.; Waked, I.; Taylor-Robinson, S.D. Hepatocellular Carcinoma: Epidemiology, Risk Factors and Pathogenesis. World J. Gastroenterol. 2008, 14, 4300–4308. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Ghavimi, S.; Apfel, T.; Azimi, H.; Persaud, A.; Pyrsopoulos, N.T. Management and Treatment of Hepatocellular Carcinoma with Immunotherapy: A Review of Current and Future Options. J. Clin. Transl. Hepatol. 2020, 8, 168–176. [Google Scholar] [CrossRef]
- National Health Commission of the People’s Republic of China. Guidelines for diagnosis and treatment of primary liver cancer (2022 edition). Chin. J. Hepatol. 2022, 30, 367–388. [Google Scholar] [CrossRef]
- Liebman, H.A.; Furie, B.C.; Tong, M.J.; Blanchard, R.A.; Lo, K.J.; Lee, S.D.; Coleman, M.S.; Furie, B. Des-Gamma-Carboxy (Abnormal) Prothrombin as a Serum Marker of Primary Hepatocellular Carcinoma. N. Engl. J. Med. 1984, 310, 1427–1431. [Google Scholar] [CrossRef]
- Feng, H.; Li, B.; Li, Z.; Wei, Q.; Ren, L. PIVKA-II Serves as a Potential Biomarker That Complements AFP for the Diagnosis of Hepatocellular Carcinoma. BMC Cancer 2021, 21, 401. [Google Scholar] [CrossRef]
- Xu, F.; Zhang, L.; He, W.; Song, D.; Ji, X.; Shao, J. The Diagnostic Value of Serum PIVKA-II Alone or in Combination with AFP in Chinese Hepatocellular Carcinoma Patients. Dis. Markers 2021, 2021, 8868370. [Google Scholar] [CrossRef]
- Qi, F.; Zhou, A.; Yan, L.; Yuan, X.; Wang, D.; Chang, R.; Zhang, Y.; Shi, F.; Han, X.; Hou, J.; et al. The Diagnostic Value of PIVKA-II, AFP, AFP-L3, CEA, and Their Combinations in Primary and Metastatic Hepatocellular Carcinoma. J. Clin. Lab. Anal. 2020, 34, e23158. [Google Scholar] [CrossRef]
- Propper, D.J.; Balkwill, F.R. Harnessing Cytokines and Chemokines for Cancer Therapy. Nat. Rev. Clin. Oncol. 2022, 19, 237–253. [Google Scholar] [CrossRef]
- Binnewies, M.; Roberts, E.W.; Kersten, K.; Chan, V.; Fearon, D.F.; Merad, M.; Coussens, L.M.; Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Hedrick, C.C.; et al. Understanding the Tumor Immune Microenvironment (TIME) for Effective Therapy. Nat. Med. 2018, 24, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Rico Montanari, N.; Anugwom, C.M.; Boonstra, A.; Debes, J.D. The Role of Cytokines in the Different Stages of Hepatocellular Carcinoma. Cancers 2021, 13, 4876. [Google Scholar] [CrossRef]
- Conlon, K.C.; Miljkovic, M.D.; Waldmann, T.A. Cytokines in the Treatment of Cancer. J. Interferon Cytokine Res. 2019, 39, 6–21. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Margolin, K. Cytokines in Cancer Immunotherapy. Cancers 2011, 3, 3856–3893. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Zhang, X.; Miao, R.; Ma, X.; Xiang, X.; Fu, Y.; Liu, C.; Niu, W.; Qu, K. A Three-Long Non-Coding RNA-Expression-Based Risk Score System Can Better Predict Both Overall and Recurrence-Free Survival in Patients with Small Hepatocellular Carcinoma. Aging 2018, 10, 1627–1639. [Google Scholar] [CrossRef]
- Dai, Y.; Qiang, W.; Lin, K.; Gui, Y.; Lan, X.; Wang, D. An Immune-Related Gene Signature for Predicting Survival and Immunotherapy Efficacy in Hepatocellular Carcinoma. Cancer Immunol. Immunother. 2021, 70, 967–979. [Google Scholar] [CrossRef]
- Wang, Y.; Ruan, Z.; Yu, S.; Tian, T.; Liang, X.; Jing, L.; Li, W.; Wang, X.; Xiang, L.; Claret, F.X.; et al. A Four-Methylated MRNA Signature-Based Risk Score System Predicts Survival in Patients with Hepatocellular Carcinoma. Aging 2019, 11, 160–173. [Google Scholar] [CrossRef]
- US Food and Drug Administration. FDA Approves Pembrolizumab for Adults and Children with TMB-H Solid Tumors. FDA 2020. Available online: https://www.fda.gov/drugs/drug-approvals-and-databases/fda-approves-pembrolizumab-adults-and-children-tmb-h-solid-tumors (accessed on 4 September 2022).
- Chalmers, Z.R.; Connelly, C.F.; Fabrizio, D.; Gay, L.; Ali, S.M.; Ennis, R.; Schrock, A.; Campbell, B.; Shlien, A.; Chmielecki, J.; et al. Analysis of 100,000 Human Cancer Genomes Reveals the Landscape of Tumor Mutational Burden. Genome Med. 2017, 9, 34. [Google Scholar] [CrossRef]
- Yarchoan, M.; Hopkins, A.; Jaffee, E.M. Tumor Mutational Burden and Response Rate to PD-1 Inhibition. N. Engl. J. Med. 2017, 377, 2500–2501. [Google Scholar] [CrossRef]
- Carbone, D.P.; Reck, M.; Paz-Ares, L.; Creelan, B.; Horn, L.; Steins, M.; Felip, E.; van den Heuvel, M.M.; Ciuleanu, T.-E.; Badin, F.; et al. First-Line Nivolumab in Stage IV or Recurrent Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2017, 376, 2415–2426. [Google Scholar] [CrossRef] [PubMed]
- Gandara, D.R.; Paul, S.M.; Kowanetz, M.; Schleifman, E.; Zou, W.; Li, Y.; Rittmeyer, A.; Fehrenbacher, L.; Otto, G.; Malboeuf, C.; et al. Blood-Based Tumor Mutational Burden as a Predictor of Clinical Benefit in Non-Small-Cell Lung Cancer Patients Treated with Atezolizumab. Nat. Med. 2018, 24, 1441–1448. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Chen, J.; Li, Y.; Wu, B.; Ye, Z.; Tian, X.; Wei, Y.; Hao, Z.; Pan, Y.; Zhou, H.; et al. 6-Phosphogluconolactonase Promotes Hepatocellular Carcinogenesis by Activating Pentose Phosphate Pathway. Front. Cell Dev. Biol. 2021, 9, 753196. [Google Scholar] [CrossRef] [PubMed]
- Aran, D.; Hu, Z.; Butte, A.J. XCell: Digitally Portraying the Tissue Cellular Heterogeneity Landscape. Genome Biol. 2017, 18, 220. [Google Scholar] [CrossRef]
- Ait-Ahmed, Y.; Lafdil, F. Impact of Cytokines in Hepatocellular Carcinoma Initiation and Progression. In Immunotherapy of Hepatocellular Carcinoma; Greten, T.F., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 137–161. ISBN 978-3-319-64958-0. [Google Scholar]
- Li, G.; Xu, W.; Zhang, L.; Liu, T.; Jin, G.; Song, J.; Wu, J.; Wang, Y.; Chen, W.; Zhang, C.; et al. Development and Validation of a CIMP-Associated Prognostic Model for Hepatocellular Carcinoma. EBioMedicine 2019, 47, 128–141. [Google Scholar] [CrossRef]
- Wang, B.; Shi, L.; Sun, X.; Wang, L.; Wang, X.; Chen, C. Production of CCL20 from Lung Cancer Cells Induces the Cell Migration and Proliferation through PI3K Pathway. J. Cell. Mol. Med. 2016, 20, 920–929. [Google Scholar] [CrossRef]
- Chen, K.-J.; Lin, S.-Z.; Zhou, L.; Xie, H.-Y.; Zhou, W.-H.; Taki-Eldin, A.; Zheng, S.-S. Selective Recruitment of Regulatory T Cell through CCR6-CCL20 in Hepatocellular Carcinoma Fosters Tumor Progression and Predicts Poor Prognosis. PLoS ONE 2011, 6, e24671. [Google Scholar] [CrossRef]
- Yuan, Q.; Zhang, J.; Liu, Y.; Chen, H.; Liu, H.; Wang, J.; Niu, M.; Hou, L.; Wu, Z.; Chen, Z.; et al. MyD88 in Myofibroblasts Regulates Aerobic Glycolysis-Driven Hepatocarcinogenesis via ERK-Dependent PKM2 Nuclear Relocalization and Activation. J. Pathol. 2022, 256, 414–426. [Google Scholar] [CrossRef]
- Duran, A.; Linares, J.F.; Galvez, A.S.; Wikenheiser, K.; Flores, J.M.; Diaz-Meco, M.T.; Moscat, J. The Signaling Adaptor P62 Is an Important NF-KappaB Mediator in Tumorigenesis. Cancer Cell 2008, 13, 343–354. [Google Scholar] [CrossRef]
- Davra, V.; Saleh, T.; Geng, K.; Kimani, S.; Mehta, D.; Kasikara, C.; Smith, B.; Colangelo, N.W.; Ciccarelli, B.; Li, H.; et al. Cyclophilin a Inhibitor Debio-025 Targets Crk, Reduces Metastasis, and Induces Tumor Immunogenicity in Breast Cancer. Mol. Cancer Res. 2020, 18, 1189–1201. [Google Scholar] [CrossRef]
- Gu, Y.; Wang, C.; Chen, S.; Tang, J.; Guo, X.; Hu, W.; Cui, A.; Zhang, D.; Yu, K.; Chen, M. A Critical Role of Peptidylprolyl Isomerase a Pseudogene 22/MicroRNA-197-3p/Peptidylprolyl Isomerase a Axis in Hepatocellular Carcinoma. Front. Genet. 2021, 12, 604461. [Google Scholar] [CrossRef] [PubMed]
- Lakshmanan, I.; Salfity, S.; Seshacharyulu, P.; Rachagani, S.; Thomas, A.; Das, S.; Majhi, P.D.; Nimmakayala, R.K.; Vengoji, R.; Lele, S.M.; et al. MUC16 Regulates TSPYL5 for Lung Cancer Cell Growth and Chemoresistance by Suppressing P53. Clin. Cancer Res. 2017, 23, 3906–3917. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Pasche, B.; Zhang, W.; Chen, K. Association of MUC16 Mutation with Tumor Mutation Load and Outcomes in Patients with Gastric Cancer. JAMA Oncol. 2018, 4, 1691–1698. [Google Scholar] [CrossRef] [PubMed]
- Akita, K.; Tanaka, M.; Tanida, S.; Mori, Y.; Toda, M.; Nakada, H. CA125/MUC16 Interacts with Src Family Kinases, and over-Expression of Its C-Terminal Fragment in Human Epithelial Cancer Cells Reduces Cell-Cell Adhesion. Eur. J. Cell Biol. 2013, 92, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Rachagani, S.; Torres-Gonzalez, M.P.; Lakshmanan, I.; Majhi, P.D.; Smith, L.M.; Wagner, K.-U.; Batra, S.K. Carboxyl-Terminal Domain of MUC16 Imparts Tumorigenic and Metastatic Functions through Nuclear Translocation of JAK2 to Pancreatic Cancer Cells. Oncotarget 2015, 6, 5772–5787. [Google Scholar] [CrossRef]
- Lu, M.; Kong, X.; Wang, H.; Huang, G.; Ye, C.; He, Z. A Novel MicroRNAs Expression Signature for Hepatocellular Carcinoma Diagnosis and Prognosis. Oncotarget 2017, 8, 8775–8784. [Google Scholar] [CrossRef]
- Perroud, B.; Lee, J.; Valkova, N.; Dhirapong, A.; Lin, P.-Y.; Fiehn, O.; Kültz, D.; Weiss, R.H. Pathway Analysis of Kidney Cancer Using Proteomics and Metabolic Profiling. Mol. Cancer 2006, 5, 64. [Google Scholar] [CrossRef]
- Kang, X.; Bai, L.; QI, X.; Wang, J. Screening and Identification of Key Genes between Liver Hepatocellular Carcinoma (LIHC) and Cholangiocarcinoma (CHOL) by Bioinformatic Analysis. Medicine 2020, 99, e23563. [Google Scholar] [CrossRef]
- Edwards, L.; Gupta, R.; Filipp, F.V. Hypermutation of DPYD Deregulates Pyrimidine Metabolism and Promotes Malignant Progression. Mol. Cancer Res. 2016, 14, 196–206. [Google Scholar] [CrossRef]






| Characteristics | High (N = 151) | Low (N = 153) | Overall (N = 304) |
|---|---|---|---|
| Age | |||
| <60 | 73 (48.3%) | 74 (48.4%) | 147 (48.4%) |
| ≥60 | 78 (51.7%) | 79 (51.6%) | 157 (51.6%) |
| Gender | |||
| Female | 45 (29.8%) | 48 (31.4%) | 93 (30.6%) |
| Male | 106 (70.2%) | 105 (68.6%) | 211 (69.4%) |
| Histologic_grade | |||
| G1 | 16 (10.6%) | 27 (17.6%) | 43 (14.1%) |
| G2 | 66 (43.7%) | 79 (51.6%) | 145 (47.7%) |
| G3 | 61 (40.4%) | 43 (28.1%) | 104 (34.2%) |
| G4 | 8 (5.3%) | 4 (2.6%) | 12 (3.9%) |
| Pathologic_T stage | |||
| T1 | 57 (37.7%) | 95 (62.1%) | 152 (50.0%) |
| T2 | 44 (29.1%) | 30 (19.6%) | 74 (24.3%) |
| T3 | 43 (28.5%) | 25 (16.3%) | 68 (22.4%) |
| T4 | 7 (4.6%) | 3 (2.0%) | 10 (3.3%) |
| Pathologic_stage | |||
| Stage I | 56 (37.1%) | 95 (62.1%) | 151 (49.7%) |
| Stage II | 43 (28.5%) | 30 (19.6%) | 73 (24.0%) |
| Stage III | 50 (33.1%) | 28 (18.3%) | 78 (25.7%) |
| Stage IV | 2 (1.3%) | 0 (0%) | 2 (0.7%) |
| Cancer_status | |||
| Tumor free | 75 (49.7%) | 96 (62.7%) | 171 (56.3%) |
| With tumor | 76 (50.3%) | 57 (37.3%) | 133 (43.8%) |
| TMB | |||
| Average | 3.9 | 3.3 | 3.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mou, L.; Jia, C.; Wu, Z.; Xin, B.; Liang Zhen, C.A.; Wang, B.; Ni, Y.; Pu, Z. Clinical and Prognostic Value of PPIA, SQSTM1, and CCL20 in Hepatocellular Carcinoma Patients by Single-Cell Transcriptome Analysis. Cells 2022, 11, 3078. https://doi.org/10.3390/cells11193078
Mou L, Jia C, Wu Z, Xin B, Liang Zhen CA, Wang B, Ni Y, Pu Z. Clinical and Prognostic Value of PPIA, SQSTM1, and CCL20 in Hepatocellular Carcinoma Patients by Single-Cell Transcriptome Analysis. Cells. 2022; 11(19):3078. https://doi.org/10.3390/cells11193078
Chicago/Turabian StyleMou, Lisha, Chenyang Jia, Zijing Wu, Boyang Xin, Carmen Alicia Liang Zhen, Bailiang Wang, Yong Ni, and Zuhui Pu. 2022. "Clinical and Prognostic Value of PPIA, SQSTM1, and CCL20 in Hepatocellular Carcinoma Patients by Single-Cell Transcriptome Analysis" Cells 11, no. 19: 3078. https://doi.org/10.3390/cells11193078
APA StyleMou, L., Jia, C., Wu, Z., Xin, B., Liang Zhen, C. A., Wang, B., Ni, Y., & Pu, Z. (2022). Clinical and Prognostic Value of PPIA, SQSTM1, and CCL20 in Hepatocellular Carcinoma Patients by Single-Cell Transcriptome Analysis. Cells, 11(19), 3078. https://doi.org/10.3390/cells11193078






