auts2 Features and Expression Are Highly Conserved during Evolution Despite Different Evolutionary Fates Following Whole Genome Duplication
Abstract
1. Introduction
2. Materials and Methods
2.1. Sequence Availability Synteny Analysis
2.2. Gene Remnants’ Identification
2.3. Phylogenetic Analysis
2.4. Read Processing and Estimation of Expression Levels
2.5. Sequence Features
2.6. Embryo Sampling
2.7. RNA Extraction
2.8. Reverse Transcription and Quantitative PCR
2.9. Statistical Analysis
2.10. RNAscope
2.10.1. Probe Design
2.10.2. Embryo Sampling and Preprocessing
2.10.3. Sample Pre-Treatment
2.10.4. RNAscope Multiplex Fluorescent V2 Assay
2.10.5. Microscopy
3. Results
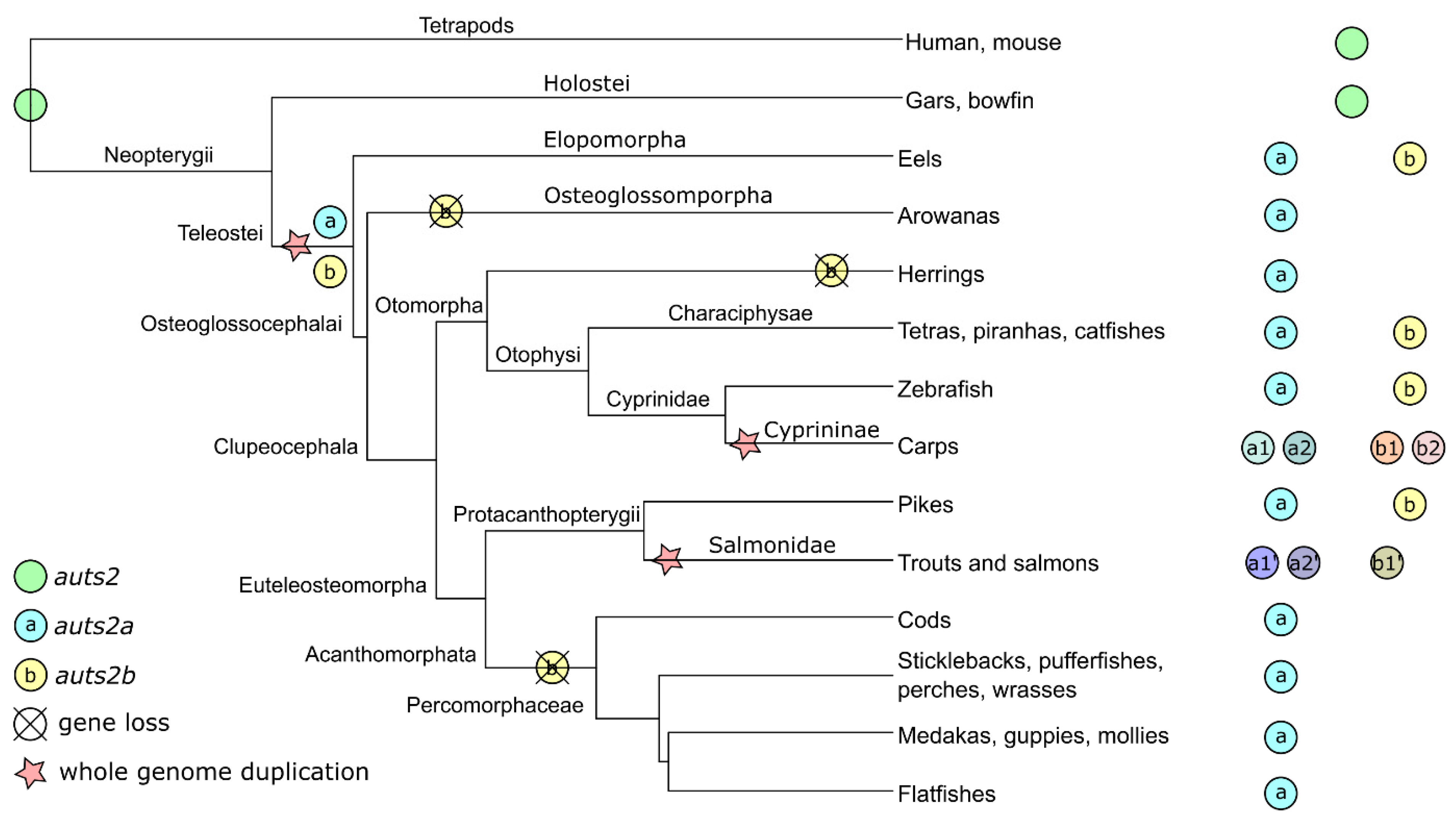
3.1. Evolution of the auts2 Gene Repertoire
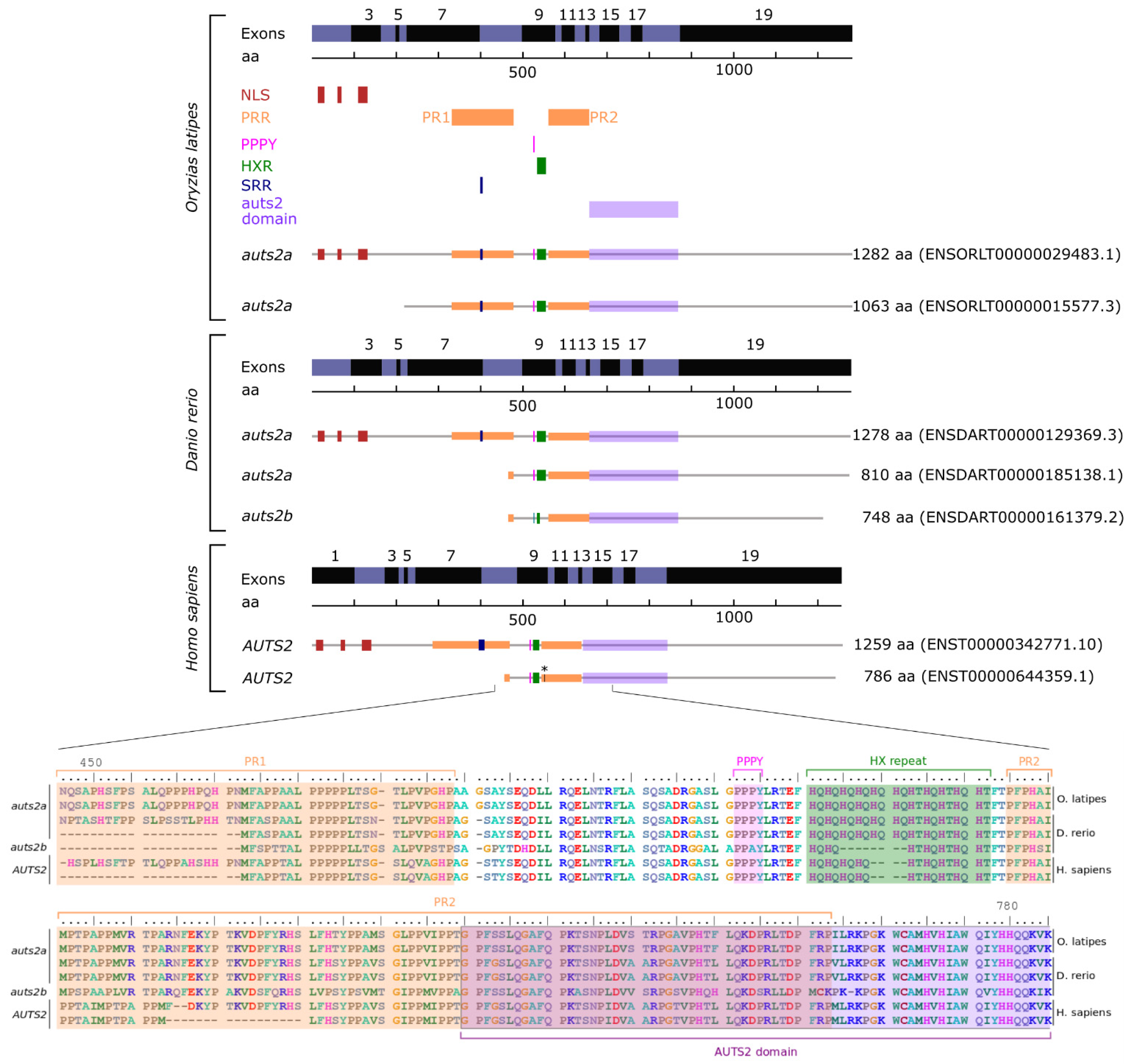
3.2. Auts2 Protein and Gene Sequence Features
3.3. Evolution of auts2 Gene Expression
3.4. Expression of auts2a during Japanese Medaka Neurodevelopment
4. Discussion
4.1. Origins and Evolution of the auts2 Gene Repertoire
4.2. A Strong Expression during Brain Formation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hori, K.; Hoshino, M. Neuronal Migration and AUTS2 Syndrome. Brain Sci. 2017, 7, 54. [Google Scholar] [CrossRef] [PubMed]
- Hori, K.; Shimaoka, K.; Hoshino, M. AUTS2 Gene: Keys to Understanding the Pathogenesis of Neurodevelopmental Disorders. Cells 2021, 11, 11. [Google Scholar] [CrossRef] [PubMed]
- Pang, W.; Yi, X.; Li, L.; Liu, L.; Xiang, W.; Xiao, L. Untangle the multi-facet functions of Auts2 as an entry point to understand neurodevelopmental disorders. Front. Psychiatry 2021, 12, 580443. [Google Scholar] [CrossRef]
- Biel, A.; Castanza, A.S.; Rutherford, R.; Fair, S.R.; Chifamba, L.; Wester, J.C.; Hester, M.E.; Hevner, R.F. AUTS2 Syndrome: Molecular Mechanisms and Model Systems. Front. Mol. Neurosci. 2022, 15, 858582. [Google Scholar] [CrossRef] [PubMed]
- Sultana, R.; Yu, C.-E.; Yu, J.; Munson, J.; Chen, D.; Hua, W.; Estes, A.; Cortes, F.; de la Barra, F.; Yu, D. Identification of a novel gene on chromosome 7q11.2 interrupted by a translocation breakpoint in a pair of autistic twins. Genomics 2002, 80, 129–134. [Google Scholar] [CrossRef]
- Beunders, G.; Voorhoeve, E.; Golzio, C.; Pardo, L.M.; Rosenfeld, J.A.; Talkowski, M.E.; Simonic, I.; Lionel, A.C.; Vergult, S.; Pyatt, R.E. Exonic deletions in AUTS2 cause a syndromic form of intellectual disability and suggest a critical role for the C terminus. Am. J. Hum. Genet. 2013, 92, 210–220. [Google Scholar] [CrossRef]
- Jolley, A.; Corbett, M.; McGregor, L.; Waters, W.; Brown, S.; Nicholl, J.; Yu, S. De novo intragenic deletion of the autism susceptibility candidate 2 (AUTS2) gene in a patient with developmental delay: A case report and literature review. Am. J. Med. Genet. A 2013, 161, 1508–1512. [Google Scholar] [CrossRef]
- Kalscheuer, V.M.; FitzPatrick, D.R.; Tommerup, N.; Bugge, M.; Niebuhr, E.; Neumann, L.M.; Tzschach, A.; Shoichet, S.A.; Menzel, C.; Erdogan, F.; et al. Mutations in autism susceptibility candidate 2 (AUTS2) in patients with mental retardation. Qual. Life Res. 2007, 121, 501–509. [Google Scholar] [CrossRef]
- Amarillo, I.E.; Li, W.L.; Li, X.; Vilain, E.; Kantarci, S. De novo single exon deletion of AUTS2 in a patient with speech and language disorder: A review of disrupted AUTS2 and further evidence for its role in neurodevelopmental disorders. Am. J. Med Genet. Part A 2014, 164, 958–965. [Google Scholar] [CrossRef]
- Schumann, G.; Coin, L.J.; Lourdusamy, A.; Charoen, P.; Berger, K.H.; Stacey, D.; Desrivières, S.; Aliev, F.A.; Khan, A.A.; Amin, N.; et al. Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proc. Natl. Acad. Sci. USA 2011, 108, 7119–7124. [Google Scholar] [CrossRef]
- Zhang, B.; Xu, Y.-H.; Wei, S.-G.; Zhang, H.-B.; Fu, D.-K.; Feng, Z.-F.; Guan, F.-L.; Zhu, Y.-S.; Li, S.-B. Association Study Identifying a New Susceptibility Gene (AUTS2) for Schizophrenia. Int. J. Mol. Sci. 2014, 15, 19406–19416. [Google Scholar] [CrossRef]
- Bedogni, F.; Hodge, R.D.; Nelson, B.R.; A Frederick, E.; Shiba, N.; Daza, R.A.; Hevner, R.F. Autism susceptibility candidate 2 (Auts2) encodes a nuclear protein expressed in developing brain regions implicated in autism neuropathology. Gene Expr. Patterns 2010, 10, 9–15. [Google Scholar] [CrossRef]
- Oksenberg, N.; E Haliburton, G.D.; Eckalbar, W.L.; Oren, I.; Nishizaki, S.; Murphy, K.; Pollard, K.S.; Birnbaum, R.Y.; Ahituv, N. Genome-wide distribution of Auts2 binding localizes with active neurodevelopmental genes. Transl. Psychiatry 2014, 4, e431. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Lee, P.; Stafford, J.; von Schimmelmann, M.; Schaefer, A.; Reinberg, D. An AUTS2-Polycomb complex activates gene expression in the CNS. Nature 2014, 516, 349–354. [Google Scholar] [CrossRef] [PubMed]
- Monderer-Rothkoff, G.; Tal, N.; Risman, M.; Shani, O.; Nissim-Rafinia, M.; Malki-Feldman, L.; Medvedeva, V.; Groszer, M.; Meshorer, E.; Shifman, S. AUTS2 isoforms control neuronal differentiation. Mol. Psychiatry 2021, 26, 666–681. [Google Scholar] [CrossRef] [PubMed]
- Castanza, A.S.; Ramirez, S.; Tripathi, P.P.; Daza, R.A.M.; Kalume, F.K.; Ramirez, J.-M.; Hevner, R.F. AUTS2 Regulates RNA Metabolism and Dentate Gyrus Development in Mice. Cereb. Cortex 2021, 31, 4808–4824. [Google Scholar] [CrossRef]
- Hori, K.; Nagai, T.; Shan, W.; Sakamoto, A.; Taya, S.; Hashimoto, R.; Hayashi, T.; Abe, M.; Yamazaki, M.; Nakao, K.; et al. Cytoskeletal Regulation by AUTS2 in Neuronal Migration and Neuritogenesis. Cell Rep. 2014, 9, 2166–2179. [Google Scholar] [CrossRef]
- Hori, K.; Nagai, T.; Shan, W.; Sakamoto, A.; Abe, M.; Yamazaki, M.; Sakimura, K.; Yamada, K.; Hoshino, M. Heterozygous Disruption of Autism susceptibility candidate 2 Causes Impaired Emotional Control and Cognitive Memory. PLoS ONE 2015, 10, e0145979. [Google Scholar] [CrossRef]
- Yamashiro, K.; Hori, K.; Lai, E.S.; Aoki, R.; Shimaoka, K.; Arimura, N.; Egusa, S.F.; Sakamoto, A.; Abe, M.; Sakimura, K.; et al. AUTS2 Governs Cerebellar Development, Purkinje Cell Maturation, Motor Function and Social Communication. iScience 2020, 23, 101820. [Google Scholar] [CrossRef]
- Gao, Z.; Zhang, J.; Bonasio, R.; Strino, F.; Sawai, A.; Parisi, F.; Kluger, Y.; Reinberg, D. PCGF Homologs, CBX Proteins, and RYBP Define Functionally Distinct PRC1 Family Complexes. Mol. Cell 2012, 45, 344–356. [Google Scholar] [CrossRef]
- Sato, S.; Hayashi, T.; Kobayashi, E. Characterization of porcine autism susceptibility candidate 2 as a candidate gene for the number of corpora lutea in pigs. Anim. Reprod. Sci. 2011, 126, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Kondrychyn, I.; Robra, L.; Thirumalai, V. Transcriptional complexity and distinct expression patterns of auts2 paralogs in Danio rerio. G3 (Bethesda) 2017, 7, 2577–2593. [Google Scholar] [CrossRef][Green Version]
- Oksenberg, N.; Stevison, L.; Wall, J.D.; Ahituv, N. Function and Regulation of AUTS2, a Gene Implicated in Autism and Human Evolution. PLoS Genet. 2013, 9, e1003221. [Google Scholar] [CrossRef] [PubMed]
- Colson, V.; Cousture, M.; Damasceno, D.; Valotaire, C.; Nguyen, T.; Le Cam, A.; Bobe, J. Maternal temperature exposure impairs emotional and cognitive responses and triggers dysregulation of neurodevelopment genes in fish. PeerJ 2019, 7, e6338. [Google Scholar] [CrossRef] [PubMed]
- Parey, E.; Louis, A.; Montfort, J.; Bouchez, O.; Roques, C.; Iampietro, C.; Lluch, J.; Castinel, A.; Donnadieu, C.; Desvignes, T. Genome structures resolve the early diversification of teleost fishes. 2022, 487469. [Google Scholar] [CrossRef]
- Pasquier, J.; Cabau, C.; Nguyen, T.; Jouanno, E.; Severac, D.; Braasch, I.; Journot, L.; Pontarotti, P.; Klopp, C.; Postlethwait, J.H.; et al. Gene evolution and gene expression after whole genome duplication in fish: The PhyloFish database. BMC Genom. 2016, 17, 368. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Iwamatsu, T. Stages of normal development in the medaka Oryzias latipes. Mech. Dev. 2004, 121, 605–618. [Google Scholar] [CrossRef]
- Wang, F.; Flanagan, J.; Su, N.; Wang, L.-C.; Bui, S.; Nielson, A.; Wu, X.; Vo, H.-T.; Ma, X.-J.; Luo, Y. RNAscope: A novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. 2012, 14, 22–29. [Google Scholar] [CrossRef]
- Gross-Thebing, T.; Paksa, A.; Raz, E. Simultaneous high-resolution detection of multiple transcripts combined with localization of proteins in whole-mount embryos. BMC. Biol. 2014, 12, 55. [Google Scholar] [CrossRef]
- Kersigo, J.; Pan, N.; Lederman, J.D.; Chatterjee, S.; Abel, T.; Pavlinkova, G.; Silos-Santiago, I.; Fritzsch, B. A RNAscope whole mount approach that can be combined with immunofluorescence to quantify differential distribution of mRNA. Cell Tissue Res. 2018, 374, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Braasch, I.; Gehrke, A.R.; Smith, J.J.; Kawasaki, K.; Manousaki, T.; Pasquier, J.; Amores, A.; Desvignes, T.; Batzel, P.; Catchen, J. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat. Genet. 2016, 48, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Hughes, L.C.; Ortí, G.; Huang, Y.; Sun, Y.; Baldwin, C.C.; Thompson, A.W.; Arcila, D.; Betancur-R, R.; Li, C.; Becker, L.; et al. Comprehensive phylogeny of ray-finned fishes (Actinopterygii) based on transcriptomic and genomic data. Proc. Natl. Acad. Sci. USA 2018, 115, 6249–6254. [Google Scholar] [CrossRef]
- Inoue, J.; Sato, Y.; Sinclair, R.; Tsukamoto, K.; Nishida, M. Rapid genome reshaping by multiple-gene loss after whole-genome duplication in teleost fish suggested by mathematical modeling. Proc. Natl. Acad. Sci. USA 2015, 112, 14918–14923. [Google Scholar] [CrossRef] [PubMed]
- Pasquier, J.; Braasch, I.; Batzel, P.; Cabau, C.; Montfort, J.; Nguyen, T.; Jouanno, E.; Berthelot, C.; Klopp, T.; Journot, L. Evolution of gene expression after whole-genome duplication: New insights from the spotted gar genome. J. Exp. Zool. B Mol. Dev. Evol. 2017, 328, 709–721. [Google Scholar] [CrossRef]
- Bertho, S.; Pasquier, J.; Pan, Q.; Le Trionnaire, G.; Bobe, J.; Postlethwait, J.H.; Pailhoux, E.; Schartl, M.; Herpin, A.; Guiguen, Y. Foxl2 and Its Relatives Are Evolutionary Conserved Players in Gonadal Sex Differentiation. Sex. Dev. 2016, 10, 111–129. [Google Scholar] [CrossRef]
- Cheung, C.T.; Pasquier, J.; Bouleau, A.; Nguyen, T.; Chesnel, F.; Guiguen, Y.; Bobe, J. Double maternal-effect: Duplicated nucleoplasmin 2 genes, npm2a and npm2b, with essential but distinct functions are shared by fish and tetrapods. BMC Evol. Biol. 2018, 18, 167. [Google Scholar] [CrossRef]
- Guiguen, Y.; Pasquier, J.; Fostier, A.; Bobe, J. Evolution of sex hormone binding globulins reveals early gene duplication at the root of vertebrates. Gen. Comp. Endocrinol. 2021, 300, 113646. [Google Scholar] [CrossRef]
- Berthelot, C.; Brunet, F.; Chalopin, D.; Juanchich, A.; Bernard, M.; Noël, B.; Bento, P.; Da Silva, C.; Labadie, K.; Alberti, A.; et al. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat. Commun. 2014, 5, 3657. [Google Scholar] [CrossRef]
- Yang, L.; Sado, T.; Vincent Hirt, M.; Pasco-Viel, E.; Arunachalam, M.; Li, J.; Wang, X.; Freyhof, J.; Saitoh, K.; Simons, A.M. Phylogeny and polyploidy: Resolving the classification of cyprinine fishes (Teleostei: Cypriniformes). Mol. Phylogenet. Evol. 2015, 85, 97–116. [Google Scholar] [CrossRef]
- Braasch, I.; Postlethwait, J.H. Polyploidy in fish and the teleost genome duplication. In Polyploidy and Genome Evolution; Soltis, P.S., Soltis, D.E., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 341–383. [Google Scholar]
- Force, A.; Lynch, M.; Pickett, F.B.; Amores, A.; Yan, Y.L.; Postlethwait, J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 1999, 151, 1531–1545. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Zhang, J. Rapid Subfunctionalization Accompanied by Prolonged and Substantial Neofunctionalization in Duplicate Gene Evolution. Genetics 2005, 169, 1157–1164. [Google Scholar] [CrossRef] [PubMed]
- Geng, Z.; Gao, Z. Mammalian PRC1 Complexes: Compositional Complexity and Diverse Molecular Mechanisms. Int. J. Mol. Sci. 2020, 21, 8594. [Google Scholar] [CrossRef] [PubMed]
- Hibi, M.; Matsuda, K.; Takeuchi, M.; Shimizu, T.; Murakami, Y. Evolutionary mechanisms that generate morphology and neural-circuit diversity of the cerebellum. Dev. Growth. Differ. 2017, 59, 228–243. [Google Scholar] [CrossRef] [PubMed]
- Ikenaga, T.; Yoshida, M.; Uematsu, K. Cerebellar efferent neurons in teleost fish. Cerebellum. 2006, 5, 268–274. [Google Scholar] [CrossRef]
- Vaz, R.; Hofmeister, W.; Lindstrand, A. Zebrafish Models of Neurodevelopmental Disorders: Limitations and Benefits of Current Tools and Techniques. Int. J. Mol. Sci. 2019, 20, 1296. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Merdrignac, C.; Clément, A.E.; Montfort, J.; Murat, F.; Bobe, J. auts2 Features and Expression Are Highly Conserved during Evolution Despite Different Evolutionary Fates Following Whole Genome Duplication. Cells 2022, 11, 2694. https://doi.org/10.3390/cells11172694
Merdrignac C, Clément AE, Montfort J, Murat F, Bobe J. auts2 Features and Expression Are Highly Conserved during Evolution Despite Different Evolutionary Fates Following Whole Genome Duplication. Cells. 2022; 11(17):2694. https://doi.org/10.3390/cells11172694
Chicago/Turabian StyleMerdrignac, Constance, Antoine Emile Clément, Jérôme Montfort, Florent Murat, and Julien Bobe. 2022. "auts2 Features and Expression Are Highly Conserved during Evolution Despite Different Evolutionary Fates Following Whole Genome Duplication" Cells 11, no. 17: 2694. https://doi.org/10.3390/cells11172694
APA StyleMerdrignac, C., Clément, A. E., Montfort, J., Murat, F., & Bobe, J. (2022). auts2 Features and Expression Are Highly Conserved during Evolution Despite Different Evolutionary Fates Following Whole Genome Duplication. Cells, 11(17), 2694. https://doi.org/10.3390/cells11172694






