Abstract
The loss-of-function conditions for an l(3)malignant brain tumour (l(3)mbt) in larvae reared at 29 °C results in malignant brain tumours and hyperplastic imaginal discs. Unlike the former that have been extensively characterised, little is known about the latter. Here we report the results of a study of the hyperplastic l(3)mbt mutant wing imaginal discs. We identify the l(3)mbt wing disc tumour transcriptome and find it to include genes involved in reactive oxygen species (ROS) metabolism. Furthermore, we show the presence of oxidative stress in l(3)mbt hyperplastic discs, even in apoptosis-blocked conditions, but not in l(3)mbt brain tumours. We also find that chemically blocking oxidative stress in l(3)mbt wing discs reduces the incidence of wing disc overgrowths. Our results reveal the involvement of oxidative stress in l(3)mbt wing discs hyperplastic growth.
1. Introduction
Research in Drosophila has led to important discoveries on the molecular mechanisms that govern tumour induction and progression [1,2]. One of the experimental tumour models reported in Drosophila is based on loss-of-function conditions for a lethal (3) malignant brain tumour (l(3)mbt). The tumour suppressor l(3)mbt encodes a conserved transcriptional regulator that is ubiquitously expressed in Drosophila. L(3)mbt harbours three MBT repeats and a zinc finger domain [3], binds to insulator sequences [4], and has a role in histone compaction [5,6]. Extensive biochemical studies show that L(3)mbt interacts with the dREAM complex, as well as the L(3)mbt-interacting (LINT) complex, which control gene expression and repress developmental genes [7,8]. L(3)mbt has been shown to repress germline genes in somatic tissues, as well as testis-specific and neuronal genes in the female germline, and also to be essential for repressing SWH target genes [4,9,10,11].
Mutations affecting the Drosophila l(3)mbt gene cause malignant brain tumours (hence referred to as mbt tumours) that originate in the neuroepithelial regions of the larval brain lobes [4,12]. Upon allografting, mbt tumours grow, invade the abdomen, and kill the host. Indeed, mbt tumours can undergo infinite rounds of allografting and are, therefore, immortal [10,12].
Gene expression profiling of brain mbt tumours reveals a tumour signature that is enriched in genes that are normally expressed only in the germline, some of which are essential for mbt tumour growth [10,13]. Notably, individuals carrying the l(3)mbt[ts1] and l(3)mbt[E2] alleles develop brain tumours that are more invasive and present a much higher rate of developing as immortal neoplasms if they originate in male larvae [14].
In addition to developing malignant brain neoplasms, l(3)mbt mutant larvae also exhibit hyperplastic imaginal discs that have not been classified as malignant because in allograft assays they are not lethal to the host and retain the capacity to differentiate [12].
Unlike mbt brain tumours that have been extensively characterised [4,10,12,14,15], little is known about mbt imaginal disc tumours. This is unfortunate because the columnar epithelium of the wing imaginal disc has become a paradigm for the study of epithelia-derived tumours [16,17]. Here we report the results of a study of the hyperplastic l(3)mbt mutant wing imaginal discs. We define the mbt wing disc tumour transcriptome and find it to include genes involved in reactive oxygen species (ROS) metabolism. Furthermore, we show the presence of oxidative stress in mbt wing discs, even in apoptosis-blocked conditions. We also find that chemically blocking oxidative stress in mbt wing discs results in partial suppression of the overgrowths. Together, our results show that the l(3)mbt is involved in preventing detrimental oxidative stress.
2. Materials and Methods
2.1. Drosophila Strains and Genotypes
For all the experiments, we used mbt tumours caused by expressing UAS-l(3)mbt-RNAi and UAS-Dcr2 from the ubiquitous Ubi-Gal4 promoter in individuals that are heterozygous for the l(3)mbtts1 allele [13], called mbt-RNAi, hereafter. Because of the temperature-sensitive condition of mbt, all crosses, including controls, were maintained at 29 °C.
The following strains were used:
(a) For expression profiling analyses: w; Ubi-Gal4 UAS-Dcr2/+; UAS-l(3)mbt-RNAi DsRed l(3)mbtts1/+ as mbt-RNAi tumours, and w; Ubi-Gal4 UAS-Dcr2/+ as control.
(b) To detect ROS in mbt tumours: w; Ubi-Gal4 UAS-Dcr2/gstD1-GFP; UAS-l(3)mbt-RNAi DsRed l(3)mbtts1/+. The strain w; gstD1-GFP/+ and the w; Ubi-Gal4 UAS-Dcr2/gstD1-GFP were used as controls.
(c) To inhibit apoptosis in mbt tumours: w; Ubi-Gal4 UAS-Dcr2/+; UAS-l(3)mbt-RNAi DsRed l(3)mbtts1/UAS-p35.
(d) To detect ROS after p35 inhibition of apoptosis: w; Ubi-Gal4 UAS-Dcr2/gstD1-GFP; UAS-l(3)mbt-RNAi DsRed l(3)mbtts1/UAS-p35.
(e) To genetically decrease ROS levels by overexpression of Superoxide dismutase 1 (Sod1) and Catalase (Cat): w; Ubi-Gal4 UAS-Dcr2/UAS-Sod1 UAS-Cat; UAS-l(3)mbt-RNAi DsRed l(3)mbtts1. The strain w; Ubi-Gal4 UAS-Dcr2/UAS-mCD8-GFP; UAS-l(3)mbt-RNAi DsRed l(3)mbtts1 was used as control.
The gstD1-GFP strain expressd GFP under control of a 2.7 kb genomic sequence upstream of gstD1, an oxidative stress response gene [18]. All other Drosophila strains used in this work were obtained from the Bloomington Drosophila Stock Center (BDSC) or the Vienna Drosophila Resource Center (VDRC), and are described in [13].
All the experiments were performed using standard food, except in the ROS scavenging experiments. To prevent ROS accumulation, we prepared standard food supplemented with N-acetyl cysteine (Sigma-Aldrich) at a final concentration of 100 μg/mL.
2.2. Wing Imaginal Disc Staining and Imaging
Nucleic acid staining was performed by incubating discs for 10 min with 1 μM TO-PRO-3 (Life Technologies, Carlsbad, CA, USA). Phalloidin-Rhodamine Red (Life Technologies) was used at 1:20 dilution for 30 min to label the F-Actin network. For the detection of apoptotic cells, we used the TUNEL assay. We employed the fluorescently labelled Alexa Fluor® 647-aha-dUTP (Thermo Fisher Scientific, Waltham, MA, USA), incorporated using terminal deoxynucleotidyl transferase (Roche, Basel, Switzerland). Immunostainings were performed using standard protocols.
For ROS detection in living tissue, we used CellROX Deep Red Reagent (Life Technologies). Third instar discs were dissected in Schneider’s medium and incubated for 15 min in medium containing 5 μM CellROX Deep Red Reagent, followed by three washes and mounted in culture medium. Samples were protected from light throughout the entire experimental procedure. Images were taken in vivo in a Leica SPE confocal microscope.
Fluorescently labelled secondary antibodies were from Thermo Fisher Scientific. Discs were mounted either in antifade supplemented with TO-PRO3 1:1000 (Thermo Fisher Scientific) to enhance labelled nuclei or in SlowFade mounting media (Thermo Fisher Scientific).
A Zeiss LSM880 and a Leica SPE confocal laser scanning microscope were used for image acquisition. Images were processed and analysed using FIJI software (version 1.53q).
Discs were dissected, labelled, and imaged. The stack images were analysed to search for possible overgrowth and were classified accordingly.
2.3. Quantification of Brain Phenotypes
For anatomy analysis, brains from larvae of 7 days after egg laying (dAEL) were dissected, fixed, and labelled with DAPI. The ratios area of neuroepithelium/area of the brain lobe (area NE/BL) and area of central brain/area of the brain lobe (area CB/BL) were calculated by using images acquired with a SP8 Leica confocal image microscope and by measuring the areas corresponding to the NE, the CB, and the brain lobe using FIJI software (version 1.53q). The results were represented in boxplots, and p-values were calculated using nonparametric Mann–Whitney U tests using GraphPad Prism 9.00 for MacOS X (GraphPad Software, La Jolla, CA, USA) (www.graphpad.com).
2.4. Larval Brain Images
Brains were dissected in phosphate-buffered saline (PBS), fixed in 4% formaldehyde, and permeabilised in PBS-0.3% Triton X-100 (PBST). DNA was stained with DAPI. Larval brains were mounted in Vectashield (Vector Laboratories, Newark, NJ, USA). Images were acquired with an SP8 Leica confocal image microscope and processed in Adobe Photoshop CS6 (Adobe, Inc., San José, CA, USA) and ImageJ.
2.5. Expression Profiling
For expression analysis, experiments were conducted in triplicates. For each replicate, 10 discs from larvae of 6 dAEL were dissected in Schneider’s Insect medium (Sigma, St. Louis, MO, USA). Discs were homogenized using the pipette in 45 µL of lysis buffer (20 mM DTT, 10 mM Tris-HCl pH 7.4, 0.5% SDS, and 0.5 µg/µL proteinase K). Samples were then incubated at 65 °C for 15 min. RNA was purified using RNA Clean XP bead suspension (Agencourt Bioscience, Beverly, MA, USA), according to manufacturer’s instructions. Microarrays were performed by the IRB Functional Genomics Facility. Briefly, cDNA was generated from 25 ng of RNA using the TransPlex® Complete Whole Transcriptome Amplification Kit (Sigma; reference WTA2) and 17 cycles of amplification. Subsequently, 8 µg of cDNA was fragmented and labelled using GeneChip Human Mapping 250K Nsp Assay Kit (Affymetrix; catalogue # 900766), according to manufacturer’s instructions. cDNA was hybridized to the GeneChip Drosophila Genome 2.0 Array (Affymetrix, (Santa Clara, CA, USA), catalogue # 520087) for 16 h at 45 °C in a GeneChip Hybridization oven 645 (Affymetrix). Washing and staining of microarrays were performed using a GeneChip Fluidics Station 450 (Affymetrix) and arrays were scanned with GeneChip scanner GSC3000 (Affymetrix). Affymetrix GeneChip Command Console software (AGCC) was used to acquire GeneChip images and generate. CEL files for analysis.
2.6. Differential Gene Expression Analysis
The microarray analysis protocol from R BioConductor [19] workflows was adapted to analyse the samples. CEL files were processed with oligo package, which performed background correction using a deconvolution method, quantile normalization, and the Robust Multichip Average (RMA) algorithm for summarization [20]. An extra quality control step was undertaken by applying the Relative Log Expression (RLE) transform [21]. Lowly expressed genes were filtered out from the resulting expression matrix. As microarray data commonly show a large number of probes in the background intensity range, combining a low variance with a low intensity, they could end up being detected as differentially expressed although they are barely above the “detection” limit and are not very informative in general. A “soft” intensity-based filtering was applied as recommended by the limma user guide [22]. The row-wise medians from the expression data were computed, as they represent the transcript medians, and a median intensity cut-off was chosen based on the data histogram; this resulted in 6624 transcripts excluded. Gene annotation was retrieved for the corresponding probe identifiers of the GeneChip Drosophila Genome 2.0 Array (Affymetrix, catalogue # 520087), from the drosophila2.db R library [23]. Here, only those probes that mapped to an annotation were kept because transcript cluster identifiers can refer to multiple gene symbols and, therefore, they cannot be unambiguously assigned. In summary, 12,043 transcripts were left for the downstream differential gene expression analysis. Different contrasts were considered when computing limma linear models, taking into account pair-wise comparisons of male versus female. Gene expression levels were considered as significant when the adjusted p-value was below 0.001 and the absolute log fold-change was greater than 1.5. Protein class annotation of those genes was conducted using the Panther web tool [24].
3. Results and Discussion
Larvae mutant for the l(3)mbt present tumours both in the brain and in imaginal discs [12] and are, therefore, well-suited to investigate the tissue context-dependent effect of tumour suppressor loss. The tumours that result from the loss of l(3)mbt function in the brain have been studied in detail [4,10,13,14]. In this study, we have sought to investigate l(3)mbt loss-of-function tumorous imaginal discs.
To this aim, we analysed discs that drive the UAS-l(3)mbt-RNAi under the Ubi-Gal4 in flies that are heterozygous for the l(3)mbt[ts1] allele. We have found that wing discs from late mbt-RNAi third instar larvae present different levels of overgrowth that can be assigned to one of three distinct phenotypic classes (Figure 1A): (1) discs without any morphological anomaly; (2) discs with mild effects consisting of small overgrowth, primarily in the hinge zone; (3) discs with strong defects consisting of large or multiple overgrowths, not only in the hinge but also in other regions of the wing disc, such as the pouch or the notum. Our results showed that 52% of the female larvae and 63% of the male larvae showed overgrown discs (Classes 2+3) (Figure 1B). The incidence of the strong overgrowth phenotype (Class 3) is also slightly higher in male (32%) than in female (18%) larvae (Figure 1B). Previous analyses have suggested that the hinge is an “oncogenic hotspot” in the wing imaginal disc [25,26,27,28]. Our observation that Class 2 and 3 phenotypes have a propensity to develop overgrowths in the hinge substantiates this hypothesis. These results confirm the presence of tumorous discs in l(3)mbt mutant larvae [12]. They also reveal that in stark contrast with the strong sex-dimorphic traits presented by mbt brain tumours in l(3)mbt[ts1] and l(3)mbt[E2] homozygous and transheterozygous larvae [14], sex dimorphism is only marginal in the case of mbt-RNAi disc tumours. To determine if such differences could be due to the mutant condition (i.e., mbt-RNAi versus classical mutant alleles), we studied mbt-RNAi brain tumours and found that sex-dependent differences are also marginal, if any exist, with most male and female mbt-RNAi brain tumours closely resembling those found in male l(3)mbt[ts1] and l(3)mbt[E2] homozygous and transheterozygous larvae (Figure 1C,D).
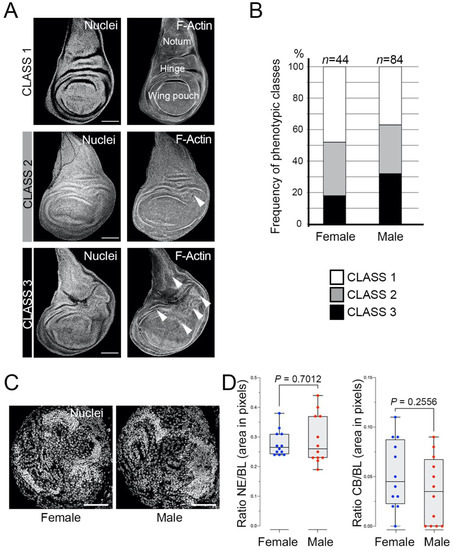
Figure 1.
mbt-RNAi mutants produce overgrowth in wing imaginal discs and brains. (A) Wing discs from mbt-RNAi mutant larvae stained with TO-PRO3 (Nuclei) and Phalloidin (F-actin), showing the three phenotypic classes: (1) absence of overgrowth; (2) mild and few overgrowths; (3) strong and multiple overgrowths. Arrowheads point to anomalous folding resulting from multiple overgrowths. The three main regions of the disc (wing pouch, hinge, and notum) are indicated. Scale bars: 50 µm. (B) Incidence of the overgrowth phenotypes in female and male mbt-RNAi wing discs. (C) Larval brain lobes from female and male mbt-RNAi larvae stained with DAPI (Nuclei). Female and male brain lobes present reduced central brains (CBs) and overgrown neuroepithelia (NE). Scale bars: 50 µm. (D) Relative sizes of NE and CB (as a fraction of brain lobe (BL) area) in female (blue) and male (red) mbt-RNAi mutant larvae. No significant differences are observed in NE and CB sizes between mbt female and male brain lobes.
To further characterise mbt tumorous wing discs we carried out a genome-wide transcriptome analysis of mbt-RNAi and control (w; Ubi-Gal4 UAS-Dcr2/+) wing discs. We identified a total of 241 and 283 genes differentially expressed (DE) in female and male mbt-RNAi samples, respectively, compared to control samples (Supplementary Table S1). Consistent with the very low extent of sex dimorphism observed in disc tumour anatomy, we found that most dysregulated genes were so in both sexes (n = 214). The major effect observed on the mbt wing disc transcriptome is the up-regulation of gene expression (Figure 2A,B).
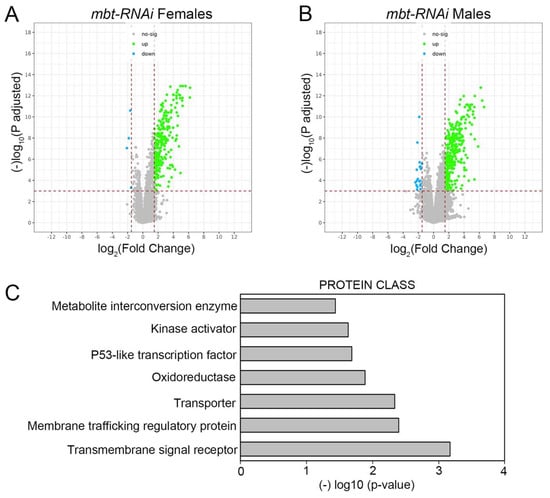
Figure 2.
Volcano plots showing the change in expression of each individual gene in females (A) and males (B) in mbt-RNAi tumours compared to control. Differentially down-regulated and up-regulated genes (|FC| > 1.5; adjusted p-value < 0.001) are indicated in cyan and green, respectively. (C) Protein class enrichment for the set of DE genes in mbt-RNAi tumours.
Our data show that among the 100 genes that belong to the mbt tumour signature (MBTS) identified in mbt brain tumours or allograft transplantations [10], 50 and 54 genes were also misregulated in female and male mbt-RNAi discs, respectively (Supplementary Table S1). A quarter of the MBTS genes are known to normally function in the germline [10]. Notably, we also found 17 and 18 germline genes up-regulated in female and male mbt wing discs, respectively (Supplementary Table S1).
Functional annotation of the dysregulated genes in our transcriptome profiling study shows an enrichment in proteins belonging to the oxidoreductase (10 DE genes) and metabolite interconversion enzyme (19 DE genes) classes (Figure 2C; Supplementary Table S1) with some genes belonging to both categories. Included among the up-regulated genes belonging to those categories are Peroxinectin-like (Pxt), the thioredoxin testis-specific Thioredoxin T (TrxT), and the ovary-specific thioredoxin deadhead (dhd). All three genes belong to the mbt brain tumour signature MBTS [10]. These results reveal that the loss of l(3)mbt in wing discs brings about the dysregulation of the transcription of genes with functions that include oxidation-reduction (redox) processes. Indeed, ROS are involved in the development and progression of cancer types and an increased ROS level is considered a hallmark of many tumours.
To investigate whether ROS is produced in mbt tumours, we monitored the expression of gstD1-GFP. The gstD1-GFP reporter expresses GFP under the control of a 2.7 kb genomic sequence upstream of the oxidative stress response gene gstD1, which is an efficient tool to track ROS in tissues [18]. In non-stressed wild-type discs, we found gstD1-GFP to be expressed in the peripodial membrane, a squamous epithelial layer which covers the apical side of the columnar epithelium of the disc and the laterals that link the columnar epithelium with the peripodial membrane [29] (Figure 3A). At the proper columnar epithelium, only the dorsoventral boundary of the wing pouch showed a GFP signal (Figure 3B). Notably, gstD1-GFP is strongly up-regulated in mbt-RNAi wing discs, with the highest GFP signal located in the hinge region (Figure 3C). In mbt larval brains, however, we found no evidence of gstD1-GFP expression in the tumour tissue that spreads over the lamina, neuroepithelium, medulla, and central brain (Figure 3E). In fact, gstD1-GFP expression levels are indistinguishable between mbt-RNAi and wild-type brain lobes, both of which present GFP fluorescence levels that are low and restricted to glial cells enveloping the lobes. We also detected ROS production in mbt-RNAi wing imaginal discs using the ROS-sensitive dye CellROX Deep Red. Wild-type discs show only low levels of CellROX in the margins. In contrast, mbt-RNAi showed extensive staining in many cells of the disc (Supplementary Figure S1).
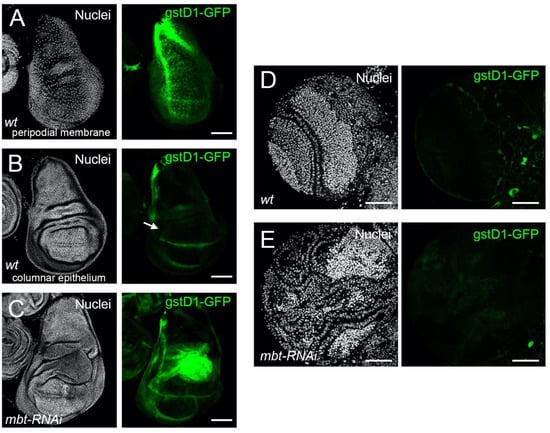
Figure 3.
Oxidative stress detected with gstD1-GFP reporter in mbt-RNAi mutant wing imaginal discs and brain lobes. (A–C) Wing imaginal discs and (D,E) brain lobes stained with DAPI (nuclei in gray) and expressing the ROS reporter gstD1-GFP (green). (A) Wing discs from wild-type larvae focusing on the peripodial membrane. (B) Same disc showing the columnar epithelium; the arrow points to the DV boundary. (C) Wing disc from mbt-RNAi mutant larvae, with strong gstD1-GFP signal in the hinge region. Brain lobes from wild-type (D) and mbt-RNAi larvae (E) showing similar background levels of gstD1-GFP signal (green). Scale bars: 50 µm.
These results reveal a significant build-up of ROS in mbt imaginal disc tumours, but not in mbt brain tumours. ROS induction, as well as a dependence upon ROS, have been extensively documented in a wide range of tumour models in larval Drosophila imaginal discs and have been considered to be common traits of epithelial tumours, independently of their origin [30,31]. The case of mbt tumours reported here is fully consistent with ROS build-up being a common trait of imaginal disc tumours, but not of epithelial tumours in general because mbt brain tumours derive from the neural epithelium [4].
Our transcriptomic profiling analysis together with the expression of gstD1-GFP observed in mbt discs suggest a role of L(3)mbt in ROS metabolism. However, it is known that cells entering apoptosis produce ROS, likely through mitochondrial disruption [32]. For this reason, we study the contribution of apoptosis to the mbt-RNAi wing phenotype. As a first step, we carried out immunofluorescence for the detection of apoptotic cells with TUNEL assay. In mbt wing discs, we detected a high concentration of apoptotic cells in the hinge, which coincides with the gstD1-GFP accumulation (Figure 4A). In very late third instar mbt discs, high levels of gstD1-GFP are present, and apoptotic cells spread through the disc, together with a general disorganisation of the epithelium (Figure 4B). Together, these results indicate that ROS is accumulated progressively in mbt-RNAi discs concomitantly with an increase in apoptosis.
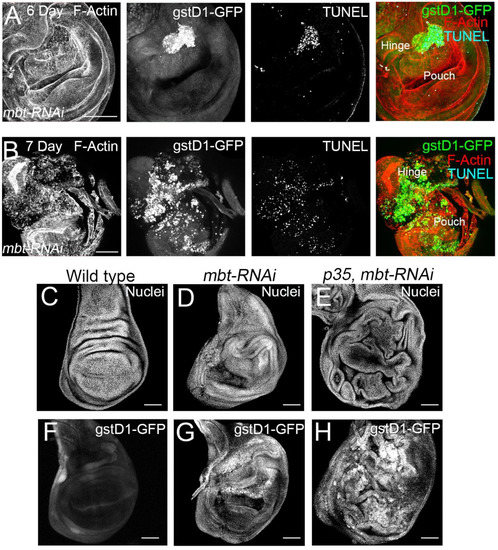
Figure 4.
Apoptosis and ROS production in mbt-RNAi wing discs. (A,B) Wing disc from mbt-RNAi mutant larvae of 6 dAEL (A) and 7 dAEL (B). Expression of gstD1-GFP is shown in green, apoptosis is revealed by TUNEL (blue), and F-actin is stained with Phalloidin (red). Note the increase in severity of the overgrowths and extension and intensity of gstD1-GFP signal. (C–H) Wing discs stained with TOPRO (Nuclei; C–E) and showing gstD1-GFP expression (F–H) from wild-type (C,F), mbt-RNAi (D,G), and mbt-RNAi co-expressing p35 larvae (E,H). Co-expression of p35 enhances the overgrowths and gstD1-GFP is still present. Scale bars: 50 µm.
We then analysed the phenotype of mbt-RNAi discs ectopically expressing the baculovirus protein p35 which blocks the function of the effector caspases [33]. Upon p35 expression, apoptotic cells that would normally be removed from the tissue remain alive in the epithelium. This assay facilitates the evaluation of the overgrowth potential. We found more extensive hyperplastic tissue overgrowth in the p35 mbt-RNAi than in the mbt-RNAi discs, with overgrowths distributed all over the wing disc epithelium (Figure 4C–E; n = 40). These findings suggest that overgrowth in mbt-RNAi discs is limited due to the loss of cells by apoptosis.
Remarkably, the analysis of gstD1-GFP accumulation in p35 mbt-RNAi discs reveals the existence of overgrowths associated with ROS production (n = 92) (Figure 4F–H). We found the gstD1-GFP expression to be highly accumulated in the entire disc, coinciding with the epithelial folding resulting from the overgrowth (Figure 4H). It has been reported that the production of ROS could be due to mitochondrial dysfunction induced by damaged cells [34]. However, our results suggest that mbt mutant cells are able to produce ROS in the absence of apoptosis. This indicates that ROS production is not only a consequence of apoptosis, but also a key feature of mbt-RNAi mutant discs. Additionally, we observed a more extensive expression of gstD1-GFP in p35 mbt-RNAi compared to mbt-RNAi, which suggests that the prevention of cell elimination by apoptosis generates a more extensive ROS-producing mutant tissue.
Having found that mbt mutant epithelial cells exhibit high ROS levels, we asked whether l(3)mbt protects from detrimental oxidative stress. To answer this question, we investigated the effect of the exposure of mbt mutant larvae to antioxidants, which should mitigate the mbt wing disc phenotype. We fed mbt-RNAi larvae with food containing the ROS scavenger N-acetyl cysteine (NAC) and checked for incidences of overgrowth in comparison to standard food. Larvae raised in NAC-supplemented food resulted in a remarkable drop in the incidences of overgrowths in the imaginal discs in comparison to mbt-RNAi discs from larvae grown in standard food (Figure 5A). Low levels of GFP were still detected in morphologically normal mbt-RNAi discs, which suggests that the blockade of oxidative stress by NAC is partial (Figure 5B,C). In those NAC-reared animals in which overgrowths are still present, the high gstD1-GFP was associated with the overgrowth (Figure 5D,E). In addition, the expression of the ROS scavengers Superoxide dismutase 1 (Sod1) and Catalase (Cat) genes in a mbt-RNAi background resulted in a partial recovery of the wing disc phenotype in female larvae (Supplementary Figure S1). Together, these observations showed that ROS scavenging is able, albeit partially, to recover the normal morphology of mbt discs.
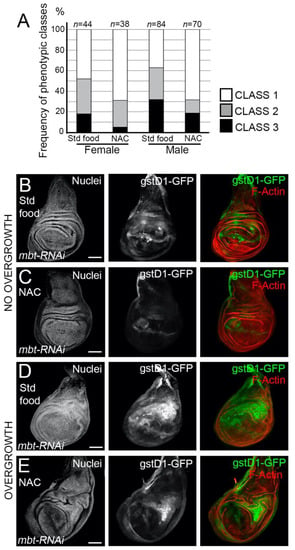
Figure 5.
Effects of the ROS scavenger NAC on the mbt-RNAi phenotype. (A) Incidence of discs with overgrowth in mbt-RNAi fed with standard food or with food supplemented with NAC in females and males. (B,C) mbt-RNAi wing discs with no folding or overgrowths, but weak gstD1-GFP activation grown in standard food (B) and grown in food supplemented with NAC (C). (D,E) mbt-RNAi wing discs with severe folding and overgrowths with strong gstD1-GFP expression grown in standard food (D) and grown in food supplemented with NAC €. Discs were stained with TO-PRO3 (Nuclei; gray) and Phalloidin (F-actin; red). Scale bars: 50 µm.
In this work, we have re-evaluated the potential of l(3)mbt mutants to generate overgrowth in wing imaginal discs as firstly observed by Gateff and colleagues [12]. We also have shown that l(3)mbt mutant wing discs have an altered expression profile, which included a set of MBTS genes. Among the dysregulated genes in mbt mutant discs, we found genes involved in the redox balance of the cell which suggests that l(3)mbt has a role in preventing damage by oxidative stress. Indeed, l(3)mbt mutant epithelia show the ROS accumulation detected by the gstD1-GFP reporter associated with overgrowth. Moreover, the reduction in the oxidative stress by feeding the animals with the NAC antioxidant results in the amelioration of the tumour progression which demonstrates the involvement of oxidative stress in l(3)mbt mutants. In addition, the different response in terms of ROS accumulation observed in mbt mutant brains and wing discs documents another instance in which a given tumorigenic event (i.e., loss of l(3)mbt function) triggers different tumour growth pathways depending on the tissular context. We do not know what the basis for the different ROS responses in mbt wing discs and brain tumours may be. The set of dysregulated redox-related genes is rather similar in both tumours, hence suggesting that the microenvironment of each tissue may be crucial to determine the stress response in each organ. Indeed, differences in ROS production have also been observed upon exposure to ionizing irradiation that induces high levels of apoptosis in imaginal discs [35,36] but not in larval brains [37]. Interestingly, our transcriptome profiling study showed two upregulated genes that could have a role in the apoptotic phenotype of mbt-stressed wing discs. The first is the Cdk5alpha subunit of Cdk5, which phosphorylates Mekk1 that activates JNK, which in turn triggers apoptosis [38]. The second is p53, the sole Drosophila member of the p53 family, that in response to stress initiates apoptosis by activating the transcription of the pro-apoptotic gene, reaper [39,40,41,42].
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cells11162542/s1, Table S1: Summary of samples and contrasts (Samples sheet) and the results for the selected probes (DE probes sheet) found significant in females and males, Figure S1: Oxidative stress detected with CellROX Deep Red and effect of the ROS scavengers Sod1 and Cat in mbt-RNAi mutant wing imaginal discs.
Author Contributions
F.S. and C.G. conceived and supervised the project. P.C.-C., C.M., P.S.-R. and C.P. performed all the experiments. J.F.A. performed the expression profiling analysis. F.S., C.G. and C.M. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the grants PGC2018-099763-B-I00 (Universidad de Barcelona) and PGC2018-097372-B-100 (IRB Barcelona), funded by the Ministry of Science, Innovation and Universities-Spanish State Research Agency.
Data Availability Statement
All data needed to evaluate the conclusion in the paper are present in the manuscript and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
Acknowledgments
We are very grateful to Manel Bosch from the CCiT-UB Imaging Facilities for support; J.I. Pons, D. Fernández, and F. De Oliveira Monteiro from the IRB-Barcelona Functional Genomics for their technical assistance; the Developmental Studies for Hybridoma Bank and the Bloomington Stock Center for antibodies and fly strains.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gonzalez, C. Drosophila melanogaster: A model and a tool to investigate malignancy and identify new therapeutics. Nat. Rev. Cancer 2013, 13, 172–183. [Google Scholar] [CrossRef] [PubMed]
- Sonoshita, M.; Cagan, R.L. Modeling Human Cancers in Drosophila. Curr. Top. Dev. Biol. 2017, 121, 287–309. [Google Scholar] [CrossRef] [PubMed]
- Bonasio, R.; Lecona, E.; Reinberg, D. MBT domain proteins in development and disease. Semin. Cell Dev. Biol. 2010, 21, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Richter, C.; Oktaba, K.; Steinmann, J.; Müller, J.; Knoblich, J.A. The tumour suppressor L(3)mbt inhibits neuroepithelial proliferation and acts on insulator elements. Nat. Cell Biol. 2011, 13, 1029–1039. [Google Scholar] [CrossRef] [PubMed]
- Grimm, C.; Matos, R.; Ly-Hartig, N.; Steuerwald, U.; Lindner, D.; Rybin, V.; Müller, J.; Müller, C.W. Molecular recognition of histone lysine methylation by the Polycomb group repressor dSfmbt. EMBO J. 2009, 28, 1965–1977. [Google Scholar] [CrossRef]
- Trojer, P.; Li, G.; Sims, R.J.; Vaquero, A.; Kalakonda, N.; Boccuni, P.; Lee, D.; Erdjument-Bromage, H.; Tempst, P.; Nimer, S.D. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell 2007, 129, 915–928. [Google Scholar] [CrossRef]
- Georlette, D.; Ahn, S.; MacAlpine, D.M.; Cheung, E.; Lewis, P.W.; Beall, E.L.; Bell, S.P.; Speed, T.; Manak, J.R.; Botchan, M.R. Genomic profiling and expression studies reveal both positive and negative activities for the Drosophila Myb MuvB/dREAM complex in proliferating cells. Genes Dev. 2007, 21, 2880–2896. [Google Scholar] [CrossRef]
- Meier, K.; Mathieu, E.-L.; Finkernagel, F.; Reuter, L.M.; Scharfe, M.; Doehlemann, G.; Jarek, M.; Brehm, A. LINT, a novel dL(3)mbt-containing complex, represses malignant brain tumour signature genes. PLoS Genet. 2012, 8, e1002676. [Google Scholar] [CrossRef]
- Coux, R.X.; Teixeira, F.K.; Lehmann, R. L(3)mbt and the LINT complex safeguard cellular identity in the Drosophila ovary. Development 2018, 145, dev160721. [Google Scholar] [CrossRef]
- Janic, A.; Mendizabal, L.; Llamazares, S.; Rossell, D.; Gonzalez, C. Ectopic expression of germline genes drives malignant brain tumor growth in Drosophila. Science 2010, 330, 1824–1827. [Google Scholar] [CrossRef]
- Sumiyoshi, T.; Sato, K.; Yamamoto, H.; Iwasaki, Y.W.; Siomi, H.; Siomi, M.C. Loss of l(3)mbt leads to acquisition of the ping-pong cycle in Drosophila ovarian somatic cells. Genes Dev. 2016, 30, 1617–1622. [Google Scholar] [CrossRef] [PubMed]
- Gateff, E.; Loffler, T.; Wismar, J. A temperature-sensitive brain tumor suppressor mutation of Drosophila melanogaster: Developmental studies and molecular localization of the gene. Mech. Dev. 1993, 41, 15–31. [Google Scholar] [CrossRef]
- Rossi, F.; Molnar, C.; Hashiyama, K.; Heinen, J.P.; Pampalona, J.; Llamazares, S.; Reina, J.; Hashiyama, T.; Rai, M.; Pollarolo, G. An in vivo genetic screen in Drosophila identifies the orthologue of human cancer/testis gene SPO11 among a network of targets to inhibit lethal(3)malignant brain tumour growth. Open Biol. 2017, 7, 170156. [Google Scholar] [CrossRef] [PubMed]
- Molnar, C.; Heinen, J.P.; Reina, J.; Llamazares, S.; Palumbo, E.; Breschi, A.; Gay, M.; Villarreal, L.; Vilaseca, M.; Pollarolo, G. The histone code reader PHD finger protein 7 controls sex-linked disparities in gene expression and malignancy in Drosophila. Sci. Adv. 2019, 5, eaaw7965. [Google Scholar] [CrossRef]
- Wismar, J.; Löffler, T.; Habtemichael, N.; Vef, O.; Geißen, M.; Zirwes, R.; Altmeyer, W.; Sass, H.; Gateff, E. The Drosophila melanogaster tumor suppressor gene lethal(3)malignant brain tumor encodes a proline-rich protein with a novel zinc finger. Mech. Dev. 1995, 53, 141–154. [Google Scholar] [CrossRef]
- Beira, J.V.; Paro, R. The legacy of Drosophila imaginal discs. Chromosoma 2016, 125, 573–592. [Google Scholar] [CrossRef] [PubMed]
- Herranz, H.; Eichenlaub, T.; Cohen, S.M. Cancer in Drosophila: Imaginal Discs as a Model for Epithelial Tumor Formation. Curr. Top. Dev. Biol. 2016, 116, 181–199. [Google Scholar] [CrossRef]
- Sykiotis, G.P.; Bohmann, D. Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev. Cell 2008, 14, 76–85. [Google Scholar] [CrossRef]
- Huber, W.; Carey, V.J.; Gentleman, R.; Anders, S.; Carlson, M.; Carvalho, B.S.; Bravo, H.C.; Davis, S.; Gatto, L.; Girke, T.; et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 2015, 12, 115–121. [Google Scholar] [CrossRef]
- Carvalho, B.S.; Irizarry, R.A. A framework for oligonucleotide microarray preprocessing. Bioinformatics 2010, 26, 2363–2367. [Google Scholar] [CrossRef]
- Gandolfo, L.C. and T.P. Speed, RLE plots: Visualizing unwanted variation in high dimensional data. PLoS ONE 2018, 13, e0191629. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Carlson, M. Drosophila2.db: Affymetrix Drosophila Genome 2.0 Array Annotation Data (Chip Drosophila2). R Package Version 3.2.3. Available online: https://bioconductor.riken.jp/packages/3.5/data/annotation/html/drosophila2.db.html (accessed on 1 January 2017).
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.-P.; Mushayamaha, T.; Thomas, P.D. PANTHER version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 2021, 49, D394–D403. [Google Scholar] [CrossRef] [PubMed]
- Tamori, Y.; Deng, W.M. Tissue-Intrinsic Tumor Hotspots: Terroir for Tumorigenesis. Trends Cancer 2017, 3, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Sander, M.; Eichenlaub, T.; Herranz, H. Oncogenic cooperation between Yorkie and the conserved microRNA miR-8 in the wing disc of Drosophila. Development 2018, 145, dev153817. [Google Scholar] [CrossRef]
- Tamori, Y.; Suzuki, E.; Deng, W.M. Epithelial Tumors Originate in Tumor Hotspots, a Tissue-Intrinsic Microenvironment. PLoS Biol. 2016, 14, e1002537. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.J.; Bajpai, A.; Alam, W.A.; Gupta, R.P.; Harsh, S.; Pandey, R.K.; Goel-Bhattacharya, S.; Nigam, A.; Mishra, A.; Sinha, P.; et al. Epithelial neoplasia in Drosophila entails switch to primitive cell states. Proc. Natl. Acad. Sci. USA 2013, 110, E2163–E2172. [Google Scholar] [CrossRef]
- Morata, G.; Calleja, M. Cell competition and tumorigenesis in the imaginal discs of Drosophila. Semin. Cancer Biol. 2020, 63, 19–26. [Google Scholar] [CrossRef]
- Diwanji, N.; Bergmann, A. Basement membrane damage by ROS- and JNK-mediated Mmp2 activation drives macrophage recruitment to overgrown tissue. Nat. Commun. 2020, 11, 3631. [Google Scholar] [CrossRef]
- Perez, E.; Lindblad, J.L.; Bergmann, A. Tumor-promoting function of apoptotic caspases by an amplification loop involving ROS, macrophages and JNK in Drosophila. Elife 2017, 6. [Google Scholar] [CrossRef]
- Liu, B.; Chen, Y.; St Clair, D.K. ROS and p53: A versatile partnership. Free Radic. Biol. Med. 2008, 44, 1529–1535. [Google Scholar] [CrossRef] [PubMed]
- Hay, B.A.; Wolff, T.; Rubin, G.M. Expression of baculovirus P35 prevents cell death in Drosophila. Development 1994, 120, 2121–2129. [Google Scholar] [CrossRef] [PubMed]
- Zorov, D.B.; Juhaszova, M.; Sollott, S.J. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev. 2014, 94, 909–950. [Google Scholar] [CrossRef] [PubMed]
- Perez-Garijo, A.; Martin, F.A.; Morata, G. Caspase inhibition during apoptosis causes abnormal signalling and developmental aberrations in Drosophila. Development 2004, 131, 5591–5598. [Google Scholar] [CrossRef] [PubMed]
- Milan, M.; Campuzano, S.; Garcia-Bellido, A. Developmental parameters of cell death in the wing disc of Drosophila. Proc. Natl. Acad. Sci. USA 1997, 94, 5691–5696. [Google Scholar] [CrossRef]
- Wagle, R.; Song, Y.H. Ionizing radiation reduces larval brain size by inducing premature differentiation of Drosophila neural stem cells. Biochem. Biophys Res. Commun. 2020, 523, 555–560. [Google Scholar] [CrossRef]
- Kang, M.J.; Chung, J.; Ryoo, H.D. CDK5 and MEKK1 mediate pro-apoptotic signalling following endoplasmic reticulum stress in an autosomal dominant retinitis pigmentosa model. Nat. Cell Biol. 2012, 14, 409–415. [Google Scholar] [CrossRef]
- Brodsky, M.H.; Nordstrom, W.; Tsang, G.; Kwan, E.; Rubin, G.M.; Abrams, J.M. Drosophila p53 binds a damage response element at the reaper locus. Cell 2000, 101, 103–113. [Google Scholar] [CrossRef]
- Fan, Y.; Lee, T.V.; Xu, D.; Chen, Z.; Lamblin, A.-F.; Steller, H.; Bergmann, A. Dual roles of Drosophila p53 in cell death and cell differentiation. Cell Death Differ. 2010, 17, 912–921. [Google Scholar] [CrossRef]
- Wells, B.S.; Yoshida, E.; Johnston, L.A. Compensatory proliferation in Drosophila imaginal discs requires Dronc-dependent p53 activity. Curr. Biol. 2006, 16, 1606–1615. [Google Scholar] [CrossRef]
- Mollereau, B.; Ma, D. The p53 control of apoptosis and proliferation: Lessons from Drosophila. Apoptosis 2014, 19, 1421–1429. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).