New Insights on the Regulation of the Insulin-Degrading Enzyme: Role of microRNAs and RBPs
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bioinformatic Analysis of miRNAs and RBPs Target Genes
2.2. Cell Culture and Treatments
2.3. Transfection of miRNA Mimics, miRNA Inhibitors and siRBPs
2.4. 3‘UTR Luciferase Reporter Assays
2.5. RNA Isolation and Quantitative Real-Time PCR
2.6. Western Blot Analysis
2.7. Mouse Studies
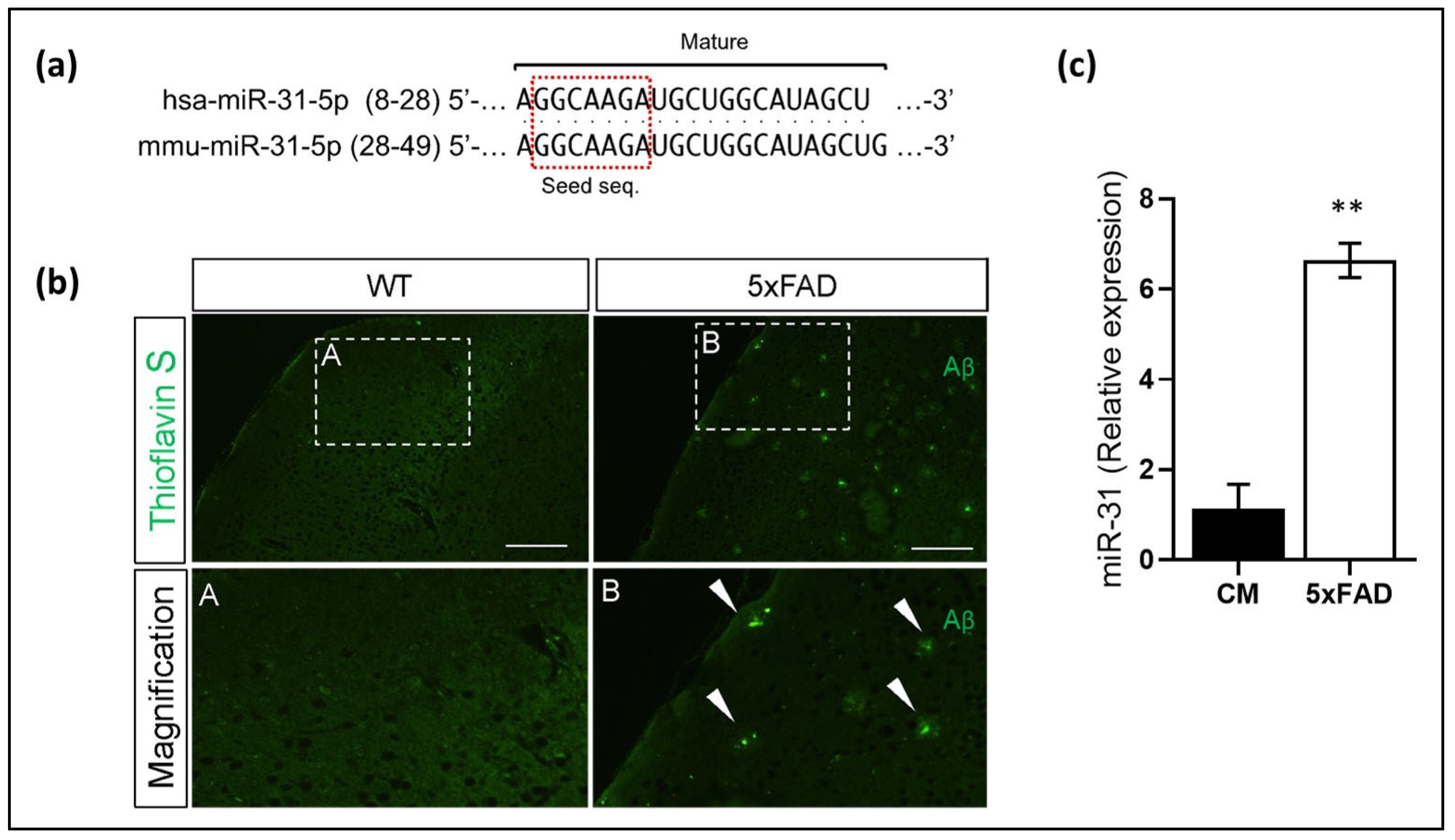
2.8. Thioflavin-S Staining
2.9. Statistical Analysis
3. Results
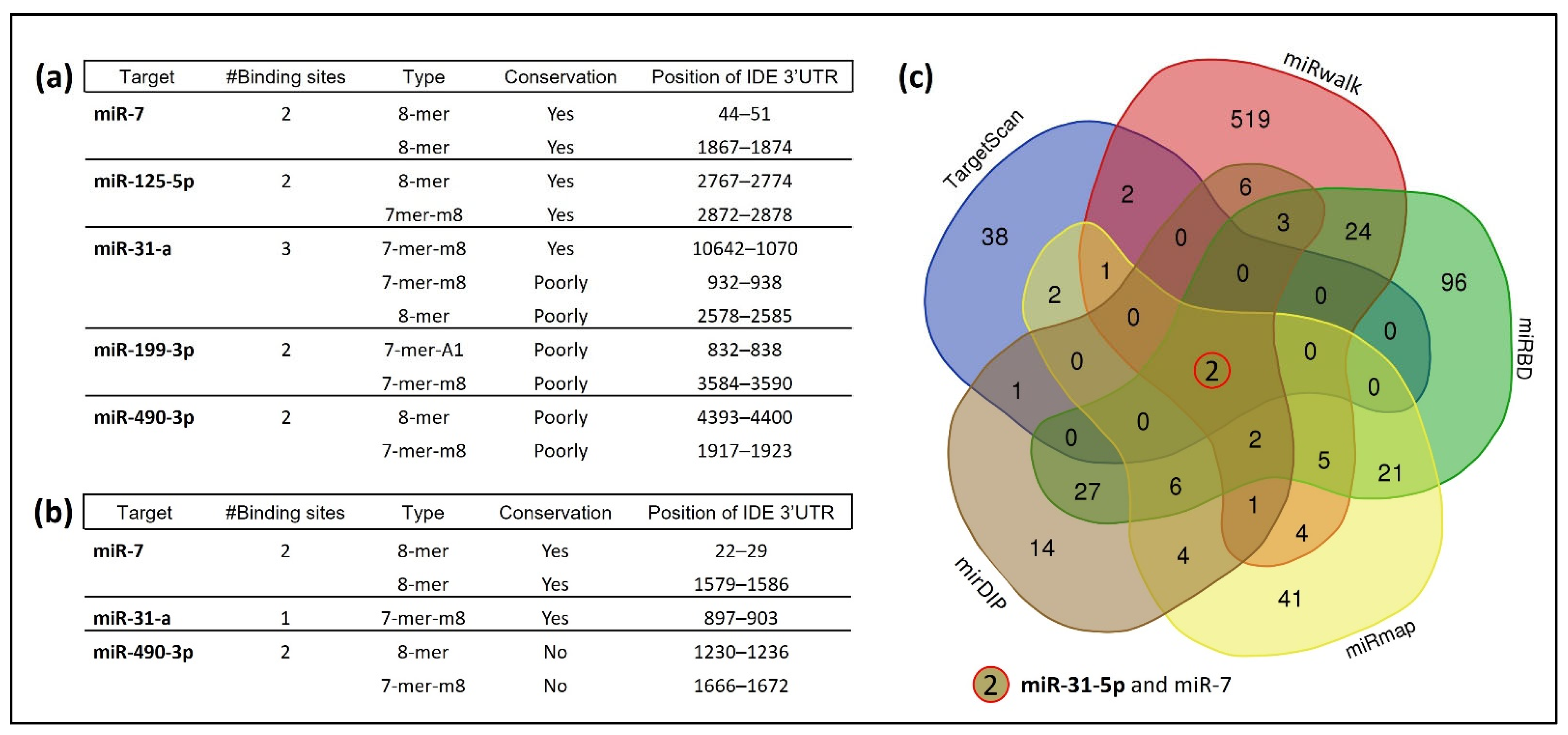
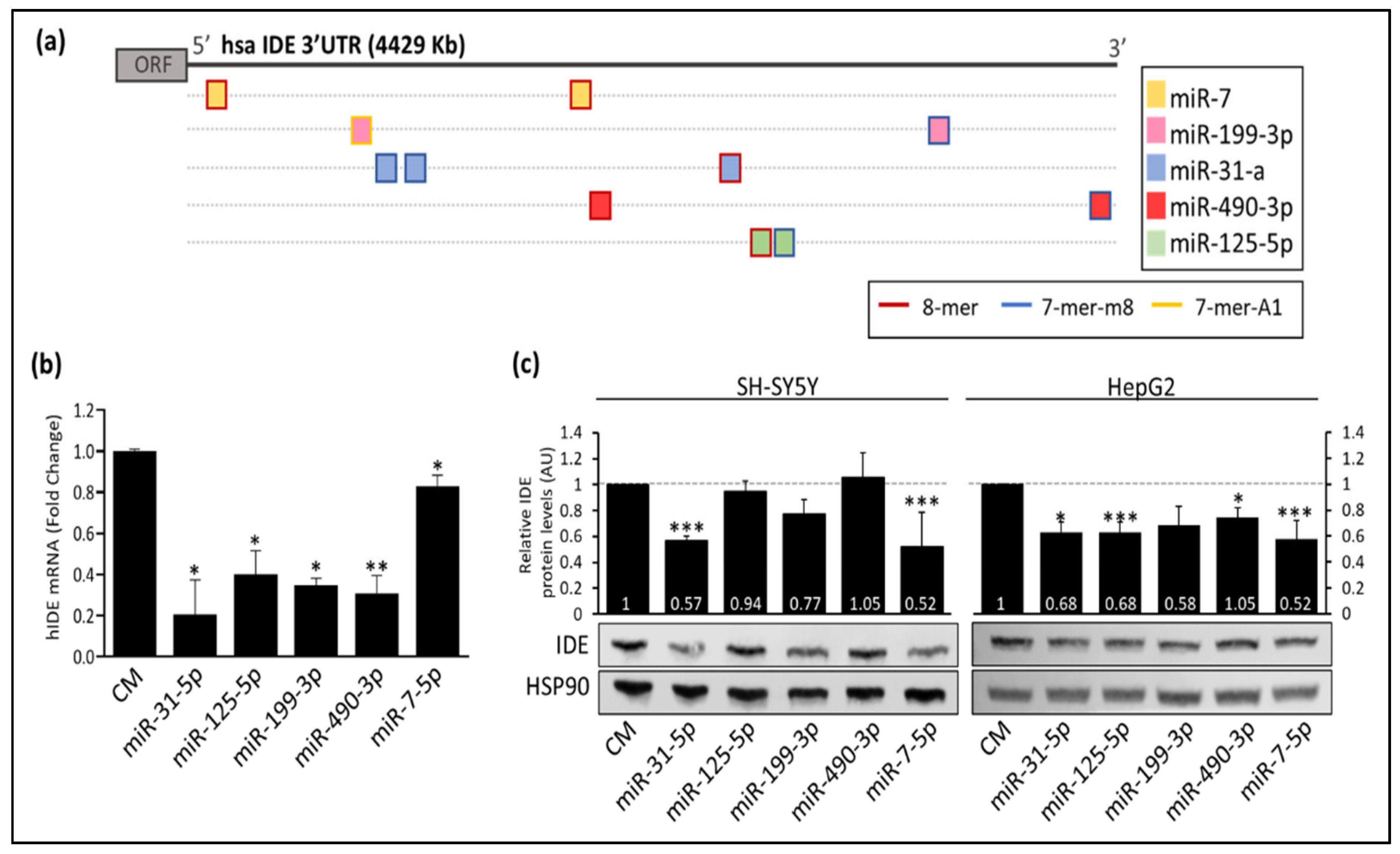
3.1. Several miRNAs Potentially Bind to hIDE 3′UTR
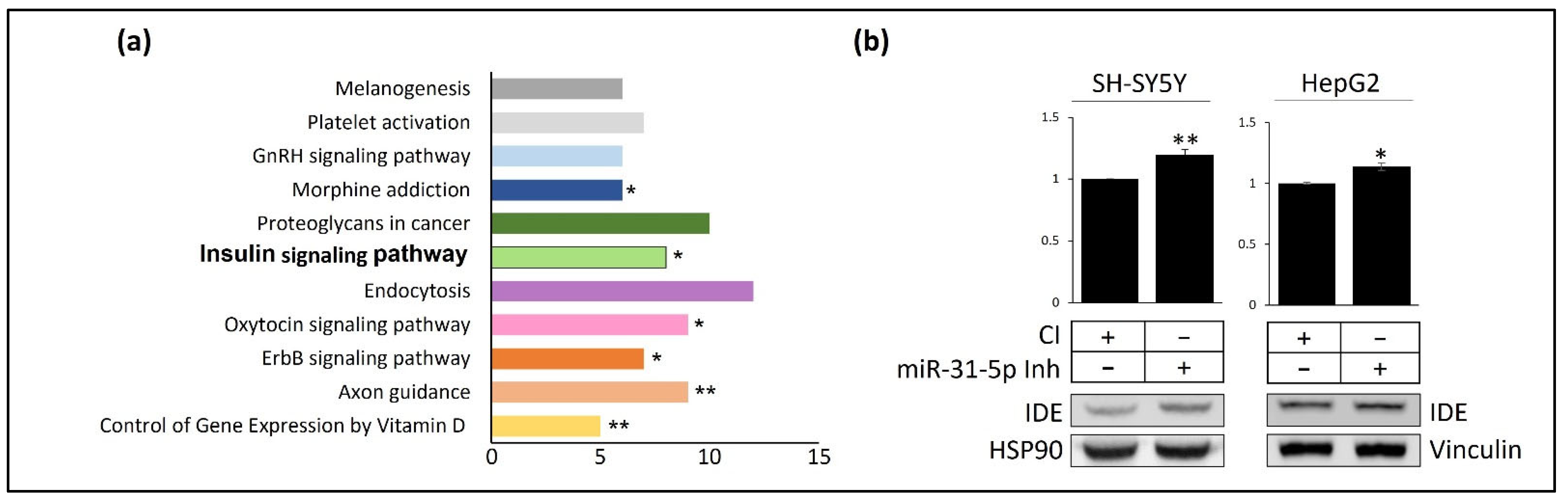
3.2. Post-Transcriptional Regulation of hIDE by miRNAs
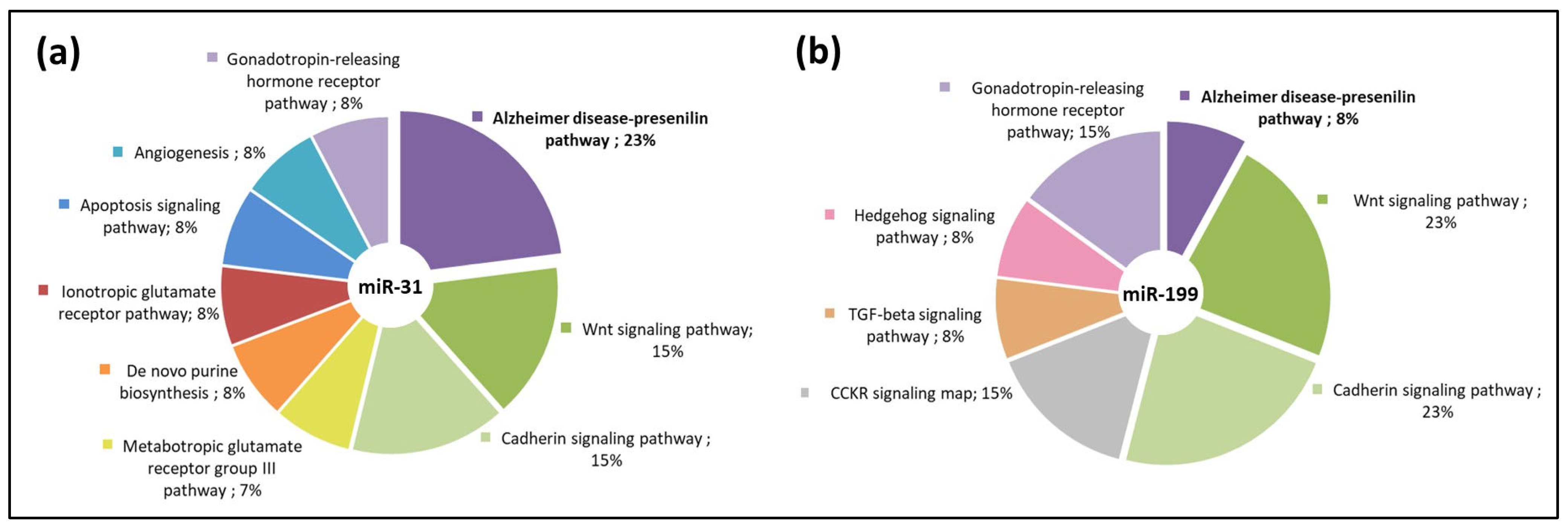
3.3. miR-31 and miR-199 Target Key Genes Involved in AD
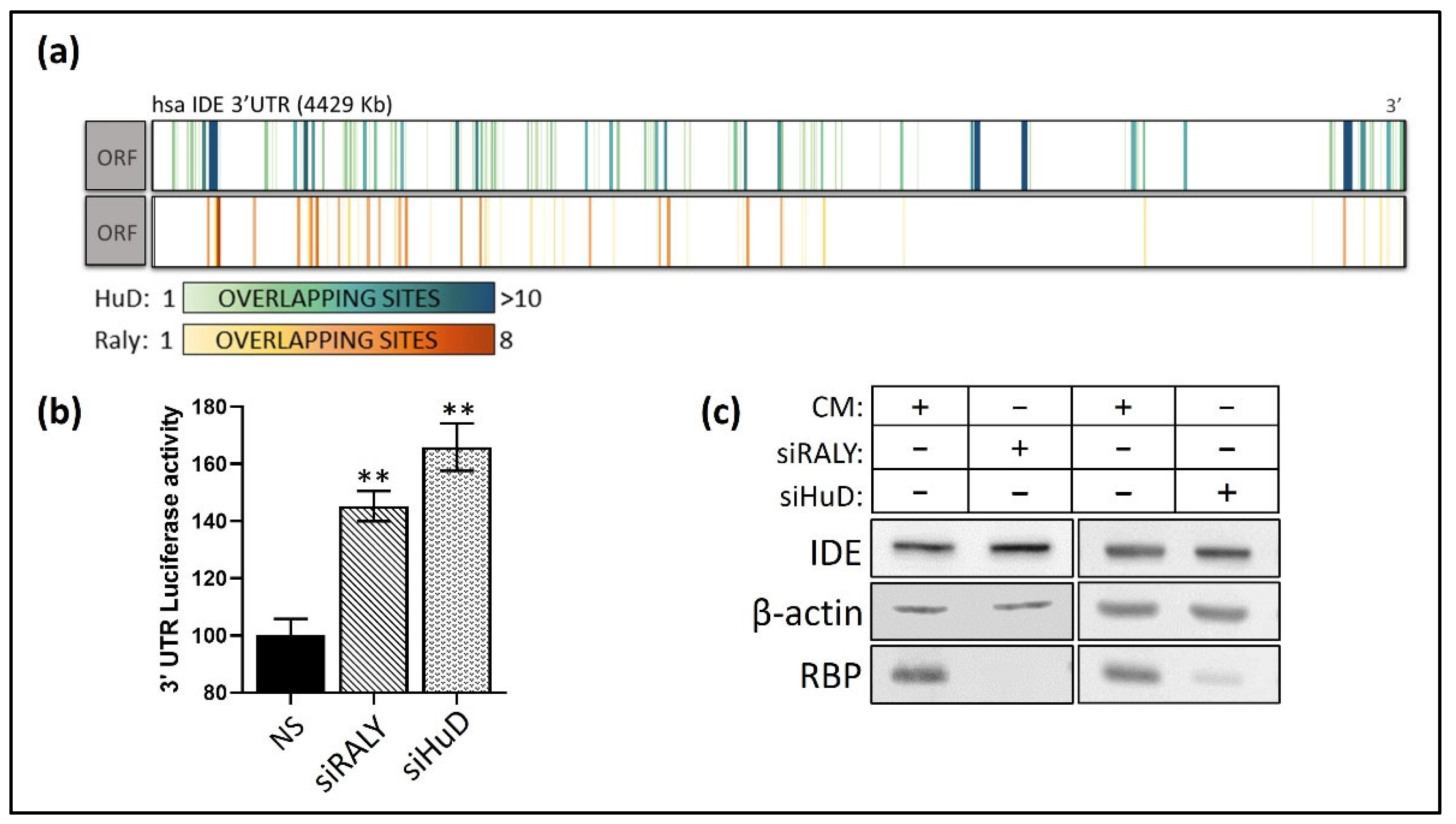
3.4. Post-Transcriptional Regulation of hIDE 3′UTR by RBPs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bedse, G.; Di Domenico, F.; Serviddio, G.; Cassano, T. Aberrant insulin signaling in Alzheimer’s disease: Current knowledge. Front. Neurosci. 2015, 9, 204. [Google Scholar] [CrossRef]
- De la Monte, S.M. Type 3 diabetes is sporadic Alzheimer’s disease: Mini-review. Eur. Neuropsychopharmacol. 2014, 24, 1954–1960. [Google Scholar] [CrossRef] [PubMed]
- Talbot, K.; Wang, H.-Y.; Kazi, H.; Han, L.-Y.; Bakshi, K.P.; Stucky, A.; Fuino, R.L.; Kawaguchi, K.R.; Samoyedny, A.J.; Wilson, R.S.; et al. Demonstrated brain insulin resistance in Alzheimer’s disease patients is associated with IGF-1 resistance, IRS-1 dysregulation, and cognitive decline. J. Clin. Investig. 2012, 122, 1316–1338. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhao, Y.; Ao, X.; Yu, W.; Zhang, L.; Wang, Y.; Chang, W. The Role of Non-coding RNAs in Alzheimer’s Disease: From Regulated Mechanism to Therapeutic Targets and Diagnostic Biomarkers. Front. Aging Neurosci. 2021, 13, 654978. [Google Scholar] [CrossRef]
- Jash, K.; Gondaliya, P.; Kirave, P.; Kulkarni, B.; Sunkaria, A.; Kalia, K. Cognitive dysfunction: A growing link between diabetes and Alzheimer’s disease. Drug Dev. Res. 2019, 81, 144–164. [Google Scholar] [CrossRef]
- Kurochkin, I.V.; Guarnera, E.; Berezovsky, I.N. Insulin-Degrading Enzyme in the Fight against Alzheimer’s Disease. Trends Pharmacol. Sci. 2018, 39, 49–58. [Google Scholar] [CrossRef]
- Sbardella, D.; Tundo, G.R.; Coletta, A.; Marcoux, J.; Koufogeorgou, E.I.; Ciaccio, C.; Santoro, A.M.; Milardi, D.; Grasso, G.; Cozza, P.; et al. The insulin-degrading enzyme is an allosteric modulator of the 20S proteasome and a potential competitor of the 19S. Cell. Mol. Life Sci. 2018, 75, 3441–3456. [Google Scholar] [CrossRef]
- Tang, W.-J. Targeting Insulin-Degrading Enzyme to Treat Type 2 Diabetes Mellitus. Trends Endocrinol. Metab. 2016, 27, 24–34. [Google Scholar] [CrossRef]
- Bennett, R.G.; Duckworth, W.C.; Hamel, F.G. Degradation of Amylin by Insulin-degrading Enzyme. J. Biol. Chem. 2000, 275, 36621–36625. [Google Scholar] [CrossRef]
- Farris, W.; Mansourian, S.; Chang, Y.; Lindsley, L.; Eckman, E.A.; Frosch, M.P.; Eckman, C.B.; Tanzi, R.E.; Selkoe, D.J.; Gué, S. Insulin-degrading enzyme regulates the levels of insulin, amyloid β-protein, and the β-amyloid precursor protein intracellular domain in vivo. Proc. Natl. Acad. Sci. USA 2003, 1, 4162–4167. [Google Scholar] [CrossRef]
- Qiu, W.Q.; Folstein, M.F. Insulin, insulin-degrading enzyme and amyloid-β peptide in Alzheimer’s disease: Review and hypothesis. Neurobiol. Aging 2006, 27, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Bian, L.; Yang, J.D.; Guo, T.W.; Sun, Y.; Duan, S.W.; Chen, W.Y.; Pan, Y.X.; Feng, G.Y.; He, L. Insulin-degrading enzyme and Alzheimer disease. Neurology 2004, 63, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Leissring, M.A. Insulin-Degrading Enzyme: Paradoxes and Possibilities. Cells 2021, 10, 2445. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Teter, B.; Morihara, T.; Lim, G.P.; Ambegaokar, S.S.; Ubeda, O.J.; Frautschy, S.A.; Cole, G.M. Insulin-Degrading Enzyme as a Downstream Target of Insulin Receptor Signaling Cascade: Implications for Alzheimer’s Disease Intervention. J. Neurosci. 2004, 24, 11120–11126. [Google Scholar] [CrossRef] [PubMed]
- Villa-Pérez, P.; Merino, B.; Fernández-Díaz, C.M.; Cidad, P.; Lobatón, C.D.; Moreno, A.; Muturi, H.T.; Ghadieh, H.E.; Najjar, S.M.; Leissring, M.A.; et al. Liver-specific ablation of insulin-degrading enzyme causes hepatic insulin resistance and glucose intolerance, without affecting insulin clearance in mice. Metabolism 2018, 88, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Steneberg, P.; Bernardo, L.; Edfalk, S.; Lundberg, L.; Backlund, F.; Östenson, C.-G.; Edlund, H. The Type 2 Diabetes—Associated Gene Ide Is Required for Insulin Secretion and Suppression of α-Synuclein Levels in β-Cells. Diabetes 2013, 62, 2004–2014. [Google Scholar] [CrossRef]
- Vionnet, N.; Hani, E.H.; Dupont, S.; Gallina, S.; Francke, S.; Dotte, S.; De Matos, F.; Durand, E.; Leprêtre, F.; Lecoeur, C.; et al. Genomewide Search for Type 2 Diabetes—Susceptibility Genes in French Whites: Evidence for a Novel Susceptibility Locus for Early-Onset Diabetes on Chromosome 3q27-qter and Independent Replication of a Type 2-Diabetes Locus on Chromosome 1q21-q24. Am. J. Hum. Genet. 2000, 67, 1470–1480. [Google Scholar] [CrossRef]
- McCarthy, M.I.; Zeggini, E. Genome-wide association studies in type 2 diabetes. Curr. Diabetes Rep. 2009, 9, 164–171. [Google Scholar] [CrossRef]
- Maianti, J.P.; Tan, G.A.; Vetere, A.; Welsh, A.J.; Wagner, B.K.; Seeliger, M.A.; Liu, D.R. Substrate-selective inhibitors that reprogram the activity of insulin-degrading enzyme. Nat. Chem. Biol. 2019, 15, 565–574. [Google Scholar] [CrossRef]
- Stargardt, A.; Gillis, J.; Kamphuis, W.; Wiemhoefer, A.; Kooijman, L.; Raspe, M.; Benckhuijsen, W.; Drijfhout, J.W.; Hol, E.M.; Reits, E. Reduced amyloid-β degradation in early Alzheimer’s disease but not in the APPswePS1dE9 and 3xTg-AD mouse models. Aging Cell 2013, 12, 499–507. [Google Scholar] [CrossRef]
- Abdul-Hay, S.O.; Kang, D.; McBride, M.; Li, L.; Zhao, J.; Leissring, M.A. Deletion of Insulin-Degrading Enzyme Elicits Antipodal, Age-Dependent Effects on Glucose and Insulin Tolerance. PLoS ONE 2011, 6, e20818. [Google Scholar] [CrossRef] [PubMed]
- Fawcett, J.; Permana, P.A.; Levy, J.L.; Duckworth, W.C. Regulation of protein degradation by insulin-degrading enzyme: Analysis by small interfering RNA-mediated gene silencing. Arch. Biochem. Biophys. 2007, 468, 128–133. [Google Scholar] [CrossRef]
- Wang, Z.Q.; Yu, Y.; Zhang, X.H.; Komorowski, J. Chromium-insulin reduces insulin clearance and enhances insulin signaling by suppressing hepatic insulin-degrading enzyme and proteasome protein expression in KKAy mice. Front. Endocrinol. 2014, 5, 99. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Fernández-Hernando, C.; Ramírez, C.M.; Goedeke, L.; Suárez, Y. MicroRNAs in Metabolic Disease. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 178–185. [Google Scholar] [CrossRef]
- Kim, M.; Zhang, X. The Profiling and Role of miRNAs in Diabetes Mellitus. J. Diabetes Clin. Res. 2019, 1, 5. [Google Scholar] [PubMed]
- Kantharidis, P.; Wang, B.; Carew, R.M.; Lan, H.Y. Diabetes Complications: The MicroRNA Perspective. Diabetes 2011, 60, 1832–1837. [Google Scholar] [CrossRef] [PubMed]
- Karolina, D.S.; Armugam, A.; Sepramaniam, S.; Jeyaseelan, K. miRNAs and diabetes mellitus. Expert Rev. Endocrinol. Metab. 2012, 7, 281–300. [Google Scholar] [CrossRef]
- Agbu, P.; Carthew, R.W. MicroRNA-mediated regulation of glucose and lipid metabolism. Nat. Rev. Mol. Cell Biol. 2021, 22, 425–438. [Google Scholar] [CrossRef]
- Frutos, M.F.-D.; Galán-Chilet, I.; Goedeke, L.; Kim, B.; Pardo-Marqués, V.; Pérez-García, A.; Herrero, J.I.; Fernández-Hernando, C.; Kim, J.; Ramírez, C.M. MicroRNA 7 Impairs Insulin Signaling and Regulates Aβ Levels through Posttranscriptional Regulation of the Insulin Receptor Substrate 2, Insulin Receptor, Insulin-Degrading Enzyme, and Liver X Receptor Pathway. Mol. Cell. Biol. 2019, 39, e00170-19. [Google Scholar] [CrossRef]
- Hug, N.; Longman, D.; Cáceres, J.F. Mechanism and regulation of the nonsense-mediated decay pathway. Nucleic Acids Res. 2016, 44, 1483–1495. [Google Scholar] [CrossRef] [PubMed]
- Re, A.; Joshi, T.; Kulberkytė, E.; Morris, Q.; Workman, C.T. RNA-Protein Interactions: An Overview. Methods Mol. Biol. 2014, 1097, 491–521. [Google Scholar] [PubMed]
- Pérez-García, A.; Torrecilla-Parra, M.; Frutos, M.F.-D.; Martín-Martín, Y.; Pardo-Marqués, V.; Ramírez, C.M. Posttranscriptional Regulation of Insulin Resistance: Implications for Metabolic Diseases. Biomolecules 2022, 12, 208. [Google Scholar] [CrossRef] [PubMed]
- McGeary, S.E.; Lin, K.S.; Shi, C.Y.; Pham, T.M.; Bisaria, N.; Kelley, G.M.; Bartel, D.P. The biochemical basis of microRNA targeting efficacy. Science 2019, 366, eaav1741. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Huang, X.; Ebert, D.; Mills, C.; Guo, X.; Thomas, P.D. Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc. 2019, 14, 703–721. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Paz, I.; Kosti, I.; Ares, M., Jr.; Cline, M.; Mandel-Gutfreund, Y. RBPmap: A web server for mapping binding sites of RNA-binding proteins. Nucleic Acids Res. 2014, 42, W361–W367. [Google Scholar] [CrossRef]
- Chakraborty, C.; Doss, C.G.P.; Bandyopadhyay, S.; Agoramoorthy, G. Influence of miRNA in insulin signaling pathway and insulin resistance: Micro-molecules with a major role in type-2 diabetes. Wiley Interdiscip. Rev. RNA 2014, 5, 697–712. [Google Scholar] [CrossRef]
- Kaur, P.; Kotru, S.; Singh, S.; Behera, B.S.; Munshi, A. Role of miRNAs in the pathogenesis of T2DM, insulin secretion, insulin resistance, and β cell dysfunction: The story so far. J. Physiol. Biochem. 2020, 76, 485–502. [Google Scholar] [CrossRef]
- Sallam, T.; Jones, M.C.; Gilliland, T.; Zhang, L.; Wu, X.; Eskin, A.; Sandhu, J.; Casero, D.; Vallim, T.Q.D.A.; Hong, C.; et al. Feedback modulation of cholesterol metabolism by the lipid-responsive non-coding RNA LeXis. Nature 2016, 534, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Wan, A.; Zhou, Z.; Chen, D.; Liang, H.; Liu, C.; Yan, S.; Niu, Y.; Lin, Z.; Zhan, S.; et al. RNA-binding protein RALY reprogrammes mitochondrial metabolism via mediating miRNA processing in colorectal cancer. Gut 2021, 70, 1698–1712. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Feng, A.-C.; Salisbury, D.; Liu, X.; Wu, X.; Kim, J.; Lapina, I.; Wang, D.; Lee, B.; Fraga, J.; et al. Collaborative interactions of heterogenous ribonucleoproteins contribute to transcriptional regulation of sterol metabolism in mice. Nat. Commun. 2020, 11, 984. [Google Scholar] [CrossRef]
- Bronicki, L.M.; Jasmin, B.J. Emerging complexity of the HuD/ELAVl4 gene; implications for neuronal development, function, and dysfunction. RNA 2013, 19, 1019–1037. [Google Scholar] [CrossRef]
- Perrone-Bizzozero, N. Role of HuD in nervous system function and pathology. Front. Biosci. 2013, 5, 554–563. [Google Scholar] [CrossRef]
- Allen, M.; Bird, C.; Feng, W.; Liu, G.; Li, W.; Perrone-Bizzozero, N.I.; Feng, Y. HuD Promotes BDNF Expression in Brain Neurons via Selective Stabilization of the BDNF Long 3′UTR mRNA. PLoS ONE 2013, 8, e55718. [Google Scholar] [CrossRef]
- Sanna, M.D.; Ghelardini, C.; Galeotti, N. HuD-mediated distinct BDNF regulatory pathways promote regeneration after nerve injury. Brain Res. 2017, 1659, 55–63. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, R.; Yang, F.; Cheng, R.; Chen, X.; Cui, S.; Gu, Y.; Sun, W.; You, C.; Liu, Z.; et al. miR-19a promotes colorectal cancer proliferation and migration by targeting TIA1. Mol. Cancer 2017, 16, 53. [Google Scholar] [CrossRef]
- Rayman, J.B.; Hijazi, J.; Li, X.; Kedersha, N.; Anderson, P.J.; Kandel, E.R. Genetic Perturbation of TIA1 Reveals a Physiological Role in Fear Memory. Cell Rep. 2019, 26, 2970–2983.e4. [Google Scholar] [CrossRef]
- Fritz, D.; Stefanovic, B. RNA-binding Protein RBMS3 Is Expressed in Activated Hepatic Stellate Cells and Liver Fibrosis and Increases Expression of Transcription Factor Prx1. J. Mol. Biol. 2007, 371, 585–595. [Google Scholar] [CrossRef]
- Lu, C.-K.; Lai, Y.-C.; Chen, H.-R.; Chiang, M.-K. Rbms3, an RNA-Binding Protein, Mediates the Expression of Ptf1a by Binding to Its 3′UTR During Mouse Pancreas Development. DNA Cell Biol. 2012, 31, 1245–1251. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Xi, P.-W.; Li, X.-X.; Sun, X.; Zhou, W.-B.; Xia, T.-S.; Shi, L.; Hu, Y.; Ding, Q.; Wei, J.-F. The RNA binding protein RBMS3 inhibits the metastasis of breast cancer by regulating Twist1 expression. J. Exp. Clin. Cancer Res. 2019, 38, 105. [Google Scholar] [CrossRef] [PubMed]
- D’Amico, G.; Di Benedetto, D.; Pezzino, F.; Giuffrida, V.; Libra, M.; Fichera, M.; Mauceri, G.; Rappazzo, G.; D’Agata, R.; Vicari, E.S.D.; et al. Quantitative evaluation of partial deletions of the DAZ gene cluster. Int. J. Mol. Med. 2006, 17, 785–789. [Google Scholar] [CrossRef]
- Yen, P.H. Putative biological functions of the DAZ family. Int. J. Androl. 2004, 27, 125–129. [Google Scholar] [CrossRef]
- Zhang, S.; Xu, L.; Yu, M.; Zhang, J. Hypomethylation of the DAZ3 promoter in idiopathic asthenospermia: A screening tool for liquid biopsy. Sci. Rep. 2020, 10, 17996. [Google Scholar] [CrossRef]
- McCloskey, A.; Taniguchi, I.; Shinmyozu, K.; Ohno, M. hnRNP C Tetramer Measures RNA Length to Classify RNA Polymerase II Transcripts for Export. Science 2012, 335, 1643–1646. [Google Scholar] [CrossRef]
- Yang, F.; Yi, F.; Han, X.; Du, Q.; Liang, Z. MALAT-1 interacts with hnRNP C in cell cycle regulation. FEBS Lett. 2013, 587, 3175–3181. [Google Scholar] [CrossRef]
- Briata, P.; Chen, C.-Y.; Ramos, A.; Gherzi, R. Functional and molecular insights into KSRP function in mRNA decay. Biochim. Biophys. Acta 2013, 1829, 689–694. [Google Scholar] [CrossRef]
- Briata, P.; Chen, C.-Y.; Giovarelli, M.; Pasero, M.; Trabucchi, M.; Ramos, A.; Gherzi, R. KSRP, many functions for a single protein. Front. Biosci. 2011, 16, 1787–1796. [Google Scholar] [CrossRef]
- González-Arzola, K.; Velázquez-Cruz, A.; Guerra-Castellano, A.; Casado-Combreras, M.; Pérez-Mejías, G.; Díaz-Quintana, A.; Díaz-Moreno, I.; De la Rosa, M.A. New moonlighting functions of mitochondrial cytochrome c in the cytoplasm and nucleus. FEBS Lett. 2019, 593, 3101–3119. [Google Scholar] [CrossRef]
- González-Arzola, K.; Guerra-Castellano, A.; Rivero-Rodríguez, F.; Casado-Combreras, M.; Pérez-Mejías, G.; Díaz-Quintana, A.; Díaz-Moreno, I.; De la Rosa, M.A. Mitochondrial cytochrome c shot towards histone chaperone condensates in the nucleus. FEBS Open Bio 2021, 11, 2418–2440. [Google Scholar] [CrossRef]
- Gao, Q.; Zhou, R.; Meng, Y.; Duan, R.; Wu, L.; Li, R.; Deng, F.; Lin, C.; Zhao, L. Long noncoding RNA CMPK2 promotes colorectal cancer progression by activating the FUBP3–c-Myc axis. Oncogene 2020, 39, 3926–3938. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, C.; Kubota, N.; Aoyama, K. Interplay of RNA-Binding Proteins and microRNAs in Neurodegenerative Diseases. Int. J. Mol. Sci. 2021, 22, 5292. [Google Scholar] [CrossRef] [PubMed]
- Ambrosio, F.A.; Coricello, A.; Costa, G.; Lupia, A.; Micaelli, M.; Marchesi, N.; Sala, F.; Pascale, A.; Rossi, D.; Vasile, F.; et al. Identification of Compounds Targeting HuD. Another Brick in the Wall of Neurodegenerative Disease Treatment. J. Med. Chem. 2021, 64, 9989–10000. [Google Scholar] [CrossRef] [PubMed]
- Jung, M.; Lee, E. RNA–Binding Protein HuD as a Versatile Factor in Neuronal and Non-Neuronal Systems. Biology 2021, 10, 361. [Google Scholar] [CrossRef]
- Yokoi, S.; Udagawa, T.; Fujioka, Y.; Honda, D.; Okado, H.; Watanabe, H.; Katsuno, M.; Ishigaki, S.; Sobue, G. 3′UTR Length-Dependent Control of SynGAP Isoform α2 mRNA by FUS and ELAV-like Proteins Promotes Dendritic Spine Maturation and Cognitive Function. Cell Rep. 2017, 20, 3071–3084. [Google Scholar] [CrossRef]
- Sims-Robinson, C.; Kim, B.; Rosko, A.; Feldman, E.L. How does diabetes accelerate Alzheimer disease pathology? Nat. Rev. Neurol. 2010, 6, 551–559. [Google Scholar] [CrossRef]
- Biessels, G.J.; Staekenborg, S.; Brunner, E.; Brayne, C.; Scheltens, P. Risk of dementia in diabetes mellitus: A systematic review. Lancet Neurol. 2006, 5, 64–74. [Google Scholar] [CrossRef]
- Björk, B.F.; Katzov, H.; Kehoe, P.; Fratiglioni, L.; Winblad, B.; Prince, J.A.; Graff, C. Positive association between risk for late-onset Alzheimer disease and genetic variation in IDE. Neurobiol. Aging 2007, 28, 1374–1380. [Google Scholar] [CrossRef]
- Pivovarova, O.; Gögebakan, Ö.; Pfeiffer, A.F.H.; Rudovich, N. Glucose inhibits the insulin-induced activation of the insulin-degrading enzyme in HepG2 cells. Diabetologia 2009, 52, 1656–1664. [Google Scholar] [CrossRef]
- Aranda, J.F.; Canfrán-Duque, A.; Goedeke, L.; Suárez, Y.; Fernández-Hernando, C. The miR-199-dynamin regulatory axis controls receptor-mediated endocytosis. J. Cell Sci. 2015, 128, 3197–3209. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Gao, Y.; Li, X.; Tian, F.; Wang, F.; Wang, Y.; Zhao, B.; Zhang, R.; Wang, C. mRNA and miRNA expression profile reveals the role of miR-31 overexpression in neural stem cell. Sci. Rep. 2020, 10, 17537. [Google Scholar] [CrossRef] [PubMed]
- Meares, G.P.; Rajbhandari, R.; Gerigk, M.; Tien, C.-L.; Chang, C.; Fehling, S.C.; Rowse, A.; Mulhern, K.C.; Nair, S.; Gray, G.K.; et al. MicroRNA-31 is required for astrocyte specification. Glia 2018, 66, 987–998. [Google Scholar] [CrossRef]
- Qian, Y.; Li, X.; Fan, R.; Li, Q.; Zhang, Y.; He, X.; Yang, W.; Sun, W.; Lv, S. MicroRNA-31 inhibits traumatic brain injury-triggered neuronal cell apoptosis by regulating hypoxia-inducible factor-1A/vascular endothelial growth factor A axis. NeuroReport 2021, 33, 1–12. [Google Scholar] [CrossRef]
- Yan, S.-T.; Li, C.-L.; Tian, H.; Li, J.; Pei, Y.; Liu, Y.; Gong, Y.-P.; Fang, F.-S.; Sun, B.-R. MiR-199a is overexpressed in plasma of type 2 diabetes patients which contributes to type 2 diabetes by targeting GLUT4. Mol. Cell. Biochem. 2014, 397, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Mellios, N.; Feldman, D.A.; Sheridan, S.D.; Ip, J.P.K.; Kwok, S.; Amoah, S.K.; Rosen, B.; Rodriguez, B.A.; Crawford, B.; Swaminathan, R.; et al. MeCP2-regulated miRNAs control early human neurogenesis through differential effects on ERK and AKT signaling. Mol. Psychiatry 2018, 23, 1051–1065. [Google Scholar] [CrossRef]
- Song, D.; Li, G.; Hong, Y.; Zhang, P.; Zhu, J.; Yang, L.; Huang, J. miR-199a decreases Neuritin expression involved in the development of Alzheimer’s disease in APP/PS1 mice. Int. J. Mol. Med. 2020, 46, 384–396. [Google Scholar] [CrossRef]
- Gugliandolo, A.; Chiricosta, L.; Boccardi, V.; Mecocci, P.; Bramanti, P.; Mazzon, E. MicroRNAs Modulate the Pathogenesis of Alzheimer’s Disease: An In Silico Analysis in the Human Brain. Genes 2020, 11, 983. [Google Scholar] [CrossRef]
- Wu, D.; Huang, H.-J.; He, C.-N.; Wang, K.-Y. MicroRNA-199a-3p Regulates Endometrial Cancer Cell Proliferation by Targeting Mammalian Target of Rapamycin (mTOR). Int. J. Gynecol. Cancer 2013, 23, 1191–1197. [Google Scholar] [CrossRef]
- Aranda, J.F.; Rathjen, S.; Johannes, L.; Fernández-Hernando, C. MicroRNA 199a-5p Attenuates Retrograde Transport and Protects against Toxin-Induced Inhibition of Protein Biosynthesis. Mol. Cell. Biol. 2018, 38, 548–565. [Google Scholar] [CrossRef]
- Ramírez, C.; Lin, C.S.; Abdelmohsen, K.; Goedeke, L.; Yoon, J.-H.; Madrigal-Matute, J.; Martin-Ventura, J.L.; Vo, D.T.; Uren, P.J.; Penalva, L.; et al. RNA binding protein HuR regulates the expression of ABCA1. J. Lipid Res. 2014, 55, 1066–1076. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Yoon, H.; Ramírez, C.M.; Lee, S.M.; Hoe, H.S.; Fernández-Hernando, C.; Kim, J. MiR-106b impairs cholesterol efflux and increases Aβ levels by repressing ABCA1 expression. Exp. Neurol. 2012, 235, 476–483. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, C.M.; Rotllan, N.; Vlassov, A.V.; Dávalos, A.; Li, M.; Goedeke, L.; Aranda, J.F.; Cirera-Salinas, D.; Araldi, E.; Salerno, A.; et al. Control of Cholesterol Metabolism and Plasma High-Density Lipoprotein Levels by microRNA-144. Circ. Res. 2013, 112, 1592–1601. [Google Scholar] [CrossRef]
- Ramirez, C.M.; Dávalos, A.; Goedeke, L.; Salerno, A.; Warrier, N.; Cirera-Salinas, D.; Suárez, Y.; Fernández-Hernando, C. MicroRNA-758 Regulates Cholesterol Efflux Through Posttranscriptional Repression of ATP-Binding Cassette Transporter A1. Arterioscler. Thromb. Vasc. Biol. 2011, 31, 2707–2714. [Google Scholar] [CrossRef] [PubMed]

| Gene | Protein Name | N° Binding Sites | Matching Motif | RNA Motif | Biological Function | Reference |
|---|---|---|---|---|---|---|
| RALY | RNA-binding protein RALY | 301 | uuuuu/uuuuuub | RRM × 1 | Cholesterol biosynthesis Mitochondrial metabolism | [41,42,43] |
| ELAVL4 (HuD) | ELAV-like protein 4 | 322 | uuauu/uaauu | RRM × 3 | mRNA metabolism regulation Post-transcriptional control of gene expression Nervous system development and plasticity | [44,45,46,47] |
| TIA1 | Nucleolysin TIA-1 isoform p40 | 322 | uuuuubk/uuuuu | RRM × 3 | Synaptic plasticity regulation Alternative pre-RNA splicing Regulation of mRNA translation Apoptosis | [48,49] |
| RBMS3 | RNA-binding motif, single-stranded-interacting protein 3 | 347 | auauau/hauaua | RRM × 2 | Tumor cell defense Gene expression regulation | [50,51,52] |
| DAZ3 | Deleted in azoospermia protein 3 | 348 | aguuu/ uuguuu | RRM × 1 | Spermatogenesis | [53,54,55] |
| HNRNPC | Heterogeneous nuclear ribonucleoproteins C1/C2 | 349 | huuuuuk/ uuuuu | RRM × 1 | Cell cycle RNA Polymerase II Transcripts for Export | [56,57] |
| KHSRP/ FUBP2 | Far upstream element-binding protein 2 | 355 | uguau/ uuuuu | KH × 4 | Transcriptional regulator Gene expression regulation | [58,59] |
| HNRNP CL1 | Heterogeneous nuclear ribonucleoprotein C-like 1 | 357 | huuuuuk/ uuuuu | RRM × 1 | Nucleosome assembly | [60,61] |
| FUBP3 | Far upstream element-binding protein 3 | 375 | uauau/ uuaau/ uuuau | KH × 4 | Gene expression | [62] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martín-Martín, Y.; Pérez-García, A.; Torrecilla-Parra, M.; Fernández-de Frutos, M.; Pardo-Marqués, V.; Casarejos, M.J.; Busto, R.; Ramírez, C.M. New Insights on the Regulation of the Insulin-Degrading Enzyme: Role of microRNAs and RBPs. Cells 2022, 11, 2538. https://doi.org/10.3390/cells11162538
Martín-Martín Y, Pérez-García A, Torrecilla-Parra M, Fernández-de Frutos M, Pardo-Marqués V, Casarejos MJ, Busto R, Ramírez CM. New Insights on the Regulation of the Insulin-Degrading Enzyme: Role of microRNAs and RBPs. Cells. 2022; 11(16):2538. https://doi.org/10.3390/cells11162538
Chicago/Turabian StyleMartín-Martín, Yolanda, Ana Pérez-García, Marta Torrecilla-Parra, Mario Fernández-de Frutos, Virginia Pardo-Marqués, María José Casarejos, Rebeca Busto, and Cristina M. Ramírez. 2022. "New Insights on the Regulation of the Insulin-Degrading Enzyme: Role of microRNAs and RBPs" Cells 11, no. 16: 2538. https://doi.org/10.3390/cells11162538
APA StyleMartín-Martín, Y., Pérez-García, A., Torrecilla-Parra, M., Fernández-de Frutos, M., Pardo-Marqués, V., Casarejos, M. J., Busto, R., & Ramírez, C. M. (2022). New Insights on the Regulation of the Insulin-Degrading Enzyme: Role of microRNAs and RBPs. Cells, 11(16), 2538. https://doi.org/10.3390/cells11162538







