Small Extracellular Vesicles: Key Forces Mediating the Development and Metastasis of Colorectal Cancer
Abstract
:1. Background
2. Tumor-Derived sEVs Reshape the CRC Tumor Microenvironment
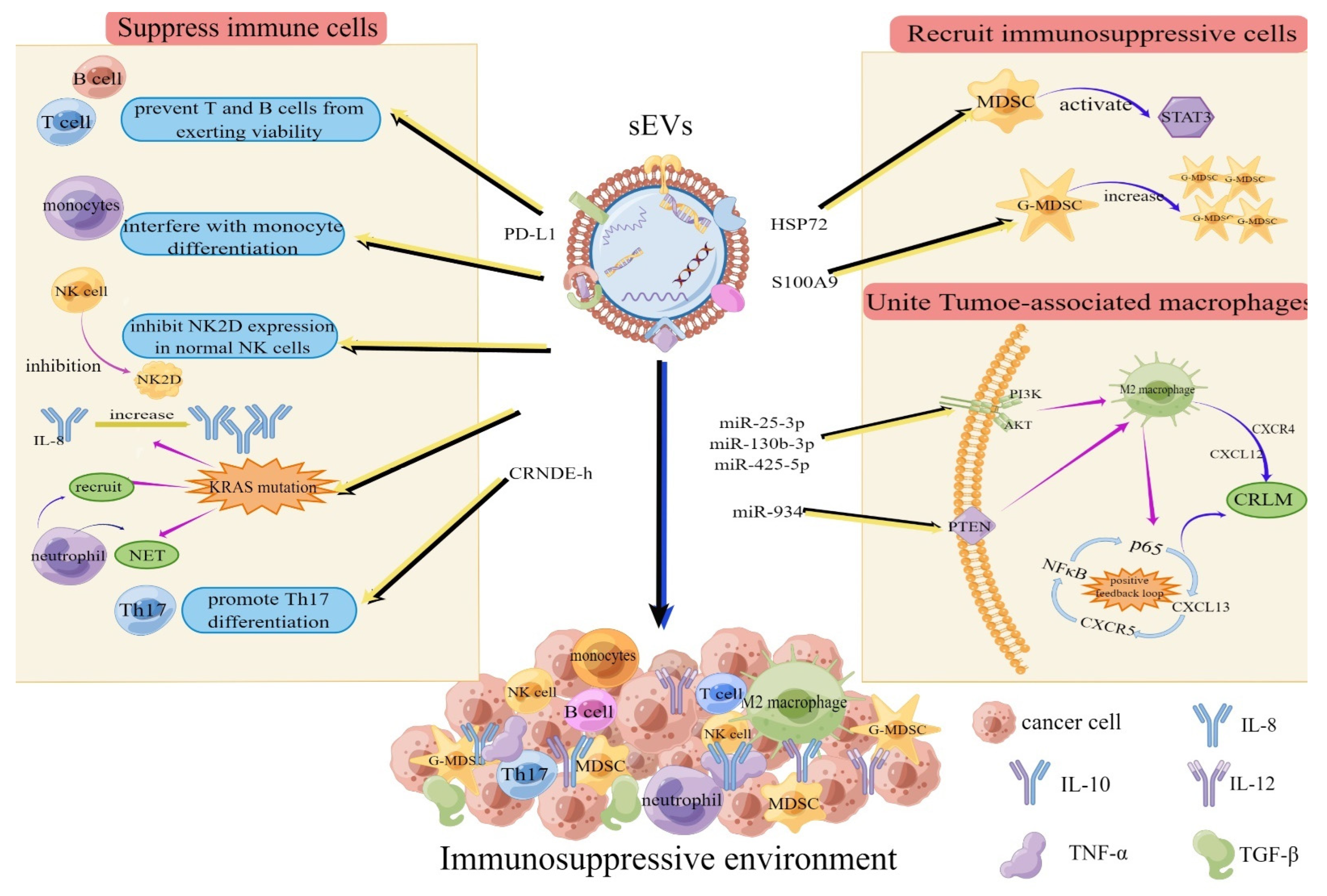
2.1. sEVs Mediate the Formation of Immunosuppressive Environments in Distant Organs
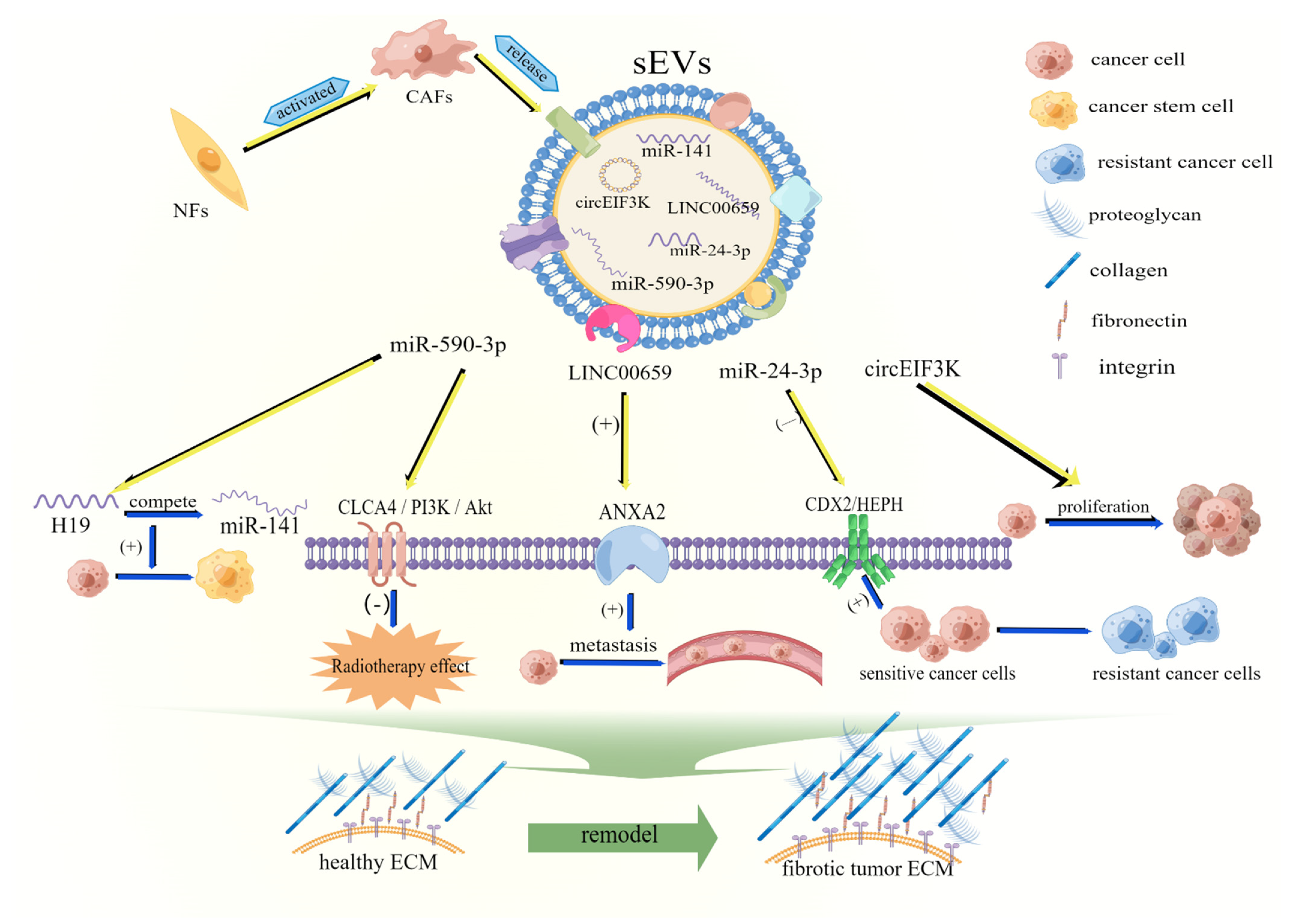
2.2. sEVs Are Involved in Remodeling the Extracellular Matrix of CRC
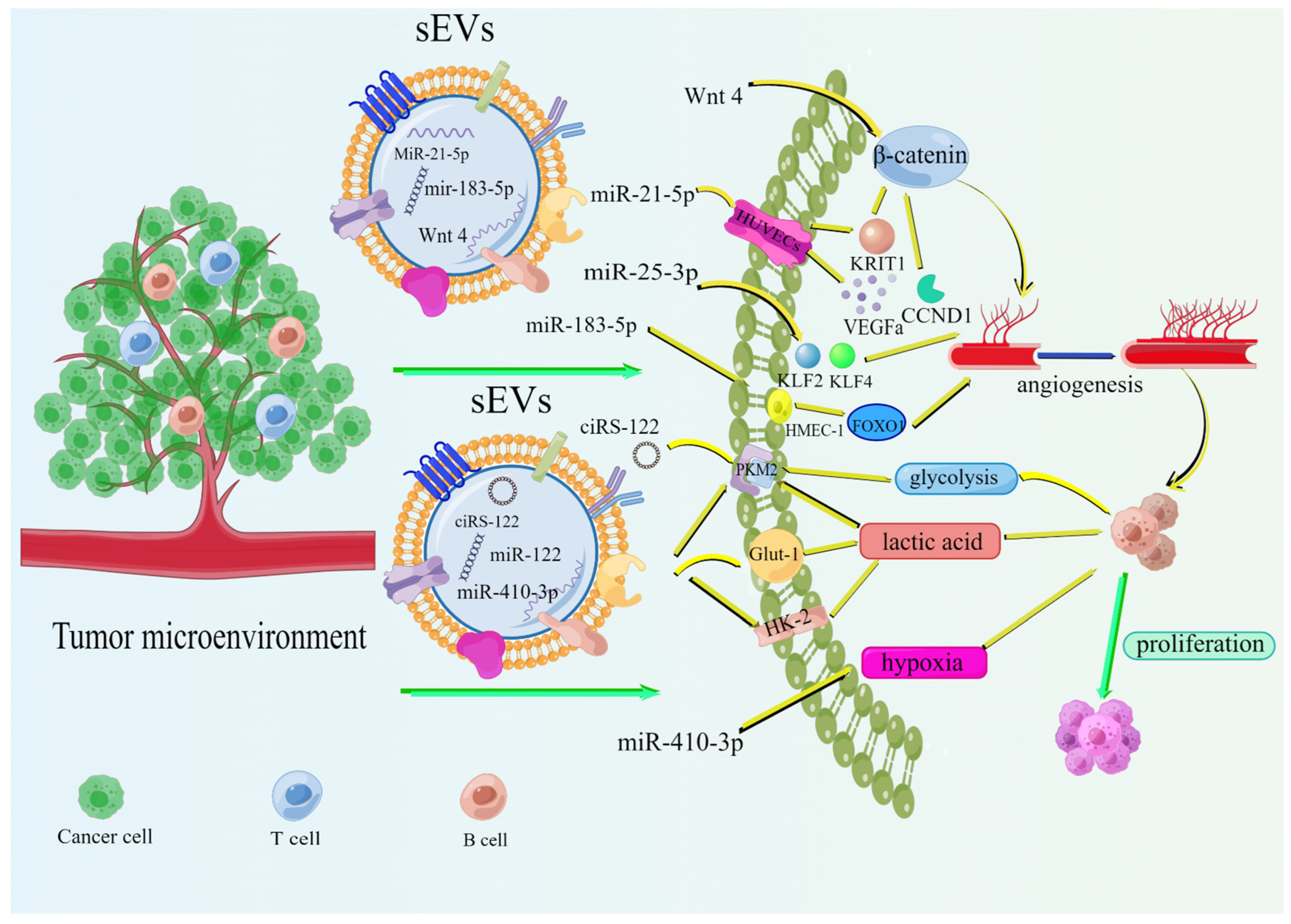
2.3. Tumor-Derived sEVs Promote the Metabolism of Promoted Tumor Cells
3. Other Cell-Derived sEVs Promote the Development and Metastasis of CRC
4. Intestinal Microbial-Derived sEVs Promote CRC Development and Metastasis
5. Application of sEVs in the Diagnosis and Treatment of CRC
5.1. sEVs as Early Markers for Colorectal Cancer Diagnosis
5.2. The Value of sEVs in CRC Treatment
5.3. Clinical Implications of sEVs as Markers and Drug Carriers in CRC Screening, Diagnosis, Treatment, and Prognosis
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CAFs | Cancer-associated fibroblasts |
| CRC | Colorectal cancer |
| sEVs | Small extracellular vesicles |
| miRNA/miR | microRNA |
| IECs | Intestinal epithelial cells |
| CDT | Cytolethal distending toxin |
| TME | Tumor microenvironment |
| TAM | Tumor-associated macrophage |
| CTLs | Cytotoxic T lymphocytes |
| PD-L1 | Programmed death-ligand 1 |
| NET | Neutrophil trap |
| MDSCs | Myeloid-derived suppressor cells |
| TNF-α | Tumor necrosis factor-alpha |
| TGF-β | Transforming growth factor-β |
| ECM | Extracellular matrix |
| ETBF | Enterotoxigenic Bacteroides fragilis |
| DSS | Dextran sodium sulfate |
| OMV | Outer membrane vesicle |
| lncRNA | Long noncoding RNA |
| HA | Hyaluronic acid |
| ICIs | Immune checkpoint inhibitors |
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terzić, J.; Grivennikov, S.; Karin, E.; Karin, M. Inflammation and colon cancer. Gastroenterology 2010, 138, 2101–2114.e2105. [Google Scholar] [CrossRef] [PubMed]
- Gallo, G.; Vescio, G.; De Paola, G. Therapeutic Targets and Tumor Microenvironment in Colorectal Cancer. J. Clin. Med. 2021, 10, 2295. [Google Scholar] [CrossRef]
- Hanus, M.; Parada-Venegas, D.; Landskron, G.; Wielandt, A.M.; Hurtado, C.; Alvarez, K.; Hermoso, M.A.; López-Köstner, F.; De la Fuente, M. Immune System, Microbiota, and Microbial Metabolites: The Unresolved Triad in Colorectal Cancer Microenvironment. Front. Immunol. 2021, 12, 612826. [Google Scholar] [CrossRef]
- Kasprzak, A. The Role of Tumor Microenvironment Cells in Colorectal Cancer (CRC) Cachexia. Int. J. Mol. Sci. 2021, 22, 1565. [Google Scholar] [CrossRef]
- Rocca, Y.S.; Roberti, M.P.; Arriaga, J.M.; Amat, M.; Bruno, L.; Pampena, M.B.; Huertas, E.; Loria, F.S.; Pairola, A.; Bianchini, M.; et al. Altered phenotype in peripheral blood and tumor-associated NK cells from colorectal cancer patients. Innate Immun. 2013, 19, 76–85. [Google Scholar] [CrossRef]
- Germann, M.; Zangger, N.; Sauvain, M.O.; Sempoux, C.; Bowler, A.D.; Wirapati, P.; Kandalaft, L.E.; Delorenzi, M.; Tejpar, S.; Coukos, G.; et al. Neutrophils suppress tumor-infiltrating T cells in colon cancer via matrix metalloproteinase-mediated activation of TGFβ. EMBO Mol. Med. 2020, 12, e10681. [Google Scholar] [CrossRef]
- Albini, A.; Bruno, A.; Noonan, D.M.; Mortara, L. Contribution to Tumor Angiogenesis From Innate Immune Cells within the Tumor Microenvironment: Implications for Immunotherapy. Front. Immunol. 2018, 9, 527. [Google Scholar] [CrossRef]
- Kowal, J.; Tkach, M.; Thery, C. Biogenesis and secretion of exosomes. Curr. Opin. Cell Biol. 2014, 29, 116–125. [Google Scholar] [CrossRef] [Green Version]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Thery, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Greening, D.W.; Gopal, S.K.; Mathias, R.A.; Liu, L.; Sheng, J.; Zhu, H.J.; Simpson, R.J. Emerging roles of exosomes during epithelial-mesenchymal transition and cancer progression. Semin. Cell Dev. Biol. 2015, 40, 60–71. [Google Scholar] [CrossRef] [PubMed]
- Mathivanan, S.; Ji, H.; Simpson, R.J. Exosomes: Extracellular organelles important in intercellular communication. J. Proteom. 2010, 73, 1907–1920. [Google Scholar] [CrossRef] [PubMed]
- Ratajczak, J.; Wysoczynski, M.; Hayek, F.; Janowska-Wieczorek, A.; Ratajczak, M.Z. Membrane-derived microvesicles: Important and underappreciated mediators of cell-to-cell communication. Leukemia 2006, 20, 1487–1495. [Google Scholar] [CrossRef]
- Li, K.; Chen, Y.; Li, A.; Tan, C.; Liu, X. Exosomes play roles in sequential processes of tumor metastasis. Int. J. Cancer 2019, 144, 1486–1495. [Google Scholar] [CrossRef]
- Wei, C.; Yang, C.; Wang, S.; Shi, D.; Zhang, C.; Lin, X.; Liu, Q.; Dou, R.; Xiong, B. Crosstalk between cancer cells and tumor associated macrophages is required for mesenchymal circulating tumor cell-mediated colorectal cancer metastasis. Mol. Cancer 2019, 18, 64. [Google Scholar] [CrossRef]
- Ge, P.; Wang, W.; Li, L.; Zhang, G.; Gao, Z.; Tang, Z.; Dang, X.; Wu, Y. Profiles of immune cell infiltration and immune-related genes in the tumor microenvironment of colorectal cancer. Biomed. Pharmacother. 2019, 118, 109228. [Google Scholar] [CrossRef]
- Kohlhapp, F.J.; Mitra, A.K.; Lengyel, E.; Peter, M.E. MicroRNAs as mediators and communicators between cancer cells and the tumor microenvironment. Oncogene 2015, 34, 5857–5868. [Google Scholar] [CrossRef] [Green Version]
- Whiteside, T.L. Exosomes and tumor-mediated immune suppression. J. Clin. Investig. 2016, 126, 1216–1223. [Google Scholar] [CrossRef] [Green Version]
- Chen, F.; Zhuang, X.; Lin, L.; Yu, P.; Wang, Y.; Shi, Y.; Hu, G.; Sun, Y. New horizons in tumor microenvironment biology: Challenges and opportunities. BMC Med. 2015, 13, 45. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Y.; Wang, Y.; Tiruthani, K. Tumor immune microenvironment and nano-immunotherapeutics in colorectal cancer. Nanomedicine 2019, 21, 102034. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Huang, A.C.; Zhang, W.; Zhang, G.; Wu, M.; Xu, W.; Yu, Z.; Yang, J.; Wang, B.; Sun, H.; et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 2018, 560, 382–386. [Google Scholar] [CrossRef] [PubMed]
- Mezzadra, R.; Sun, C.; Jae, L.T.; Gomez-Eerland, R.; de Vries, E.; Wu, W.; Logtenberg, M.E.W.; Slagter, M.; Rozeman, E.A.; Hofland, I.; et al. Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature 2017, 549, 106–110. [Google Scholar] [CrossRef] [PubMed]
- Vafaei, S.; Roudi, R.; Madjd, Z.; Aref, A.R.; Ebrahimi, M. Potential theranostics of circulating tumor cells and tumor-derived exosomes application in colorectal cancer. Cancer Cell Int. 2020, 20, 288. [Google Scholar] [CrossRef]
- Sun, J.; Jia, H.; Bao, X.; Wu, Y.; Zhu, T.; Li, R.; Zhao, H. Tumor exosome promotes Th17 cell differentiation by transmitting the lncRNA CRNDE-h in colorectal cancer. Cell Death Dis. 2021, 12, 123. [Google Scholar] [CrossRef]
- Liu, Y.; Cao, X. Characteristics and Significance of the Pre-Metastatic Niche. Cancer Cell 2016, 30, 668–681. [Google Scholar] [CrossRef] [Green Version]
- Ji, D.; Song, C.; Li, Y.; Xia, J.; Wu, Y.; Jia, J.; Cui, X.; Yu, S.; Gu, J. Combination of radiotherapy and suppression of Tregs enhances abscopal antitumor effect and inhibits metastasis in rectal cancer. J. Immunother. Cancer 2020, 8, e000826. [Google Scholar] [CrossRef]
- Liu, J.; Ren, L.; Li, S.; Li, W.; Zheng, X.; Yang, Y.; Fu, W.; Yi, J.; Wang, J.; Du, G. The biology, function, and applications of exosomes in cancer. Acta Pharm. Sin. B 2021, 11, 2783–2797. [Google Scholar] [CrossRef]
- Wang, Y.; Yin, K.; Tian, J.; Xia, X.; Ma, J.; Tang, X.; Xu, H.; Wang, S. Granulocytic Myeloid-Derived Suppressor Cells Promote the Stemness of Colorectal Cancer Cells through Exosomal S100A9. Adv. Sci. (Weinh) 2019, 6, 1901278. [Google Scholar] [CrossRef] [Green Version]
- Komohara, Y.; Fujiwara, Y.; Ohnishi, K.; Takeya, M. Tumor-associated macrophages: Potential therapeutic targets for anti-cancer therapy. Adv. Drug Deliv. Rev. 2016, 99, 180–185. [Google Scholar] [CrossRef]
- Liang, Z.-X.; Liu, H.-S.; Wang, F.-W.; Xiong, L.; Zhou, C.; Hu, T.; He, X.-W.; Wu, X.-J.; Xie, D.; Wu, X.-R.; et al. LncRNA RPPH1 promotes colorectal cancer metastasis by interacting with TUBB3 and by promoting exosomes-mediated macrophage M2 polarization. Cell Death Dis. 2019, 10, 829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mills, C.D. M1 and M2 Macrophages: Oracles of Health and Disease. Crit. Rev. Immunol. 2012, 32, 463–488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Wang, X.; Si, M.; Yang, J.; Sun, S.; Wu, H.; Cui, S.; Qu, X.; Yu, X. Exosome-encapsulated miRNAs contribute to CXCL12/CXCR4-induced liver metastasis of colorectal cancer by enhancing M2 polarization of macrophages. Cancer Lett. 2020, 474, 36–52. [Google Scholar] [CrossRef]
- Zhao, S.; Mi, Y.; Guan, B.; Zheng, B.; Wei, P.; Gu, Y.; Zhang, Z.; Cai, S.; Xu, Y.; Li, X.; et al. Tumor-derived exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer. J. Hematol. Oncol. 2020, 13, 156. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Cao, F.; Huang, S.; Zheng, Y. Follistatin-Like 3 Correlates with Lymph Node Metastasis and Serves as a Biomarker of Extracellular Matrix Remodeling in Colorectal Cancer. Front. Immunol. 2021, 12, 717505. [Google Scholar] [CrossRef] [PubMed]
- Mouw, J.K.; Ou, G.; Weaver, V.M. Extracellular matrix assembly: A multiscale deconstruction. Nat. Rev. Mol. Cell Biol. 2014, 15, 771–785. [Google Scholar] [CrossRef] [PubMed]
- Au Yeung, C.L.; Co, N.N.; Tsuruga, T.; Yeung, T.L.; Kwan, S.Y.; Leung, C.S.; Li, Y.; Lu, E.S.; Kwan, K.; Wong, K.K.; et al. Exosomal transfer of stroma-derived miR21 confers paclitaxel resistance in ovarian cancer cells through targeting APAF1. Nat. Commun. 2016, 7, 11150. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.-P.; Xu, K.; Cui, J.; Yuan, D.-Y.; Zou, B.; Li, J.; Liu, J.-L.; Li, K.-Y.; Meng, Z.; Zhang, B. Cancer-associated fibroblast-derived exosomal miR-382-5p promotes the migration and invasion of oral squamous cell carcinoma. Oncol. Rep. 2019, 42, 1319–1328. [Google Scholar] [CrossRef]
- Chen, X.; Liu, Y.; Zhang, Q.; Liu, B.; Cheng, Y.; Zhang, Y.; Sun, Y.; Liu, J. Exosomal miR-590-3p derived from cancer-associated fibroblasts confers radioresistance in colorectal cancer. Mol. Ther. Nucleic Acids 2021, 24, 113–126. [Google Scholar] [CrossRef]
- Piersma, B.; Hayward, M.K.; Weaver, V.M. Fibrosis and cancer: A strained relationship. Biochim. Biophys. Acta Rev. Cancer 2020, 1873, 188356. [Google Scholar] [CrossRef]
- Albacete-Albacete, L.; Navarro-Lérida, I.; López, J.A.; Martín-Padura, I.; Astudillo, A.M.; Ferrarini, A.; Van-Der-Heyden, M.; Balsinde, J.; Orend, G.; Vázquez, J.; et al. ECM deposition is driven by caveolin-1-dependent regulation of exosomal biogenesis and cargo sorting. J. Cell Biol. 2020, 219, e202006178. [Google Scholar] [CrossRef] [PubMed]
- Herrera, M.; Llorens, C.; Rodríguez, M.; Herrera, A.; Ramos, R.; Gil, B.; Candia, A.; Larriba, M.J.; Garre, P.; Earl, J.; et al. Differential distribution and enrichment of non-coding RNAs in exosomes from normal and Cancer-associated fibroblasts in colorectal cancer. Mol. Cancer 2018, 17, 114. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Ding, L.; Zhang, D.; Shi, G.; Xu, Q.; Shen, S.; Wang, Y.; Wang, T.; Hou, Y. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics 2018, 8, 3932–3948. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Zhang, J.; Bao, C. Exosomal circEIF3K from cancer-associated fibroblast promotes colorectal cancer (CRC) progression via miR-214/PD-L1 axis. BMC Cancer 2021, 21, 933. [Google Scholar] [CrossRef]
- Zhou, L.; Li, J.; Tang, Y.; Yang, M. Exosomal LncRNA LINC00659 transferred from cancer-associated fibroblasts promotes colorectal cancer cell progression via miR-342-3p/ANXA2 axis. J. Transl. Med. 2021, 19, 8. [Google Scholar] [CrossRef]
- Zhang, H.W.; Shi, Y.; Liu, J.B.; Wang, H.M.; Wang, P.Y.; Wu, Z.J.; Li, L.; Gu, L.P.; Cao, P.S.; Wang, G.R.; et al. Cancer-associated fibroblast-derived exosomal microRNA-24-3p enhances colon cancer cell resistance to MTX by down-regulating CDX2/HEPH axis. J. Cell. Mol. Med. 2021, 25, 3699–3713. [Google Scholar] [CrossRef]
- Guo, Y.; Ji, X.; Liu, J.; Fan, D.; Zhou, Q.; Chen, C.; Wang, W.; Wang, G.; Wang, H.; Yuan, W.; et al. Effects of exosomes on pre-metastatic niche formation in tumors. Mol. Cancer 2019, 18, 39. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Yu, D. Exosomes in cancer development, metastasis, and immunity. Biochim. Biophys. Acta Rev. Cancer 2019, 1871, 455–468. [Google Scholar] [CrossRef]
- Wang, W.; Zhu, N.; Yan, T.; Shi, Y.N.; Chen, J.; Zhang, C.J.; Xie, X.J.; Liao, D.F.; Qin, L. The crosstalk: Exosomes and lipid metabolism. Cell Commun. Signal. 2020, 18, 119. [Google Scholar] [CrossRef]
- Peinado, H.; Zhang, H.; Matei, I.R.; Costa-Silva, B.; Hoshino, A.; Rodrigues, G.; Psaila, B.; Kaplan, R.N.; Bromberg, J.F.; Kang, Y.; et al. Pre-metastatic niches: Organ-specific homes for metastases. Nat. Rev. Cancer 2017, 17, 302–317. [Google Scholar] [CrossRef]
- Tai, Y.L.; Chen, K.C.; Hsieh, J.T.; Shen, T.L. Exosomes in cancer development and clinical applications. Cancer Sci. 2018, 109, 2364–2374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Des Guetz, G.; Uzzan, B.; Nicolas, P.; Cucherat, M.; Morere, J.F.; Benamouzig, R.; Breau, J.L.; Perret, G.Y. Microvessel density and VEGF expression are prognostic factors in colorectal cancer. Meta-analysis of the literature. Br. J. Cancer 2006, 94, 1823–1832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kather, J.N.; Zöllner, F.G.; Schad, L.R.; Melchers, S.M.; Sinn, H.P.; Marx, A.; Gaiser, T.; Weis, C.A. Identification of a characteristic vascular belt zone in human colorectal cancer. PLoS ONE 2017, 12, e0171378. [Google Scholar] [CrossRef]
- Olejarz, W.; Kubiak-Tomaszewska, G.; Chrzanowska, A.; Lorenc, T. Exosomes in Angiogenesis and Anti-Angiogenic Therapy in Cancers. Int. J. Mol. Sci. 2020, 21, 5840. [Google Scholar] [CrossRef]
- Zeng, Z.; Li, Y.; Pan, Y.; Lan, X.; Song, F.; Sun, J.; Zhou, K.; Liu, X.; Ren, X.; Wang, F.; et al. Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat. Commun. 2018, 9, 5395. [Google Scholar] [CrossRef] [Green Version]
- He, Q.; Ye, A.; Ye, W.; Liao, X.; Qin, G.; Xu, Y.; Yin, Y.; Luo, H.; Yi, M.; Xian, L.; et al. Cancer-secreted exosomal miR-21-5p induces angiogenesis and vascular permeability by targeting KRIT1. Cell Death Dis. 2021, 12, 576. [Google Scholar] [CrossRef]
- Shang, A.; Wang, X.; Gu, C.; Liu, W.; Sun, J.; Zeng, B.; Chen, C.; Ji, P.; Wu, J.; Quan, W.; et al. Exosomal miR-183-5p promotes angiogenesis in colorectal cancer by regulation of FOXO1. Aging 2020, 12, 8352–8371. [Google Scholar] [CrossRef]
- Huang, Z.; Feng, Y. Exosomes Derived from Hypoxic Colorectal Cancer Cells Promote Angiogenesis through Wnt4-Induced β-Catenin Signaling in Endothelial Cells. Oncol. Res. 2017, 25, 651–661. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, H.; Yang, H.; Bai, M.; Ning, T.; Deng, T.; Liu, R.; Fan, Q.; Zhu, K.; Li, J.; et al. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol. Oncol. 2020, 14, 539–555. [Google Scholar] [CrossRef]
- Annese, T.; Tamma, R.; Ruggieri, S.; Ribatti, D. Angiogenesis in Pancreatic Cancer: Pre-Clinical and Clinical Studies. Cancers 2019, 11, 381. [Google Scholar] [CrossRef] [Green Version]
- Hu, X.; Mu, Y.; Liu, J.; Mu, X.; Gao, F.; Chen, L.; Wu, H.; Wu, H.; Liu, W.; Zhao, Y. Exosomes Derived from Hypoxic Colorectal Cancer Cells Transfer miR-410-3p to Regulate Tumor Progression. J. Cancer 2020, 11, 4724–4735. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yang, P.; Chen, F.; Tan, Y.; Huang, C.; Shen, H.; Peng, C.; Feng, Y.; Sun, Y. Hypoxic colorectal cancer-derived extracellular vesicles deliver microRNA-361-3p to facilitate cell proliferation by targeting TRAF3 via the noncanonical NF-κB pathways. Clin. Transl. Med. 2021, 11, e349. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Li, H.; Xie, H.; Wu, X.; Lan, P. The malignant role of exosomes in the communication among colorectal cancer cell, macrophage and microbiome. Carcinogenesis 2019, 40, 601–610. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, B.; Ren, C.; Chen, Y.; Guo, X.; Zhou, L.; Peng, Z.; Tang, Y.; Chen, Y.; Liu, W.; et al. The clinical significance of circulating GPC1 positive exosomes and its regulative miRNAs in colon cancer patients. Oncotarget 2017, 8, 101189–101202. [Google Scholar] [CrossRef]
- Shabo, I.; Svanvik, J. Expression of macrophage antigens by tumor cells. Adv. Exp. Med. Biol. 2011, 714, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.; Sun, L.; Xu, F.; Liu, L.; Hu, F.; Song, D.; Hou, Z.; Wu, W.; Luo, X.; Wang, J.; et al. M2 Macrophage-Derived Exosomes Promote Cell Migration and Invasion in Colon Cancer. Cancer Res. 2019, 79, 146–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Su, Z.; Amoah Barnie, P. Crosstalk among colon cancer-derived exosomes, fibroblast-derived exosomes, and macrophage phenotypes in colon cancer metastasis. Int. Immunopharmacol. 2020, 81, 106298. [Google Scholar] [CrossRef]
- Hu, J.L.; Wang, W.; Lan, X.L.; Zeng, Z.C.; Liang, Y.S.; Yan, Y.R.; Song, F.Y.; Wang, F.F.; Zhu, X.H.; Liao, W.J.; et al. CAFs secreted exosomes promote metastasis and chemotherapy resistance by enhancing cell stemness and epithelial-mesenchymal transition in colorectal cancer. Mol. Cancer 2019, 18, 91. [Google Scholar] [CrossRef] [Green Version]
- Chronopoulos, A.; Kalluri, R. Emerging role of bacterial extracellular vesicles in cancer. Oncogene 2020, 39, 6951–6960. [Google Scholar] [CrossRef]
- Kim, J.H.; Lee, J.; Park, J.; Gho, Y.S. Gram-negative and Gram-positive bacterial extracellular vesicles. Semin. Cell Dev. Biol. 2015, 40, 97–104. [Google Scholar] [CrossRef]
- Zhou, L.; Srisatjaluk, R.; Justus, D.E.; Doyle, R.J. On the origin of membrane vesicles in gram-negative bacteria. FEMS Microbiol. Lett. 1998, 163, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.J.; Wang, X.H.; Fan, G.C. Versatile effects of bacterium-released membrane vesicles on mammalian cells and infectious/inflammatory diseases. Acta Pharmacol. Sin. 2018, 39, 514–533. [Google Scholar] [CrossRef] [PubMed]
- Celluzzi, A.; Masotti, A. How Our Other Genome Controls Our Epi-Genome. Trends Microbiol. 2016, 24, 777–787. [Google Scholar] [CrossRef] [PubMed]
- Brenchley, J.M.; Douek, D.C. Microbial translocation across the GI tract. Annu. Rev. Immunol. 2012, 30, 149–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Catalioto, R.M.; Maggi, C.A.; Giuliani, S. Intestinal epithelial barrier dysfunction in disease and possible therapeutical interventions. Curr. Med. Chem. 2011, 18, 398–426. [Google Scholar] [CrossRef]
- Sultan, S.; Mottawea, W.; Yeo, J.; Hammami, R. Gut Microbiota Extracellular Vesicles as Signaling Molecules Mediating Host-Microbiota Communications. Int. J. Mol. Sci. 2021, 22, 13166. [Google Scholar] [CrossRef]
- Elmi, A.; Nasher, F.; Jagatia, H.; Gundogdu, O.; Bajaj-Elliott, M.; Wren, B.; Dorrell, N. Campylobacter jejuni outer membrane vesicle-associated proteolytic activity promotes bacterial invasion by mediating cleavage of intestinal epithelial cell E-cadherin and occludin. Cell Microbiol. 2016, 18, 561–572. [Google Scholar] [CrossRef]
- Hickey, C.A.; Kuhn, K.A.; Donermeyer, D.L.; Porter, N.T.; Jin, C.; Cameron, E.A.; Jung, H.; Kaiko, G.E.; Wegorzewska, M.; Malvin, N.P.; et al. Colitogenic Bacteroides thetaiotaomicron Antigens Access Host Immune Cells in a Sulfatase-Dependent Manner via Outer Membrane Vesicles. Cell Host Microbe 2015, 17, 672–680. [Google Scholar] [CrossRef] [Green Version]
- Cañas, M.A.; Fábrega, M.J.; Giménez, R.; Badia, J.; Baldomà, L. Outer Membrane Vesicles from Probiotic and Commensal Escherichia coli Activate NOD1-Mediated Immune Responses in Intestinal Epithelial Cells. Front. Microbiol. 2018, 9, 498. [Google Scholar] [CrossRef] [Green Version]
- Patten, D.A.; Hussein, E.; Davies, S.P.; Humphreys, P.N.; Collett, A. Commensal-derived OMVs elicit a mild proinflammatory response in intestinal epithelial cells. Microbiology (Reading) 2017, 163, 702–711. [Google Scholar] [CrossRef]
- Lee, H.S.; Boulton, I.C.; Reddin, K.; Wong, H.; Halliwell, D.; Mandelboim, O.; Gorringe, A.R.; Gray-Owen, S.D. Neisserial outer membrane vesicles bind the coinhibitory receptor carcinoembryonic antigen-related cellular adhesion molecule 1 and suppress CD4+ T lymphocyte function. Infect. Immun. 2007, 75, 4449–4455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsatsaronis, J.A.; Franch-Arroyo, S.; Resch, U.; Charpentier, E. Extracellular Vesicle RNA: A Universal Mediator of Microbial Communication? Trends Microbiol. 2018, 26, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Shah, B.; Sullivan, C.J.; Lonergan, N.E.; Stanley, S.; Soult, M.C.; Britt, L.D. Circulating bacterial membrane vesicles cause sepsis in rats. Shock 2012, 37, 621–628. [Google Scholar] [CrossRef] [PubMed]
- van Bergenhenegouwen, J.; Kraneveld, A.D.; Rutten, L.; Kettelarij, N.; Garssen, J.; Vos, A.P. Extracellular vesicles modulate host-microbe responses by altering TLR2 activity and phagocytosis. PLoS ONE 2014, 9, e89121. [Google Scholar] [CrossRef] [PubMed]
- Kok, V.C.; Yu, C.C. Cancer-Derived Exosomes: Their Role in Cancer Biology and Biomarker Development. Int. J. Nanomed. 2020, 15, 8019–8036. [Google Scholar] [CrossRef]
- Wang, J.; Yan, F.; Zhao, Q.; Zhan, F.; Wang, R.; Wang, L.; Zhang, Y.; Huang, X. Circulating exosomal miR-125a-3p as a novel biomarker for early-stage colon cancer. Sci. Rep. 2017, 7, 4150. [Google Scholar] [CrossRef] [Green Version]
- Yan, S.; Jiang, Y.; Liang, C.; Cheng, M.; Jin, C.; Duan, Q.; Xu, D. Exosomal miR-6803-5p as potential diagnostic and prognostic marker in colorectal cancer. J. Cell Biochem. 2018, 119, 4113–4119. [Google Scholar] [CrossRef]
- Min, L.; Chen, L.; Liu, S.; Yu, Y.; Guo, Q.; Li, P.; Zhu, S. Loss of Circulating Exosomal miR-92b is a Novel Biomarker of Colorectal Cancer at Early Stage. Int. J. Med. Sci. 2019, 16, 1231–1237. [Google Scholar] [CrossRef] [Green Version]
- Dong, S.J.; Cai, X.J.; Li, S.J. The Clinical Significance of MiR-429 as a Predictive Biomarker in Colorectal Cancer Patients Receiving 5-Fluorouracil Treatment. Med. Sci. Monit. 2016, 22, 3352–3361. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Zhao, J.; Zhang, R. Prognostic significance of serum miR-193b in colorectal cancer. Int. J. Clin. Exp. Pathol. 2017, 10, 9509–9514. [Google Scholar]
- Dai, M.; Chen, X.; Mo, S.; Li, J.; Huang, Z.; Huang, S.; Xu, J.; He, B.; Zou, Y.; Chen, J.; et al. Meta-signature LncRNAs serve as novel biomarkers for colorectal cancer: Integrated bioinformatics analysis, experimental validation and diagnostic evaluation. Sci. Rep. 2017, 7, 46572. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Zhan, Y.; Zhu, K.; Bai, M.; Han, J.; Si, Y.; Zhang, H.; Kong, D. Plasma Exosomal Long Non-Coding RNAs Serve as Biomarkers for Early Detection of Colorectal Cancer. Cell. Physiol. Biochem. 2018, 51, 2704–2715. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhang, X.; Gao, S.; Jing, F.; Yang, Y.; Du, L.; Zheng, G.; Li, P.; Li, C.; Wang, C. Exosomal long noncoding RNA CRNDE-h as a novel serum-based biomarker for diagnosis and prognosis of colorectal cancer. Oncotarget 2016, 7, 85551–85563. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Meng, T.; Yang, X.H.; Sayim, P.; Lei, C.; Jin, B.; Ge, L.; Wang, H.J. Prognostic and predictive value of long non-coding RNA GAS5 and mircoRNA-221 in colorectal cancer and their effects on colorectal cancer cell proliferation, migration and invasion. Cancer Biomark. 2018, 22, 283–299. [Google Scholar] [CrossRef] [PubMed]
- Ling, H.; Spizzo, R.; Atlasi, Y.; Nicoloso, M.; Shimizu, M.; Redis, R.S.; Nishida, N.; Gafà, R.; Song, J.; Guo, Z.; et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013, 23, 1446–1461. [Google Scholar] [CrossRef] [Green Version]
- Paul, D.; Roy, A.; Nandy, A.; Datta, B.; Borar, P.; Pal, S.K.; Senapati, D.; Rakshit, T. Identification of Biomarker Hyaluronan on Colon Cancer Extracellular Vesicles Using Correlative AFM and Spectroscopy. J. Phys. Chem. Lett. 2020, 11, 5569–5576. [Google Scholar] [CrossRef]
- Tian, Y.; Ma, L.; Gong, M.; Su, G.; Zhu, S.; Zhang, W.; Wang, S.; Li, Z.; Chen, C.; Li, L.; et al. Protein Profiling and Sizing of Extracellular Vesicles from Colorectal Cancer Patients via Flow Cytometry. ACS Nano 2018, 12, 671–680. [Google Scholar] [CrossRef]
- Chen, Y.; Xie, Y.; Xu, L.; Zhan, S.; Xiao, Y.; Gao, Y.; Wu, B.; Ge, W. Protein content and functional characteristics of serum-purified exosomes from patients with colorectal cancer revealed by quantitative proteomics. Int. J. Cancer 2017, 140, 900–913. [Google Scholar] [CrossRef]
- Campanella, C.; Rappa, F.; Sciumè, C.; Marino Gammazza, A.; Barone, R.; Bucchieri, F.; David, S.; Curcurù, G.; Caruso Bavisotto, C.; Pitruzzella, A.; et al. Heat shock protein 60 levels in tissue and circulating exosomes in human large bowel cancer before and after ablative surgery. Cancer 2015, 121, 3230–3239. [Google Scholar] [CrossRef]
- Li, J.; Chen, Y.; Guo, X.; Zhou, L.; Jia, Z.; Peng, Z.; Tang, Y.; Liu, W.; Zhu, B.; Wang, L.; et al. GPC1 exosome and its regulatory miRNAs are specific markers for the detection and target therapy of colorectal cancer. J. Cell. Mol. Med. 2017, 21, 838–847. [Google Scholar] [CrossRef]
- Sun, B.; Li, Y.; Zhou, Y.; Ng, T.K.; Zhao, C.; Gan, Q.; Gu, X.; Xiang, J. Circulating exosomal CPNE3 as a diagnostic and prognostic biomarker for colorectal cancer. J. Cell. Physiol. 2019, 234, 1416–1425. [Google Scholar] [CrossRef] [PubMed]
- Zhong, M.E.; Chen, Y.; Xiao, Y.; Xu, L.; Zhang, G.; Lu, J.; Qiu, H.; Ge, W.; Wu, B. Serum extracellular vesicles contain SPARC and LRG1 as biomarkers of colon cancer and differ by tumour primary location. EBioMedicine 2019, 50, 211–223. [Google Scholar] [CrossRef] [Green Version]
- Yokoyama, S.; Takeuchi, A.; Yamaguchi, S.; Mitani, Y.; Watanabe, T.; Matsuda, K.; Hotta, T.; Shively, J.E.; Yamaue, H. Clinical implications of carcinoembryonic antigen distribution in serum exosomal fraction-Measurement by ELISA. PLoS ONE 2017, 12, e0183337. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Li, S.; Song, J.; Ji, T.; Zhu, M.; Anderson, G.J.; Wei, J.; Nie, G. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials 2014, 35, 2383–2390. [Google Scholar] [CrossRef] [PubMed]
- Zaharie, F.; Muresan, M.S.; Petrushev, B.; Berce, C.; Gafencu, G.A.; Selicean, S.; Jurj, A.; Cojocneanu-Petric, R.; Lisencu, C.I.; Pop, L.A.; et al. Exosome-Carried microRNA-375 Inhibits Cell Progression and Dissemination via Bcl-2 Blocking in Colon Cancer. J. Gastrointestin. Liver Dis. 2015, 24, 435–443. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.L.; He, G.Y.; Lan, X.L.; Zeng, Z.C.; Guan, J.; Ding, Y.; Qian, X.L.; Liao, W.T.; Ding, Y.Q.; Liang, L. Inhibition of ATG12-mediated autophagy by miR-214 enhances radiosensitivity in colorectal cancer. Oncogenesis 2018, 7, 16. [Google Scholar] [CrossRef]
- Roberts, R.; Moreno, G.; Bottero, D.; Gaillard, M.E.; Fingermann, M.; Graieb, A.; Rumbo, M.; Hozbor, D. Outer membrane vesicles as acellular vaccine against pertussis. Vaccine 2008, 26, 4639–4646. [Google Scholar] [CrossRef]
- Wang, S.; Gao, J.; Wang, Z. Outer membrane vesicles for vaccination and targeted drug delivery. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2019, 11, e1523. [Google Scholar] [CrossRef]
- Sierra, G.V.; Campa, H.C.; Varcacel, N.M.; Garcia, I.L.; Izquierdo, P.L.; Sotolongo, P.F.; Casanueva, G.V.; Rico, C.O.; Rodriguez, C.R.; Terry, M.H. Vaccine against group B Neisseria meningitidis: Protection trial and mass vaccination results in Cuba. NIPH Ann. 1991, 14, 195–207; discussion 208–110. [Google Scholar]
- Kim, O.Y.; Park, H.T.; Dinh, N.T.H.; Choi, S.J.; Lee, J.; Kim, J.H.; Lee, S.W.; Gho, Y.S. Bacterial outer membrane vesicles suppress tumor by interferon-γ-mediated antitumor response. Nat. Commun. 2017, 8, 626. [Google Scholar] [CrossRef]
- Ahmadi Badi, S.; Khatami, S.H.; Irani, S.H.; Siadat, S.D. Induction Effects of Bacteroides fragilis Derived Outer Membrane Vesicles on Toll Like Receptor 2, Toll Like Receptor 4 Genes Expression and Cytokines Concentration in Human Intestinal Epithelial Cells. Cell J. 2019, 21, 57–61. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, V.; Jalan, R. Acute-on-Chronic Liver Failure: Definition, Diagnosis, and Clinical Characteristics. Semin Liver Dis 2016, 36, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, V.; Helmink, B.A.; Spencer, C.N.; Reuben, A.; Wargo, J.A. The Influence of the Gut Microbiome on Cancer, Immunity, and Cancer Immunotherapy. Cancer Cell 2018, 33, 570–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baothman, O.A.; Zamzami, M.A.; Taher, I.; Abubaker, J.; Abu-Farha, M. The role of Gut Microbiota in the development of obesity and Diabetes. Lipids Health Dis. 2016, 15, 108. [Google Scholar] [CrossRef] [Green Version]
- Marin, I.A.; Goertz, J.E.; Ren, T.; Rich, S.S.; Onengut-Gumuscu, S.; Farber, E.; Wu, M.; Overall, C.C.; Kipnis, J.; Gaultier, A. Microbiota alteration is associated with the development of stress-induced despair behavior. Sci. Rep. 2017, 7, 43859. [Google Scholar] [CrossRef]
- Liu, S.; da Cunha, A.P.; Rezende, R.M.; Cialic, R.; Wei, Z.; Bry, L.; Comstock, L.E.; Gandhi, R.; Weiner, H.L. The Host Shapes the Gut Microbiota via Fecal MicroRNA. Cell Host Microbe 2016, 19, 32–43. [Google Scholar] [CrossRef] [Green Version]
- Ren, D.; Lin, B.; Zhang, X.; Peng, Y.; Ye, Z.; Ma, Y.; Liang, Y.; Cao, L.; Li, X.; Li, R.; et al. Maintenance of cancer stemness by miR-196b-5p contributes to chemoresistance of colorectal cancer cells via activating STAT3 signaling pathway. Oncotarget 2017, 8, 49807–49823. [Google Scholar] [CrossRef] [Green Version]
- Verbanac, D.; Čeri, A. Profiling Colorectal Cancer in the Landscape Personalized Testing-Advantages of Liquid Biopsy. Int. J. Mol. Sci. 2021, 22, 4327. [Google Scholar] [CrossRef]
- Min, L.; Zhu, S.; Chen, L.; Liu, X.; Wei, R.; Zhao, L.; Yang, Y.; Zhang, Z.; Kong, G.; Li, P.; et al. Evaluation of circulating small extracellular vesicles derived miRNAs as biomarkers of early colon cancer: A comparison with plasma total miRNAs. J. Extracell. Vesicles 2019, 8, 1643670. [Google Scholar] [CrossRef] [Green Version]
- Ogata-Kawata, H.; Izumiya, M.; Kurioka, D.; Honma, Y.; Yamada, Y.; Furuta, K.; Gunji, T.; Ohta, H.; Okamoto, H.; Sonoda, H.; et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS ONE 2014, 9, e92921. [Google Scholar] [CrossRef]
- Tan, H.Y.; Zheng, Y.B.; Liu, J. Serum miR-199a as a potential diagnostic biomarker for detection of colorectal cancer. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 8657–8663. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Zhang, X.; Wang, H.; Wang, L.; Liu, T.; Du, L.; Yang, Y.; Wang, C. MALAT1 Is Associated with Poor Response to Oxaliplatin-Based Chemotherapy in Colorectal Cancer Patients and Promotes Chemoresistance through EZH2. Mol. Cancer Ther. 2017, 16, 739–751. [Google Scholar] [CrossRef] [Green Version]
- Agrawal, A.K.; Aqil, F.; Jeyabalan, J.; Spencer, W.A.; Beck, J.; Gachuki, B.W.; Alhakeem, S.S.; Oben, K.; Munagala, R.; Bondada, S.; et al. Milk-derived exosomes for oral delivery of paclitaxel. Nanomedicine 2017, 13, 1627–1636. [Google Scholar] [CrossRef] [PubMed]
- Kamerkar, S.; LeBleu, V.S.; Sugimoto, H.; Yang, S.; Ruivo, C.F.; Melo, S.A.; Lee, J.J.; Kalluri, R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature 2017, 546, 498–503. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; Zhu, Y.; Ali, D.J.; Tian, T.; Xu, H.; Si, K.; Sun, B.; Chen, B.; Xiao, Z. Engineered exosomes for targeted co-delivery of miR-21 inhibitor and chemotherapeutics to reverse drug resistance in colon cancer. J. Nanobiotechnology 2020, 18, 10. [Google Scholar] [CrossRef]
| Category | Name | Expression | Use in Colorectal Cancer | References |
|---|---|---|---|---|
| miRNA lncRNA | miR-92b | Downward adjustment | Distinguishing CRC from CA | [88] |
| miR-125a-3p | Upward adjustment | Inhibition of migration and invasion | [86] | |
| miR-6803-5p miR-27a miR-130a | Upward adjustment Upward adjustment Upward adjustment | Suggests poor prognosis Differentiating CRC patients Differentiating CRC patients | [81] [83] [84] | |
| LOC344887 LINC00675 DPP10-AS HAGLR GAS5 | Upward adjustment Upward adjustment Upward adjustment Upward adjustment Upward adjustment | Differentiating between CRC patients Differentiating between CRC patients Differentiating between CRC patients Differentiating between CRC patients Inhibits cell proliferation, migration, and invasion | [85] [85] [85] [85] [94] | |
| CCAT2 | Upward adjustment | Enhanced invasion and metastasis | [95] | |
| CRNDE-h | Upward adjustment | Differentiating between patients with CRC and those with benign colorectal disease | [93] | |
| Protein | LNCV6_116109 | Upward adjustment | Differentiating between CRC patients | [92] |
| LNCV6_98390 | Upward adjustment | Differentiating between CRC patients | [92] | |
| LNCV6_84003 | Upward adjustment | Differentiating between CRC patients | [92] | |
| CD147 Hsp60 | Upward adjustment Upward adjustment | Differentiating between CRC patients Induced anti-tumor response | [91] [99] | |
| GPC1 | Upward adjustment | Differentiating between CRC patients | [100] | |
| CPNE3 | Upward adjustment | Differentiating colorectal cancer from other tumors | [101] | |
| SPARC | Upward adjustment | Promotes invasion and induces angiogenesis | [102] | |
| CEA | Upward adjustment | Monitoring for CRC recurrence | [103] |
| NCT Number | Title | Status | Study Results | Interventions | Characteristics |
|---|---|---|---|---|---|
| NCT04523389 | Contents of Circulating Extracellular Vesicles: Biomarkers in Colorectal Cancer Patients | Recruiting | No Results Available |
| |
| NCT04298398 | Impact of Group Psychological Interventions on Extracellular Vesicles in People Who Had Cancer | Not yet recruiting | No Results Available |
| Phase: Not Applicable |
| NCT04394572 | Identification of New Diagnostic Protein Markers for Colorectal Cancer | Recruiting | No Results Available |
| |
| NCT01294072 | Study Investigating the Ability of Plant Exosomes to Deliver Curcumin to Normal and Colon Cancer Tissue | Recruiting | No Results Available |
| Phase: Phase 1 |
| NCT04852653 | A Prospective Feasibility Study Evaluating Extracellular Vesicles Obtained by Liquid Biopsy for Neoadjuvant Treatment Response Assessment in Rectal Cancer | Not yet recruiting | No Results Available |
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, W.; Hu, X.; Jiang, Z. Small Extracellular Vesicles: Key Forces Mediating the Development and Metastasis of Colorectal Cancer. Cells 2022, 11, 1780. https://doi.org/10.3390/cells11111780
Zhang W, Hu X, Jiang Z. Small Extracellular Vesicles: Key Forces Mediating the Development and Metastasis of Colorectal Cancer. Cells. 2022; 11(11):1780. https://doi.org/10.3390/cells11111780
Chicago/Turabian StyleZhang, Wenjie, Xiaoxue Hu, and Zhengting Jiang. 2022. "Small Extracellular Vesicles: Key Forces Mediating the Development and Metastasis of Colorectal Cancer" Cells 11, no. 11: 1780. https://doi.org/10.3390/cells11111780





