Genetic Structure and Molecular Mechanisms Underlying the Formation of Tassel, Anther, and Pollen in the Male Inflorescence of Maize (Zea mays L.)
Abstract
:1. Introduction
2. Cellular Developmental Process of Maize Male Inflorescence
3. Genes and the Molecular Regulation Mechanisms of Male Inflorescence Development and Tassel Morphology
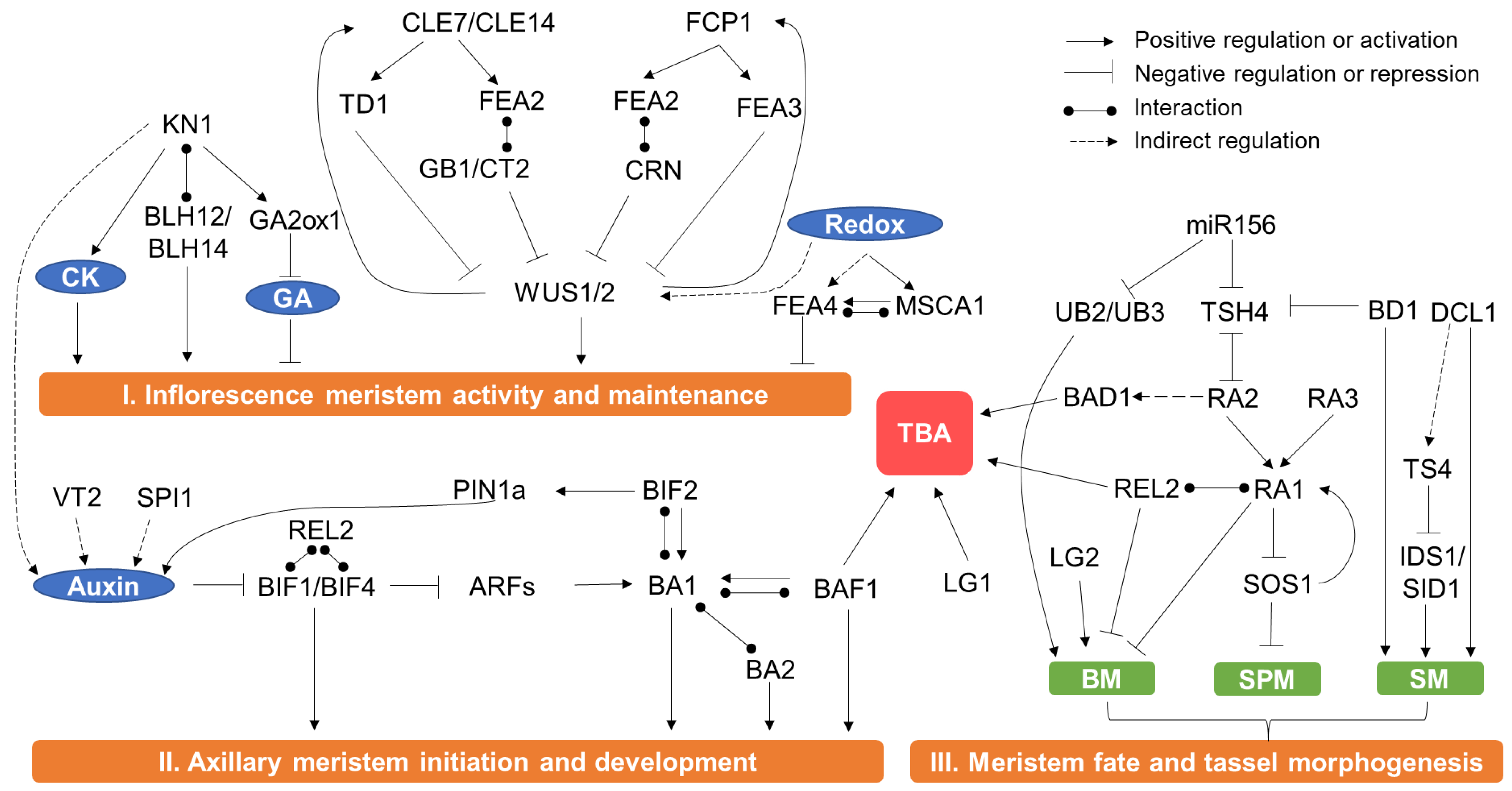
3.1. Meristem Activity and Maintenance
3.2. Axillary Meristem Initiation and Formation in the Reproductive Phase
3.3. Regulation of Meristem Fate
3.4. Regulation of TBA
4. Genetic Basis of Tassel Traits Revealed by Quantitative Genetics Methodologies
5. Application of the Quantitative Genetic Variation Regulating Tassel Traits in Maize Breeding
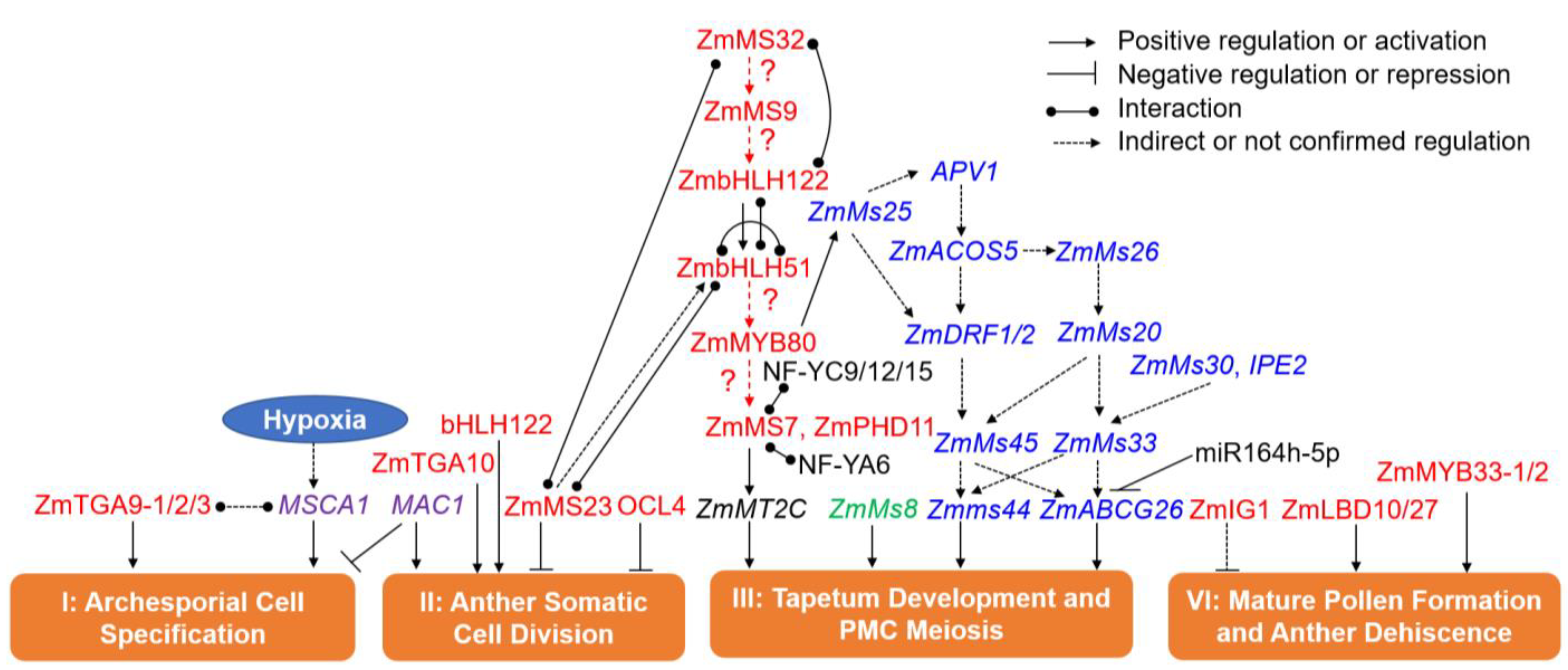
6. The Cloned Maize GMS Genes and Their Roles in Anther and Pollen Development
| Categories | Gene Names | Encoded Proteins | Expression Stages | Molecular Functions | Orthologs | References |
|---|---|---|---|---|---|---|
| Transcription Factors | OCL4 | HD-ZIP IV transcription factor | S5 | Relates to anther wall development in early stage | - | [152] |
| ZmTGA9-1 | bZIP family transcription factor | S5 | Involve in the Ubisch bodies and cuticle formation of anther wall | AtTGA9 | [68,150,153] | |
| ZmTGA9-2 | S5 | |||||
| ZmTGA9-3 | S5–S6 | |||||
| ZmTGA10 | S5 | Controls anther dehiscence | OsTGA10, AtTGA10 | [68,153,154] | ||
| ZmMs32 | bHLH transcription factor | S6 | Controls periclinal division and tapetum differentiation | OsUDT1, AtDYT1 | [155,156,157] | |
| ZmbHLH122 | S5–6 and S8 | Relate to the formation of anther cuticle and Ubisch bodies | OsEAT1 | [68] | ||
| ZmbHLH51 | S6 to S9 | OsTDR, AtAMS | [68,158] | |||
| ZmMs23 | S5 to S8 | Controls tapetal specification and maturation | OsTIP2/ bHLH142 | [159,160,161] | ||
| ZmMs9 | R2/R3 MYB transcription factor | S6 and S9 | The control point for the entry into meiosis | OsTDF1, AtTDF1 | [162,163] | |
| ZmMYB33-1 | MYB transcription factor | S7 and S10 | Involve in the Ubisch bodies and cuticle formation of anther wall and pollen development | OsGAMYB, AtMYB33/65 | [68,164] | |
| ZmMYB33-2 | S7 and S10 | OsGAMYB, AtMYB33/65 | ||||
| ZmMYB80 | S8b-S9 | Involves in the Ubisch bodies formation and pollen development | OsMYB80, AtMYB80 | [68,165,166] | ||
| ZmPHD11 | PHD transcription factor | S8a-S8b | Involves in the Ubisch bodies and cuticle formation of anther wall | OsTIP3, AtMMD1 | [68,167,168] | |
| ZmMs7 | S8b-S9 | Involves in tapetum development and pollen exine formation | OsPTC1, AtMs1 | [169,170] | ||
| IG1 | LBD transcription factor | S9 | Involves in leaf and embryo sac development | OsIG1, AS2 | [171] | |
| ZmLBD27 | S7–S8b, S11–S12 | Important for viable pollen formation | AtLBD10 | [68,172] | ||
| ZmLBD10 | S11–S12 | AtLBD10 | ||||
| Lipid Metabolism | ZmMs33 | Glycerol-3-phosphate acyltransferase (GPAT) | S6 | Plays important roles in anther cuticle, exine and Ubisch bodies formation | OsGPAT3 | [173,174,175] |
| ZmMs26 | Cytochrome P450 monooxygenase | S6 and S9 | Regulates pollen exine formation | OsCYP704B2, AtCYP704B1 | [176] | |
| ZmMs10/ APV1 | P450 subfamily protein | S6 and S9 | Specifically expresses in the tapetum and involve in the hydroxylation of lauric acid | OsCYP704B2, AtCYP704B1 | [177] | |
| ZmMs30 | GDSL lipase | S7–S8 | Regulates exine formation | - | [178] | |
| Zmms44 | Lipid transfer protein | S7–8 | Involves in postmeiotic tapetum secretion of proteins from tapetal cells into locule | - | [179] | |
| IPE1/ZmMs20 | Putative glucose-methanolcholine oxidoreductase | S8b–S9 | Controls anther cuticle and exine development | OsNP1 | [180,181,182] | |
| IPE2 | GDSL lipase | S8b–S9 | Essential for anther cuticle and exine formation | - | [183] | |
| ZmMs6021/ ZmMs25/ ZmFAR1 | Acyl-coenzyme A (CoA)-acyl carrier protein | S9 | Affects anther cuticle and Ubisch body formation as well as microspore development | OsDPW, AtMs2 | [184,185,186] | |
| ZmMs45 | Strictosidine synthase | S9 | Involves in pollen wall initiation after tetrad stage | OsSTRL2, AtLAP3 | [42,187] | |
| ZmMs2/ ZmABCG26 | ABCG transporter | S9 | Transports lipid molecules to exine | OsABCG15, AtABCG26 | [185,188] | |
| AmACOS5-2 | Acyl-CoA Synthetase 5 | - | Play a role in sporopollenin synthesis | OsACOS12, AtACOS5 | [150,189,190,191] | |
| ZmDFR1 | Dihydroflavonol-4-reductase 1 | OsTKPR1, AtTKPR1 | [150,192,193] | |||
| ZmDFR2 | Dihydroflavonoid reductase 2 | OsTKPR1, AtTKPR1 | ||||
| Polysaccharide Metabolism | ZmMs8 | β-1,3-galactosyltransferases | S7–S8 | Affects the epidermal and tapetal cells of maize anthers and meiosis at dyad stage | AtKNS4 | [194,195,196] |
| Other Processes | ZmMs22/ MSCA1 | Plant-specific CC-type GRX | S5 | Controls the redox state and the initiation of archesporial cells | OsMIL1, AtROXY1/2 | [197,198,199] |
| MAC1 | Small secreted protein | S6 | Suppresses excess AR proliferation, triggers periclinal division of subepidermal cells | OsTDL1A | [200] |
6.1. GMS Genes in Phase I (Archesporial Cell Specification)
6.2. GMS Genes in Phase II (Anther Somatic Cell Division)
6.3. GMS Genes in Phase III (Tapetum Development and PMC Formation)
6.4. GMS Genes in Phase IV (Mature Pollen Formation and Anther Dehiscence)
7. Application of GMS Genes in Maize Heterosis Utilization
8. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ranum, P.; Peña-Rosas, J.P.; Garcia-Casal, M.N. Global maize production, utilization, and consumption. Ann. N. Y. Acad. Sci. 2014, 1312, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Rosegrant, M.; Ringler, C.; Sulser, T.B.; Ewing, M.; Palazzo, A.; Zhu, T.; Nelson, G.C.; Koo, J.; Robertson, R.; Msangi, S. Agriculture and Food Security under Global Change: Prospects for 2025/2050; International Food Policy Research Institute: Washington, DC, USA, 2009. [Google Scholar]
- Group, G.; Barker, N.; Clark, L.; Davis, J.; Duvall, M.; Guala, G.; Hsiao, C.; Kellogg, E.; Linder, H. Phylogeny and subfamilial classification of the grasses (Poaceae). Ann. Mo. Bot. Gard. 2002, 88, 373–457. [Google Scholar] [CrossRef]
- Vollbrecht, E.; Schmidt, R.J. Development of the Inflorescences; Springer: New York, NY, USA, 2009. [Google Scholar]
- Dellaporta, S.L.; Calderon-Urrea, A. The sex determination process in maize. Science 1994, 266, 1501–1505. [Google Scholar] [CrossRef] [PubMed]
- Acosta, I.F.; Laparra, H.; Romero, S.P.; Schmelz, E.; Hamberg, M.; Mottinger, J.P.; Moreno, M.A.; Dellaporta, S.L. tasselseed1 is a lipoxygenase affecting jasmonic acid signaling in sex determination of maize. Science 2009, 323, 262–265. [Google Scholar] [CrossRef] [Green Version]
- Berke, T.G.; Rocheford, T.R. Quantitative trait loci for tassel traits in maize. Crop Sci. 1999, 39, 1439–1443. [Google Scholar] [CrossRef]
- Upadyayula, N.; da Silva, H.S.; Bohn, M.O.; Rocheford, T.R. Genetic and QTL analysis of maize tassel and ear inflorescence architecture. Theor. Appl. Genet. 2006, 112, 592–606. [Google Scholar] [CrossRef]
- Xu, G.H.; Wang, X.F.; Huang, C.; Xu, D.Y.; Li, D.; Tian, J.G.; Chen, Q.Y.; Wang, C.L.; Liang, Y.M.; Wu, Y.Y.; et al. Complex genetic architecture underlies maize tassel domestication. New Phytol. 2017, 214, 852–864. [Google Scholar] [CrossRef] [Green Version]
- Matsuoka, Y.; Vigouroux, Y.; Goodman, M.M.; Sanchez, G.J.; Buckler, E.; Doebley, J. A single domestication for maize shown by multilocus microsatellite genotyping. Proc. Natl. Acad. Sci. USA 2002, 99, 6080–6084. [Google Scholar] [CrossRef] [Green Version]
- Piperno, D.R.; Ranere, A.J.; Holst, I.; Iriarte, J.; Dickau, R. Starch grain and phytolith evidence for early ninth millennium B.P. Maize from the Central Balsas River Valley, Mexico. Proc. Natl. Acad. Sci. USA 2009, 106, 5019–5024. [Google Scholar] [CrossRef] [Green Version]
- Doebley, J.; Stec, A. Genetic analysis of the morphological differences between maize and teosinte. Genetics 1991, 129, 285–295. [Google Scholar] [CrossRef]
- Doebley, J.; Stec, A.; Wendel, J.; Edwards, M. Genetic and morphological analysis of a maize-teosinte F2 population: Implications for the origin of maize. Proc. Natl. Acad. Sci. USA 1990, 87, 9888–9892. [Google Scholar] [CrossRef] [Green Version]
- Doebley, J.F. The maize and teosinte male inflorescence: A numerical taxonomic study. Ann. Mo. Bot. Gard. 1983, 70, 32–70. [Google Scholar] [CrossRef]
- Guei, R.G.; Wassom, C.E. Genetic analysis of tassel size and leaf senescence and their relationships with yield in two tropical lowland maize populations. Afr. Crop Sci. J. 1996, 4, 275–281. [Google Scholar]
- Brewbaker, J.L. Diversity and genetics of tassel branch numbers in maize. Crop Sci. 2015, 55, 65–78. [Google Scholar] [CrossRef]
- Duvick, D.N.; Cassman, K.G. Post–green revolution trends in yield potential of temperate maize in the north-central united states. Crop Sci. 1999, 39, 1622–1630. [Google Scholar] [CrossRef]
- Wang, B.; Lin, Z.; Li, X.; Zhao, Y.; Zhao, B.; Wu, G.; Ma, X.; Wang, H.; Xie, Y.; Li, Q.; et al. Genome-wide selection and genetic improvement during modern maize breeding. Nat. Genet. 2020, 52, 565–571. [Google Scholar] [CrossRef]
- Gao, S.B.; Zhao, M.J.; Hai, L.; Zhang, Z.M. Identification of QTL associated with tassel branch number and total tassel length in maize. Hereditas 2007, 29, 1013–1017. [Google Scholar] [CrossRef]
- Mickelson, S.M.; Stuber, C.S.; Senior, L.; Kaeppler, S.M. Quantitative trait loci controlling leaf and tassel traits in a B73 x Mo17 population of maize. Crop Sci. 2002, 42, 1902–1909. [Google Scholar] [CrossRef]
- Upadyayula, N.; Wassom, J.; Bohn, M.O.; Rocheford, T.R. Quantitative trait loci analysis of phenotypic traits and principal components of maize tassel inflorescence architecture. Theor. Appl. Genet. 2006, 113, 1395–1407. [Google Scholar] [CrossRef]
- Li, Y.; Dong, Y.; Niu, S.; Cui, D.; Wang, Y.; Liu, Y.; Wei, M.; Li, X. Identification of agronomically favorable quantitative trait loci alleles from a dent corn inbred Dan232 using advanced backcross QTL analysis and comparison with the F2:3 population in popcorn. Mol. Breed. 2008, 21, 1–14. [Google Scholar] [CrossRef]
- Nikoli, A.; Anelkovi, V.; Dodig, D.; Mici-Ignjatovi, D. Quantitative trait loci for yield and morphological traits in maize under drought stress. Genetika 2011, 43, 263–276. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, B.; Dong, X.; Liu, H.; Ren, L.; Chen, J.; Hauck, A.; Song, W.; Lai, J. An ultra-high density bin-map for rapid QTL mapping for tassel and ear architecture in a large F2 maize population. BMC Genom. 2014, 15, 433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rice, B.R.; Fernandes, S.B.; Lipka, A.E. Multi-trait genome-wide association studies reveal loci associated with maize inflorescence and leaf architecture. Plant Cell Physiol. 2020, 61, 1427–1437. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Wang, X.; Ren, X.; Yang, X.; Zhao, R. A SNP-based high-density genetic map reveals reproducible QTLs for tassel-related traits in maize (zea mays l.). Trop. Plant Biol. 2019, 12, 244–254. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, J.; Guan, Z.; Zhang, X.; Zhang, Y.; Ma, L.; Yao, Y.; Peng, H.; Zhang, Q.; Zhang, B.; et al. Combination of multi-locus genome-wide association study and QTL mapping reveals genetic basis of tassel architecture in maize. Mol. Genet. Genom. 2019, 294, 1421–1440. [Google Scholar] [CrossRef]
- Wu, X.; Li, Y.X.; Shi, Y.S.; Song, Y.C.; Zhang, D.F.; Li, C.H.; Buckler, E.S.; Li, Y.; Zhang, Z.W.; Wang, T.Y. Joint-linkage mapping and GWAS reveal extensive genetic loci that regulate male inflorescence size in maize. Plant Biotechnol. J. 2016, 14, 1551–1562. [Google Scholar] [CrossRef] [Green Version]
- Yang, N.; Lu, Y.L.; Yang, X.H.; Huang, J.; Zhou, Y.; Ali, F.; Wen, W.W.; Liu, J.; Li, J.S.; Yan, J.B. Genome wide association studies using a new nonparametric model reveal the genetic architecture of 17 agronomic traits in an enlarged maize association panel. PLoS Genet. 2014, 10, e1004573. [Google Scholar] [CrossRef] [Green Version]
- Gage, J.L.; White, M.R.; Edwards, J.W.; Kaeppler, S.; de Leon, N. Selection signatures underlying dramatic male inflorescence transformation during modern hybrid maize breeding. Genetics 2018, 210, 1125–1138. [Google Scholar] [CrossRef] [Green Version]
- Pan, Q.C.; Xu, Y.C.; Li, K.; Peng, Y.; Zhan, W.; Li, W.Q.; Li, L.; Yan, J.B. The genetic basis of plant architecture in 10 maize recombinant inbred line populations. Plant Physiol. 2017, 175, 858–873. [Google Scholar] [CrossRef] [Green Version]
- Yi, Q.; Liu, Y.; Zhang, X.; Hou, X.; Zhang, J.; Liu, H.; Hu, Y.; Yu, G.; Huang, Y. Comparative mapping of quantitative trait loci for tassel-related traits of maize in F2:3 and ril populations. J. Genet. 2018, 97, 253–266. [Google Scholar] [CrossRef]
- Brown, P.J.; Upadyayula, N.; Mahone, G.S.; Tian, F.; Bradbury, P.J.; Myles, S.; Holland, J.B.; Flint-Garcia, S.; McMullen, M.D.; Buckler, E.S.; et al. Distinct genetic architectures for male and female inflorescence traits of maize. PLoS Genet. 2011, 7, e1002383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Hao, L.; Kou, S.; Su, E.; Zhou, Y.; Wang, R.; Mohamed, A.; Gao, C.; Zhang, D.; Li, Y.; et al. High-density quantitative trait locus mapping revealed genetic architecture of leaf angle and tassel size in maize. Mol. Breed. 2018, 39, 7. [Google Scholar] [CrossRef]
- Bouchet, S.; Bertin, P.; Presterl, T.; Jamin, P.; Coubriche, D.; Gouesnard, B.; Laborde, J.; Charcosset, A. Association mapping for phenology and plant architecture in maize shows higher power for developmental traits compared with growth influenced traits. Heredity 2017, 118, 249–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Hou, X.; Xiao, Q.; Yi, Q.; Bian, S.; Hu, Y.; Liu, H.; Zhang, J.; Hao, X.; Cheng, W.; et al. Genetic analysis in maize foundation parents with mapping population and testcross population: Ye478 carried more favorable alleles and using QTL information could improve foundation parents. Front. Plant Sci. 2016, 7, 1417. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Peng, Y.; Zhang, J.; Fang, P.; Wu, B. Mapping QTLs and meta-QTLs for two inflorescence architecture traits in multiple maize populations under different watering environments. Mol. Breed. 2017, 37, 91. [Google Scholar] [CrossRef]
- Wilson, Z.A.; Zhang, D.B. From Arabidopsis to rice: Pathways in pollen development. J. Exp. Bot. 2009, 60, 1479–1492. [Google Scholar] [CrossRef] [Green Version]
- Yuan, L. Purification and production of foundation seed of rice PGMS and TGMS lines. Rice 1994, 6, 1–3. [Google Scholar] [CrossRef]
- Cheng, S.H.; Zhuang, J.Y.; Fan, Y.Y.; Du, J.H.; Cao, L.Y. Progress in research and development on hybrid rice: A super-domesticate in china. Ann. Bot. 2007, 100, 599–660. [Google Scholar] [CrossRef]
- Wise, R.P.; Gobelman-Werner, K.; Pei, D.; Dill, C.L.; Schnable, P.S. Mitochondrial transcript processing and restoration of male fertility in T-cytoplasm maize. J. Hered. 1999, 90, 380–385. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Fox, T.W.; Trimnell, M.R.; Wang, L.; Xu, R.; Cigan, A.M.; Huffman, G.A.; Garnaat, C.W.; Hershey, H.; Albertsen, M.C. Development of a novel recessive genetic male sterility system for hybrid seed production in maize and other cross-pollinating crops. Plant Biotechnol. J. 2016, 14, 1046–1054. [Google Scholar] [CrossRef] [Green Version]
- Wan, X.; Wu, S.; Li, Z.; Dong, Z.; Li, J. Maize genic male-sterility genes and their applications in hybrid breeding: Progress and perspectives. Mol. Plant 2019, 12, 321–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, X.; Wu, S.; Li, X. Breeding with dominant genic male-sterility genes to boost crop grain yield in the post-heterosis utilization era. Mol. Plant 2021, 14, 531–534. [Google Scholar] [CrossRef] [PubMed]
- Ma, H. Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu. Rev. Plant Biol. 2005, 56, 393–434. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Luo, X.; Zhu, L. Cytological analysis and genetic control of rice anther development. J. Genet. Genom. 2011, 38, 379–390. [Google Scholar] [CrossRef]
- van der Linde, K.; Walbot, V. Pre-meiotic anther development. Curr. Top. Dev. Biol. 2019, 131, 239–256. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; Wu, S.; Li, Z.; An, X.; Tian, Y. Lipid metabolism: Critical roles in male fertility and other aspects of reproductive development in plants. Mol. Plant 2020, 13, 955–983. [Google Scholar] [CrossRef]
- Liu, S.; Li, Z.; Wu, S.; Wan, X. The essential roles of sugar metabolism for pollen development and male fertility in plants. Crop J. 2021, 9, 1223–1236. [Google Scholar] [CrossRef]
- Gómez, J.F.; Talle, B.; Wilson, Z.A. Anther and pollen development: A conserved developmental pathway. J. Integr. Plant Biol. 2015, 57, 876–891. [Google Scholar] [CrossRef] [Green Version]
- Chaudhury, A.M. Nuclear genes controlling male fertility. Plant Cell 1993, 5, 1277–1283. [Google Scholar] [CrossRef] [Green Version]
- Ariizumi, T.; Toriyama, K. Genetic regulation of sporopollenin synthesis and pollen exine development. Annu. Rev. Plant Biol. 2011, 62, 437–460. [Google Scholar] [CrossRef]
- Li, Z.; An, X.; Zhu, T.; Yan, T.; Wu, S.; Tian, Y.; Li, J.; Wan, X. Discovering and constructing cerna-mirna-target gene regulatory networks during anther development in maize. Int. J. Mol. Sci. 2019, 20, 3480. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Skibbe, D.S.; Fernandes, J.; Walbot, V. Male reproductive development: Gene expression profiling of maize anther and pollen ontogeny. Genome Biol. 2008, 9, R181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelms, B.; Walbot, V. Defining the developmental program leading to meiosis in maize. Science 2019, 364, 52–56. [Google Scholar] [CrossRef]
- Nelms, B.; Walbot, V. Gametophyte genome activation occurs at pollen mitosis i in maize. Science 2022, 375, 424–429. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Yang, X.; Li, G. Molecular insights into inflorescence meristem specification for yield potential in cereal crops. Int. J. Mol. Sci. 2021, 22, 3508. [Google Scholar] [CrossRef]
- Du, Y.; Wu, B.; Xing, Y.; Zhang, Z. Conservation and divergence: Regulatory networks underlying reproductive branching in rice and maize. J. Adv. Res. 2022, in press. [Google Scholar] [CrossRef]
- Zhang, D.; Yuan, Z. Molecular control of grass inflorescence development. Annu. Rev. Plant Biol. 2014, 65, 553–578. [Google Scholar] [CrossRef]
- Li, M.; Zhong, W.; Yang, F.; Zhang, Z. Genetic and molecular mechanisms of quantitative trait loci controlling maize inflorescence architecture. Plant Cell Physiol. 2018, 59, 448–457. [Google Scholar] [CrossRef] [Green Version]
- Gaillochet, C.; Daum, G.; Lohmann, J.U. O cell, where art thou? The mechanisms of shoot meristem patterning. Curr. Opin. Plant Biol. 2015, 23, 91–97. [Google Scholar] [CrossRef] [Green Version]
- Tucker, M.R.; Laux, T. Connecting the paths in plant stem cell regulation. Trends Cell Biol. 2007, 17, 403–410. [Google Scholar] [CrossRef]
- Hake, S. Inflorescence architecture: The transition from branches to flowers. Curr. Biol. 2008, 18, 1106–1108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, H.; Skirpan, A.; Wardell, B.; Matthes, M.S.; Best, N.B.; McCubbin, T.; Durbak, A.; Smith, T.; Malcomber, S.; McSteen, P. The barren stalk2 gene is required for axillary meristem development in maize. Mol. Plant 2019, 12, 374–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chongloi, G.L.; Prakash, S.; Vijayraghavan, U. Regulation of meristem maintenance and organ identity during rice reproductive development. J. Exp. Bot. 2019, 70, 1719–1736. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.T.; Neuffer, M.G. Chromosomal Behavior during Microsporogenesis; Springer: New York, NY, USA, 1994. [Google Scholar]
- Huang, A. Transcriptomes of the anther sporophyte: Availability and uses. Plant Cell Physiol. 2011, 52, 1459–1466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.; An, X.; Li, Z.; Yan, T.; Zhu, T.; Xie, K.; Liu, S.; Hou, Q.; Zhao, L.; Wu, S.; et al. CRISPR/Cas9-based discovery of maize transcription factors regulating male sterility and their functional conservation in plants. Plant Biotechnol. J. 2021, 19, 1769–1784. [Google Scholar] [CrossRef]
- Zhou, L.-Z.; Juranić, M.; Dresselhaus, T. Germline development and fertilization mechanisms in maize. Mol. Plant 2017, 10, 389–401. [Google Scholar] [CrossRef] [Green Version]
- Bommert, P.; Je, B.I.; Goldshmidt, A.; Jackson, D. The maize Galpha gene COMPACT PLANT2 functions in CLAVATA signalling to control shoot meristem size. Nature 2013, 502, 555–558. [Google Scholar] [CrossRef]
- Taguchi-Shiobara, F.; Yuan, Z.; Hake, S.; Jackson, D. The fasciated ear2 gene encodes a leucine-rich repeat receptor-like protein that regulates shoot meristem proliferation in maize. Genes Dev. 2001, 15, 2755–2766. [Google Scholar] [CrossRef] [Green Version]
- Je, B.I.; Gruel, J.; Lee, Y.K.; Bommert, P.; Arevalo, E.D.; Eveland, A.L.; Wu, Q.; Goldshmidt, A.; Meeley, R.; Bartlett, M.; et al. Signaling from maize organ primordia via FASCIATED EAR3 regulates stem cell proliferation and yield traits. Nat. Genet. 2016, 48, 785–791. [Google Scholar] [CrossRef] [Green Version]
- Pautler, M.; Eveland, A.L.; LaRue, T.; Yang, F.; Weeks, R.; Lunde, C.; Il Je, B.; Meeley, R.; Komatsu, M.; Vollbrecht, E.; et al. FASCIATED EAR4 encodes a bZIP transcription factor that regulates shoot meristem size in maize. Plant Cell 2015, 27, 104–120. [Google Scholar] [CrossRef] [Green Version]
- Bommert, P.; Lunde, C.; Nardmann, J.; Vollbrecht, E.; Running, M.; Jackson, D.; Hake, S.; Werr, W. thick tassel dwarf1 encodes a putative maize ortholog of the Arabidopsis CLAVATA1 leucine-rich repeat receptor-like kinase. Development 2005, 132, 1235–1245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, S.; Luo, H.; Huang, Y.; Wang, Y.; Ru, W.; Shi, Y.; Huang, W.; Wang, H.; Dong, Z.; Jin, W. A missense mutation in a large subunit of ribonucleotide reductase confers temperature-gated tassel formation. Plant Physiol. 2020, 184, 1979–1997. [Google Scholar] [CrossRef]
- Wu, Q.; Xu, F.; Liu, L.; Char, S.N.; Ding, Y.; Je, B.I.; Schmelz, E.; Yang, B.; Jackson, D. The maize heterotrimeric G protein β subunit controls shoot meristem development and immune responses. Proc. Natl. Acad. Sci. USA 2020, 117, 1799–1805. [Google Scholar] [CrossRef] [PubMed]
- Nardmann, J.; Werr, W. The shoot stem cell niche in angiosperms: Expression patterns of WUS orthologues in rice and maize imply major modifications in the course of mono- and dicot evolution. Mol. Biol. Evol. 2006, 23, 2492–2504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ritter, M.K.; Padilla, C.M.; Schmidt, R.J. The maize mutant barren stalk1 is defective in axillary meristem development. Am. J. Bot. 2002, 89, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Gallavotti, A.; Zhao, Q.; Kyozuka, J.; Meeley, R.B.; Ritter, M.; Doebley, J.F.; Pe, M.E.; Schmidt, R.J. The role of barren stalk1 in the architecture of maize. Nature 2004, 432, 630–635. [Google Scholar] [CrossRef] [PubMed]
- Barazesh, S.; McSteen, P. Barren inflorescence1 functions in organogenesis during vegetative and inflorescence development in maize. Genetics 2008, 179, 389–401. [Google Scholar] [CrossRef] [Green Version]
- Galli, M.; Liu, Q.; Moss, B.L.; Malcomber, S.; Li, W.; Gaines, C.; Federici, S.; Roshkovan, J.; Meeley, R.; Nemhauser, J.L.; et al. Auxin signaling modules regulate maize inflorescence architecture. Proc. Natl. Acad. Sci. USA 2015, 112, 13372–13377. [Google Scholar] [CrossRef] [Green Version]
- McSteen, P.; Hake, S. barren inflorescence2 regulates axillary meristem development in the maize inflorescence. Development 2001, 128, 2881–2891. [Google Scholar] [CrossRef]
- McSteen, P.; Malcomber, S.; Skirpan, A.; Lunde, C.; Wu, X.T.; Kellogg, E.; Hake, S. barren inflorescence2 encodes a co-ortholog of the PINOID serine/threonine kinase and is required for organogenesis during inflorescence and vegetative development in maize. Plant Physiol. 2007, 144, 1000–1011. [Google Scholar] [CrossRef] [Green Version]
- Skirpan, A.; Wu, X.T.; McSteen, P. Genetic and physical interaction suggest that BARREN STALK1 is a target of BARREN INFLORESCENCE2 in maize inflorescence development. Plant J. 2008, 55, 787–797. [Google Scholar] [CrossRef] [PubMed]
- Vollbrecht, E.; Reiser, L.; Hake, S. Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene, knotted1. Development 2000, 127, 3161–3172. [Google Scholar] [CrossRef]
- Bolduc, N.; Yilmaz, A.; Mejia-Guerra, M.K.; Morohashi, K.; O’Connor, D.; Grotewold, E.; Hake, S. Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev. 2012, 26, 1685–1690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolduc, N.; Hake, S. The maize transcription factor KNOTTED1 directly regulates the gibberellin catabolism gene ga2ox1. Plant Cell 2009, 21, 1647–1658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walsh, J.; Freeling, M. The liguleless2 gene of maize functions during the transition from the vegetative to the reproductive shoot apex. Plant J. 1999, 19, 489–495. [Google Scholar] [CrossRef] [Green Version]
- Qin, X.; Tian, S.; Zhang, W.; Dong, X.; Ma, C.; Wang, Y.; Yan, J.; Yue, B. Qdtbn1, an F-box gene affecting maize tassel branch number by a dominant model. Plant Biotechnol. J. 2020, 19, 1183–1194. [Google Scholar] [CrossRef]
- Chatterjee, M.; Tabi, Z.; Galli, M.; Malcomber, S.; Buck, A.; Muszynski, M.; Gallavotti, A. The boron efflux transporter ROTTEN EAR is required for maize inflorescence development and fertility. Plant Cell 2014, 26, 2962–2977. [Google Scholar] [CrossRef] [Green Version]
- Gallavotti, A.; Barazesh, S.; Malcomber, S.; Hall, D.; Jackson, D.; Schmidt, R.J.; McSteen, P. sparse inflorescence1 encodes a monocot-specific YUCCA-like gene required for vegetative and reproductive development in maize. Proc. Natl. Acad. Sci. USA 2008, 105, 15196–15201. [Google Scholar] [CrossRef] [Green Version]
- Leonard, A.; Holloway, B.; Guo, M.; Rupe, M.; Yu, G.X.; Beatty, M.; Zastrow-Hayes, G.; Meeley, R.; Llaca, V.; Butler, K.; et al. tassel-less1 encodes a boron channel protein required for inflorescence development in maize. Plant Cell Physiol. 2014, 55, 1044–1054. [Google Scholar] [CrossRef] [Green Version]
- Chuck, G.; Whipple, C.; Jackson, D.; Hake, S. The maize SBP-box transcription factor encoded by tasselsheath4 regulates bract development and the establishment of meristem boundaries. Development 2010, 137, 1243–1250. [Google Scholar] [CrossRef] [Green Version]
- Han, J.J.; Jackson, D.; Martienssen, R. Pod corn is caused by rearrangement at the Tunicate1 locus. Plant Cell 2012, 24, 2733–2744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chuck, G.S.; Brown, P.J.; Meeley, R.; Hake, S. Maize SBP-box transcription factors unbranched2 and unbranched3 affect yield traits by regulating the rate of lateral primordia initiation. Proc. Natl. Acad. Sci. USA 2014, 111, 18775–18780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, Y.; Liu, L.; Li, M.; Fang, S.; Shen, X.; Chu, J.; Zhang, Z. UNBRANCHED3 regulates branching by modulating cytokinin biosynthesis and signaling in maize and rice. New Phytol. 2017, 214, 721–733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillips, K.A.; Skirpan, A.L.; Liu, X.; Christensen, A.; Slewinski, T.L.; Hudson, C.; Barazesh, S.; Cohen, J.D.; Malcomber, S.; McSteen, P. vanishing tassel2 encodes a grass-specific tryptophan aminotransferase required for vegetative and reproductive development in maize. Plant Cell 2011, 23, 550–566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bomblies, K.; Wang, R.L.; Ambrose, B.A.; Schmidt, R.J.; Meeley, R.B.; Doebley, J. Duplicate FLORICAULA/LEAFY homologs zfl1 and zfl2 control inflorescence architecture and flower patterning in maize. Development 2003, 130, 2385–2395. [Google Scholar] [CrossRef] [Green Version]
- Chuck, G.; Muszynski, M.; Kellogg, E.; Hake, S.; Schmidt, R.J. The control of spikelet meristem identity by the branched silkless1 gene in maize. Science 2002, 298, 1238–1241. [Google Scholar] [CrossRef]
- Kaplinsky, N.J. Combinatorial control of meristem identity in maize inflorescences. Development 2003, 130, 1149–1158. [Google Scholar] [CrossRef] [Green Version]
- Chuck, G.; Meeley, R.; Hake, S. Floral meristem initiation and meristem cell fate are regulated by the maize AP2 genes ids1 and sid1. Development 2008, 135, 3013–3019. [Google Scholar] [CrossRef] [Green Version]
- Hartwig, T.; Chuck, G.S.; Fujioka, S.; Klempien, A.; Weizbauer, R.; Potluri, D.P.V.; Choe, S.; Johal, G.S.; Schulz, B. Brassinosteroid control of sex determination in maize. Proc. Natl. Acad. Sci. USA 2011, 108, 19814–19819. [Google Scholar] [CrossRef] [Green Version]
- Vollbrecht, E.; Springer, P.S.; Goh, L.; Buckler, E.S.; Martienssen, R. Architecture of floral branch systems in maize and related grasses. Nature 2005, 436, 1119–1126. [Google Scholar] [CrossRef]
- Bortiri, E.; Chuck, G.; Vollbrecht, E.; Rocheford, T.; Martienssen, R.; Hake, S. ramosa2 encodes a LATERAL ORGAN BOUNDARY domain protein that determines the fate of stem cells in branch meristems of maize. Plant Cell 2006, 18, 574–585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Satoh-Nagasawa, N.; Nagasawa, N.; Malcomber, S.; Sakai, H.; Jackson, D. A trehalose metabolic enzyme controls inflorescence architecture in maize. Nature 2006, 441, 227–230. [Google Scholar] [CrossRef]
- Gallavotti, A.; Long, J.A.; Stanfield, S.; Yang, X.; Jackson, D.; Vollbrecht, E.; Schmidt, R.J. The control of axillary meristem fate in the maize ramosa pathway. Development 2010, 137, 2849–2856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, H.; Meng, D.; Liu, H.; Xie, M.; Yin, C.; Liu, F.; Dong, Z.; Jin, W. Ectopic expression of the transcriptional regulator silky3 causes pleiotropic meristem and sex determination defects in maize inflorescences. Plant Cell 2020, 32, 3750–3773. [Google Scholar] [CrossRef] [PubMed]
- DeLong, A.; Calderon-Urrea, A.; Dellaporta, S.L. Sex determination gene TASSELSEED2 of maize encodes a short-chain alcohol dehydrogenase required for stage-specific floral organ abortion. Cell 1993, 74, 757–768. [Google Scholar] [CrossRef]
- Chuck, G.; Cigan, A.M.; Saeteurn, K.; Hake, S. The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat. Genet. 2007, 39, 544–549. [Google Scholar] [CrossRef]
- Thompson, B.E.; Basham, C.; Hammond, R.; Ding, Q.Y.; Kakrana, A.; Lee, T.F.; Simon, S.A.; Meeley, R.; Meyers, B.C.; Hake, S. The dicer-like1 Homolog fuzzy tassel is required for the regulation of meristem determinacy in the inflorescence and vegetative growth in maize. Plant Cell 2014, 26, 4702–4717. [Google Scholar] [CrossRef] [Green Version]
- Chuck, G.; Meeley, R.; Irish, E.; Sakai, H.; Hake, S. The maize tasselseed4 microRNA controls sex determination and meristem cell fate by targeting Tasselseed6/indeterminate spikelet1. Nat. Genet. 2007, 39, 1517–1521. [Google Scholar] [CrossRef]
- Bai, F.; Reinheimer, R.; Durantini, D.; Kellogg, E.A.; Schmidt, R.J. TCP transcription factor, BRANCH ANGLE DEFECTIVE 1 (BAD1), is required for normal tassel branch angle formation in maize. Proc. Natl. Acad. Sci. USA 2012, 109, 12225–12230. [Google Scholar] [CrossRef] [Green Version]
- Gallavotti, A.; Malcomber, S.; Gaines, C.; Stanfield, S.; Whipple, C.; Kellogg, E.; Schmidt, R.J. BARREN STALK FASTIGIATE1 is an AT-Hook protein required for the formation of maize ears. Plant Cell 2011, 23, 1756–1771. [Google Scholar] [CrossRef] [Green Version]
- Lewis, M.W.; Bolduc, N.; Hake, K.; Htike, Y.; Hay, A.; Candela, H.; Hake, S. Gene regulatory interactions at lateral organ boundaries in maize. Development 2014, 141, 4590–4597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whipple, C.J.; Hall, D.H.; DeBlasio, S.; Taguchi-Shiobara, F.; Schmidt, R.J.; Jackson, D.P. A conserved mechanism of bract suppression in the grass family. Plant Cell 2010, 22, 565–578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schoof, H.; Lenhard, M.; Haecker, A.; Mayer, K.F.X.; Jürgens, G.; Laux, T. The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 2000, 100, 635–644. [Google Scholar] [CrossRef] [Green Version]
- Galli, M.; Gallavotti, A. Expanding the regulatory network for meristem size in plants. Trends Genet. 2016, 32, 372–383. [Google Scholar] [CrossRef]
- Brand, U.; Grunewald, M.; Hobe, M.; Simon, R. Regulation of CLV3 expression by two homeobox genes in Arabidopsis. Plant Physiol. 2002, 129, 565–575. [Google Scholar] [CrossRef] [Green Version]
- Trotochaud, A.E.; Jeong, S.; Clark, S.E. CLAVATA3, a multimeric ligand for the CLAVATA1 receptor-kinase. Science 2000, 289, 613–617. [Google Scholar] [CrossRef]
- Fletcher, J.C.; Brand, U.; Running, M.P.; Simon, R.; Meyerowitz, E.M. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 1999, 283, 1911–1914. [Google Scholar] [CrossRef]
- Hirakawa, Y. CLAVATA3, a plant peptide controlling stem cell fate in the meristem. Peptides 2021, 142, 170579. [Google Scholar] [CrossRef]
- Brand, U.; Fletcher, J.C.; Hobe, M.; Meyerowitz, E.M.; Simon, R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 2000, 289, 617–619. [Google Scholar] [CrossRef]
- Nimchuk, Z.L.; Zhou, Y.; Tarr, P.T.; Peterson, B.A.; Meyerowitz, E.M. Plant stem cell maintenance by transcriptional cross-regulation of related receptor kinases. Development 2015, 142, 1043–1049. [Google Scholar] [CrossRef] [Green Version]
- Hu, C.; Zhu, Y.; Cui, Y.; Cheng, K.; Liang, W.; Wei, Z.; Zhu, M.; Yin, H.; Zeng, L.; Xiao, Y.; et al. A group of receptor kinases are essential for CLAVATA signalling to maintain stem cell homeostasis. Nat. Plants 2018, 4, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Muller, R.; Bleckmann, A.; Simon, R. The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell 2008, 20, 934–946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.; Li, W.; Gaines, C.; Buck, A.; Galli, M.; Gallavotti, A. Structural variation at the maize WUSCHEL1 locus alters stem cell organization in inflorescences. Nat. Commun. 2021, 12, 2378. [Google Scholar] [CrossRef] [PubMed]
- Bommert, P.; Nagasawa, N.S.; Jackson, D. Quantitative variation in maize kernel row number is controlled by the FASCIATED EAR2 locus. Nat. Genet. 2013, 45, 334–337. [Google Scholar] [CrossRef] [PubMed]
- Je, B.I.; Xu, F.; Wu, Q.; Liu, L.; Meeley, R.; Gallagher, J.P.; Corcilius, L.; Payne, R.J.; Bartlett, M.E.; Jackson, D. The CLAVATA receptor FASCIATED EAR2 responds to distinct CLE peptides by signaling through two downstream effectors. eLife 2018, 7, e35673. [Google Scholar] [CrossRef]
- Tsuda, K.; Abraham-Juarez, M.J.; Maeno, A.; Dong, Z.; Aromdee, D.; Meeley, R.; Shiroishi, T.; Nonomura, K.I.; Hake, S. KNOTTED1 cofactors, BLH12 and BLH14, regulate internode patterning and vein anastomosis in maize. Plant Cell 2017, 29, 1105–1118. [Google Scholar] [CrossRef] [Green Version]
- Yang, R.S.; Xu, F.; Wang, Y.M.; Zhong, W.S.; Dong, L.; Shi, Y.N.; Tang, T.J.; Sheng, H.J.; Jackson, D.; Yang, F. Glutaredoxins regulate maize inflorescence meristem development via redox control of TGA transcriptional activity. Nat. Plants 2021, 7, 1589–1601. [Google Scholar] [CrossRef]
- Yang, F.; Bui, H.T.; Pautler, M.; Llaca, V.; Johnston, R.; Lee, B.H.; Kolbe, A.; Sakai, H.; Jackson, D. A maize glutaredoxin gene, abphyl2, regulates shoot meristem size and phyllotaxy. Plant Cell 2015, 27, 121–131. [Google Scholar] [CrossRef] [Green Version]
- Zeng, J.; Dong, Z.; Wu, H.; Tian, Z.; Zhao, Z. Redox regulation of plant stem cell fate. EMBO J. 2017, 36, 2844–2855. [Google Scholar] [CrossRef]
- Salehin, M.; Bagchi, R.; Estelle, M. SCFTIR1/AFB-based auxin perception: Mechanism and role in plant growth and development. Plant Cell 2015, 27, 9–19. [Google Scholar] [CrossRef] [Green Version]
- Skirpan, A.; Culler, A.H.; Gallavotti, A.; Jackson, D.; Cohen, J.D.; McSteen, P. BARREN INFLORESCENCE2 interaction with ZmPIN1a suggests a role in auxin transport during maize inflorescence development. Plant Cell Physiol. 2009, 50, 652–657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friml, J.; Yang, X.; Michniewicz, M.; Weijers, D.; Quint, A.; Tietz, O. A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 2004, 306, 862–865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tabuchi, H.; Zhang, Y.; Hattori, S.; Omae, M.; Shimizu-Sato, S.; Oikawa, T.; Qian, Q.; Nishimura, M.; Kitano, H.; Xie, H.; et al. LAX PANICLE2 of rice encodes a novel nuclear protein and regulates the formation of axillary meristems. Plant Cell 2011, 23, 3276–3287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walsh, J.; Waters, C.A.; Freeling, M. The maize gene liguleless2 encodes a basic leucine zipper protein involved in the establishment of the leaf blade-sheath boundary. Genes Dev. 1998, 12, 208–218. [Google Scholar] [CrossRef] [Green Version]
- Mcsteen, P. Branching out: The ramosa pathway and the evolution of grass inflorescence morphology. Plant Cell 2006, 18, 518–522. [Google Scholar] [CrossRef] [Green Version]
- Kellogg, E.A. Floral displays: Genetic control of grass inflorescences. Curr. Opin. Plant Biol. 2007, 10, 26–31. [Google Scholar] [CrossRef]
- Wu, X.; Skirpan, A.; McSteen, P. Suppressor of sessile spikelets1 functions in the ramosa pathway controlling meristem determinacy in maize. Plant Physiol. 2009, 149, 205–219. [Google Scholar] [CrossRef] [Green Version]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef]
- Schauer, S.E.; Jacobsen, S.E.; Meinke, D.W.; Ray, A. DICER-LIKE1: Blind men and elephants in Arabidopsis development. Trends Plant Sci. 2002, 7, 487–491. [Google Scholar] [CrossRef]
- Liu, B.; Li, P.; Li, X.; Liu, C.; Cao, S.; Chu, C.; Cao, X. Loss of function of OsDCL1 affects microRNA accumulation and causes developmental defects in rice. Plant Physiol. 2005, 139, 296–305. [Google Scholar] [CrossRef] [Green Version]
- Moreno, M.A.; Harper, L.C.; Krueger, R.W.; Dellaporta, S.L.; Freeling, M. Liguleless1 encodes a nuclear-localized protein required for induction of ligules and auricles during maize leaf organogenesis. Genes Dev. 1997, 11, 616–628. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Y.J.; Liu, H.J.; Wu, L.J.; Warburton, M.; Yan, J.B. Genome-wide association studies in maize: Praise and stargaze. Mol. Plant 2017, 10, 359–374. [Google Scholar] [CrossRef] [Green Version]
- Maher, B. Personal genomes: The case of the missing heritability. Nature 2008, 456, 18–21. [Google Scholar] [CrossRef] [PubMed]
- Eichler, E.E.; Flint, J.; Gibson, G.; Kong, A.; Leal, S.M.; Moore, J.H.; Nadeau, J.H. Missing heritability and strategies for finding the underlying causes of complex disease. Nat. Rev. Genet. 2010, 11, 446–450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Gallagher, J.; Arevalo, E.D.; Chen, R.; Skopelitis, T.; Wu, Q.; Bartlett, M.; Jackson, D. Enhancing grain-yield-related traits by CRISPR-Cas9 promoter editing of maize CLE genes. Nat. Plants 2021, 7, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Timofejeva, L.; Skibbe, D.; Lee, S.; Golubovskaya, I.; Wang, C.-J.; Harper, L.; Walbot, V.; Cande, W. Cytological characterization and allelism testing of anther developmental mutants identified in a screen of maize male sterile lines. G3-Genes Genom Genet. 2013, 3, 231–249. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Zhang, S.; Jiang, Y.; Yan, T.; Fang, C.; Hou, Q.; Wu, S.; Xie, K.; An, X.; Wan, X. Use of CRISPR/Cas9-based gene editing to simultaneously mutate multiple homologous genes required for pollen development and male fertility in maize. Cells 2022, 11, 439. [Google Scholar] [CrossRef]
- Nan, G.-L.; Teng, C.; Fernandes, J.; O’Connor, L.; Meyers, B.C.; Walbot, V. A cascade of bHLH-regulated pathways programs maize anther development. Plant Cell 2022, 34, 1207–1225. [Google Scholar] [CrossRef]
- Vernoud, V.; Laigle, G.; Rozier, F.; Meeley, R.B.; Perez, P.; Rogowsky, P.M. The HD-ZIP IV transcription factor OCL4 is necessary for trichome patterning and anther development in maize. Plant J. 2009, 59, 883–894. [Google Scholar] [CrossRef]
- Murmu, J.; Bush, M.J.; Delong, C.; Li, S.; Xu, M.; Khan, M.; Malcolmson, C.; Fobert, P.R.; Zachgo, S.; Hepworth, S.R. Arabidopsis basic leucine-zipper transcription factors TGA9 and TGA10 interact with floral glutaredoxins ROXY1 and ROXY2 and are redundantly required for anther development. Plant Physiol. 2010, 154, 1492–1504. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.S.; Liu, X.F.; Wang, D.H.; Chen, R.; Zhang, X.; Xu, Z.H.; Bai, S.N. Transcription factor OsTGA10 is a target of the MADS protein OsMADS8 and is required for tapetum development. Plant Physiol. 2017, 176, 819–835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moon, J.; Skibbe, D.; Timofejeva, L.; Wang, C.J.; Kelliher, T.; Kremling, K.; Walbot, V.; Cande, W.Z. Regulation of cell divisions and differentiation by MALE STERILITY32 is required for anther development in maize. Plant J. 2013, 76, 592–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, K.H.; Han, M.J.; Lee, Y.S.; Kim, Y.W.; An, G. Rice Undeveloped Tapetum1 is a major regulator of early tapetum development. Plant Cell 2005, 17, 2705–2722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, Z.; Sun, Y.; Timofejeva, L.; Chen, C.; Ma, H. Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development 2006, 133, 3085–3095. [Google Scholar] [CrossRef] [Green Version]
- Na, L.; Zhang, D.S.; Liu, H.S.; Yin, C.S.; Zhang, D.B. The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 2006, 18, 2999–3014. [Google Scholar] [CrossRef] [Green Version]
- Nan, G.L.; Zhai, J.; Arikit, S.; Morrow, D.; Fernandes, J.; Mai, L.; Nguyen, N.; Meyers, B.C.; Walbot, V. MS23, a master basic helix-loop-helix factor, regulates the specification and development of the tapetum in maize. Development 2017, 144, 163–172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, Z.; Yu, J.; Cheng, X.; Zong, X.; Xu, J.; Chen, M.; Li, Z.; Zhang, D.; Liang, W. The rice basic helix-loop-helix transcription factor TDR interacting protein2 is a central switch in early anther development. Plant Cell 2014, 26, 1512–1514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ko, S.S.; Li, M.J.; Ku, S.B.; Ho, Y.C.; Lin, Y.J.; Chuang, M.H.; Hsing, H.X.; Lien, Y.C.; Yang, H.T.; Chang, H.C. The bHLH142 transcription factor coordinates with TDR1 to modulate the expression of EAT1 and regulate pollen development in rice. Plant Cell 2014, 26, 2486–2504. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.; Chen, H.; Li, H.; Gao, J.F.; Jiang, H.; Wang, C.; Guan, Y.F.; Yang, Z.N. Defective in tapetal development and function 1 is essential for anther development and tapetal function for microspore maturation in Arabidopsis. Plant J. 2008, 55, 266–277. [Google Scholar] [CrossRef]
- Cai, C.-F.; Zhu, J.; Lou, Y.; Guo, Z.-L.; Xiong, S.-X.; Wang, K.; Yang, Z.-N. The functional analysis of OsTDF1 reveals a conserved genetic pathway for tapetal development between rice and Arabidopsis. Sci. Bull. 2015, 60, 1073–1082. [Google Scholar] [CrossRef] [Green Version]
- Millar, A.A.; Gubler, F. The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 2005, 17, 705–721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phan, H.A.; Li, S.F.; Parish, R.W. MYB80, a regulator of tapetal and pollen development, is functionally conserved in crops. Plant Mol. Biol. 2012, 78, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Yan, W.; Chang, Z.; Xu, Y.; Luo, M.; Xu, C.; Chen, Z.; Wu, J.; Tang, X. OsMYB80 regulates anther development and pollen fertility by targeting multiple biological pathways. Plant Cell Physiol. 2020, 61, 988–1004. [Google Scholar] [CrossRef]
- Yang, X.; Makaroff, C.A.; Ma, H. The Arabidopsis MALE MEIOCYTE DEATH1 gene encodes a PHD-finger protein that is required for male meiosis. Plant Cell 2003, 15, 1281–1295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.; Sun, L.; Zhang, P.; Zhang, Y.; Yu, P.; Liu, L.; Abbas, A.; Xiang, X.; Wu, W.; Zhan, X.; et al. TDR interacting protein 3, encoding a PHD-finger transcription factor, regulates ubisch bodies and pollen wall formation in rice. Plant J. 2019, 99, 844–861. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Wu, S.; An, X.; Xie, K.; Dong, Z.; Zhou, Y.; Xu, L.; Fang, W.; Liu, S.; Liu, S. Construction of a multicontrol sterility system for a maize male-sterile line and hybrid seed production based on the ZmMs7 gene encoding a PHD-finger transcription factor. Plant Biotechnol. J. 2018, 16, 459–471. [Google Scholar] [CrossRef] [Green Version]
- An, X.; Ma, B.; Duan, M.; Dong, Z.; Liu, R.; Yuan, D.; Hou, Q.; Wu, S.; Zhang, D.; Liu, D.; et al. Molecular regulation of ZmMs7 required for maize male fertility and development of a dominant male-sterility system in multiple species. Proc. Natl. Acad. Sci. USA 2020, 117, 23499–23509. [Google Scholar] [CrossRef]
- Evans, M.M. The indeterminate gametophyte1 gene of maize encodes a LOB domain protein required for embryo sac and leaf development. Plant Cell 2007, 19, 46–62. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.J.; Kim, M.; Lee, M.R.; Park, S.K.; Kim, J. LATERAL ORGAN BOUNDARIES DOMAIN (LBD)10 interacts with SIDECAR POLLEN/LBD27 to control pollen development in Arabidopsis. Plant J. 2015, 81, 794–809. [Google Scholar] [CrossRef]
- Xie, K.; Wu, S.; Li, Z.; Zhou, Y.; Zhang, D.; Dong, Z.; An, X.; Zhu, T.; Zhang, S.; Liu, S.; et al. Map-based cloning and characterization of zea mays male sterility33 (ZmMs33) gene, encoding a glycerol-3-phosphate acyltransferase. Theor. Appl. Genet. 2018, 131, 1363–1378. [Google Scholar] [CrossRef] [Green Version]
- Zhu, T.; Wu, S.; Zhang, D.; Li, Z.; Xie, K.; An, X.; Ma, B.; Hou, Q.; Dong, Z.; Tian, Y.; et al. Genome-wide analysis of maize GPAT gene family and cytological characterization and breeding application of ZmMs33/ZmGPAT6 gene. Theor. Appl. Genet. 2019, 132, 2137–2154. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Li, Z.; An, X.; Long, Y.; Xue, X.; Xie, K.; Ma, B.; Zhang, D.; Guan, Y.; Niu, C.; et al. Normal structure and function of endothecium chloroplasts maintained by ZmMs33-mediated lipid biosynthesis in tapetal cells are critical for anther development in maize. Mol. Plant 2020, 13, 1624–1643. [Google Scholar] [CrossRef] [PubMed]
- Djukanovic, V.; Smith, J.; Lowe, K.; Yang, M.; Gao, H.; Jones, S.; Nicholson, M.G.; West, A.; Lape, J.; Bidney, D.; et al. Male-sterile maize plants produced by targeted mutagenesis of the cytochrome p450-like gene (MS26) using a re-designed i-crei homing endonuclease. Plant J. 2013, 76, 888–899. [Google Scholar] [CrossRef] [PubMed]
- Somaratne, Y.; Tian, Y.; Zhang, H.; Wang, M.; Huo, Y.; Cao, F.; Zhao, L.; Chen, H. ABNORMAL POLLEN VACUOLATION1 (APV1) is required for male fertility by contributing to anther cuticle and pollen exine formation in maize. Plant J. 2017, 90, 96–110. [Google Scholar] [CrossRef]
- An, X.; Dong, Z.; Tian, Y.; Xie, K.; Wu, S.; Zhu, T.; Zhang, D.; Zhou, Y.; Niu, C.; Ma, B.; et al. ZmMs30 encoding a novel GDSL lipase is essential for male fertility and valuable for hybrid breeding in maize. Mol. Plant 2019, 12, 343–359. [Google Scholar] [CrossRef] [Green Version]
- Fox, T.; DeBruin, J.; Haug Collet, K.; Trimnell, M.; Clapp, J.; Leonard, A.; Li, B.; Scolaro, E.; Collinson, S.; Glassman, K.; et al. A single point mutation in Ms44 results in dominant male sterility and improves nitrogen use efficiency in maize. Plant Biotechnol. J. 2017, 15, 942–952. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Zhang, H.; Sun, H.; Luo, H.; Zhao, L.; Dong, Z.; Yan, S.; Zhao, C.; Liu, R.; Xu, C.; et al. IRREGULAR POLLEN EXINE1 is a novel factor in anther cuticle and pollen exine formation. Plant Physiol. 2017, 173, 307–325. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Liu, D.; Tian, Y.; Wu, S.; An, X.; Dong, Z.; Zhang, S.; Bao, J.; Li, Z.; Li, J.; et al. Map-based cloning, phylogenetic, and microsynteny analyses of ZmMs20 gene regulating male fertility in maize. Int. J. Mol. Sci. 2019, 20, 1411. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Lin, S.; Shi, J.; Yu, J.; Zhu, L.; Yang, X.; Zhang, D.; Liang, W. Rice no pollen 1 (NP1) is required for anther cuticle formation and pollen exine patterning. Plant J. 2017, 91, 263–277. [Google Scholar] [CrossRef] [Green Version]
- Huo, Y.; Pei, Y.; Tian, Y.; Zhang, Z.; Li, K.; Liu, J.; Xiao, S.; Chen, H.; Liu, J. IRREGULAR POLLEN EXINE2 encodes a GDSL lipase essential for male fertility in maize. Plant Physiol. 2020, 184, 1438–1454. [Google Scholar] [CrossRef]
- Tian, Y.; Xiao, S.; Liu, J.; Somaratne, Y.; Zhang, H.; Wang, M.; Zhang, H.; Zhao, L.; Chen, H. MALE STERILE6021 (MS6021) is required for the development of anther cuticle and pollen exine in maize. Sci. Rep. 2017, 7, 16736. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Li, Z.; Liu, X.; Zhu, T.; Xie, K.; Hou, Q.; Yan, T.; Niu, C.; Zhang, S.; Yang, M.; et al. ZmFAR1 and ZmABCG26 regulated by microRNA are essential for lipid metabolism in maize anther. Int. J. Mol. Sci. 2021, 22, 7916. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wu, S.; Niu, C.; Liu, D.; Yan, T.; Tian, Y.; Liu, S.; Xie, K.; Li, Z.; Wang, Y.; et al. ZmMs25 encoding a plastid-localized fatty acyl reductase is critical for anther and pollen development in maize. J. Exp. Bot. 2021, 72, 4298–4318. [Google Scholar] [CrossRef] [PubMed]
- Cigan, A.M.; Unger, E.; Xu, R.J.; Kendall, T.; Fox, T.W. Phenotypic complementation of ms45 maize requires tapetal expression of MS45. Sex. Plant Reprod. 2001, 14, 135–142. [Google Scholar] [CrossRef]
- Xu, Q.; Yang, L.; Kang, D.; Ren, Z.; Liu, Y. Maize MS2 encodes an ATP-binding cassette transporter that is essential for anther development. Crop J. 2021, 9, 1301–1308. [Google Scholar] [CrossRef]
- de Azevedo Souza, C.; Kim, S.S.; Koch, S.; Kienow, L.; Schneider, K.; McKim, S.M.; Haughn, G.W.; Kombrink, E.; Douglas, C.J. A novel fatty acyl-CoA synthetase is required for pollen development and sporopollenin biosynthesis in Arabidopsis. Plant Cell 2009, 21, 507–525. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Liang, W.; Chen, M.; Zhang, D.; Zhao, X.; Shi, J. Rice fatty acyl-CoA synthetase OsACOS12 is required for tapetum programmed cell death and male fertility. Planta 2017, 246, 105–122. [Google Scholar] [CrossRef]
- Li, Y.; Li, D.; Guo, Z.; Shi, Q.; Xiong, S.; Zhang, C.; Zhu, J.; Yang, Z. OsACOS12, an orthologue of Arabidopsis acyl-CoA synthetase5, plays an important role in pollen exine formation and anther development in rice. BMC Plant Biol. 2016, 16, 256. [Google Scholar] [CrossRef] [Green Version]
- Grienenberger, E.; Kim, S.S.; Lallemand, B.; Geoffroy, P.; Heintz, D.; Souza Cde, A.; Heitz, T.; Douglas, C.J.; Legrand, M. Analysis of TETRAKETIDE α-PYRONE REDUCTASE function in Arabidopsis thaliana reveals a previously unknown, but conserved, biochemical pathway in sporopollenin monomer biosynthesis. Plant Cell 2010, 22, 4067–4083. [Google Scholar] [CrossRef] [Green Version]
- Xu, D.; Qu, S.; Tucker, M.R.; Zhang, D.; Liang, W.; Shi, J. Ostkpr1 functions in anther cuticle development and pollen wall formation in rice. BMC Plant Biol. 2019, 19, 104. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Oses-Prieto, J.A.; Li, K.H.; Fernandes, J.F.; Burlingame, A.L.; Walbot, V. The male sterile 8 mutation of maize disrupts the temporal progression of the transcriptome and results in the mis-regulation of metabolic functions. Plant J. 2010, 63, 939–951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Skibbe, D.S.; Walbot, V. Maize Male sterile 8 (Ms8), a putative beta-1,3-galactosyltransferase, modulates cell division, expansion, and differentiation during early maize anther development. Plant Reprod 2013, 26, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Narciso, J.O.; Zeng, W.; van de Meene, A.; Yasutomi, M.; Takemura, S.; Lampugnani, E.R.; Doblin, M.S.; Bacic, A.; Ishiguro, S. Kns4/upex1: A type II arabinogalactan β-(1,3)-galactosyltransferase required for pollen exine development. Plant Physiol. 2016, 173, 183–205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaubal, R.; Anderson, J.R.; Trimnell, M.R.; Fox, T.W.; Albertsen, M.C.; Bedinger, P. The transformation of anthers in the msca1 mutant of maize. Planta 2003, 216, 778–788. [Google Scholar] [CrossRef] [PubMed]
- Hong, L.; Tang, D.; Shen, Y.; Hu, Q.; Wang, K.; Li, M.; Lu, T.; Cheng, Z. MIL2 (MICROSPORELESS2) regulates early cell differentiation in the rice anther. New Phytol. 2012, 196, 402–413. [Google Scholar] [CrossRef]
- Xing, S.; Zachgo, S. ROXY1 and ROXY2, two Arabidopsis glutaredoxin genes, are required for anther development. Plant J. 2010, 53, 790–801. [Google Scholar] [CrossRef]
- Wang, C.J.; Nan, G.L.; Kelliher, T.; Timofejeva, L.; Vernoud, V.; Golubovskaya, I.N.; Harper, L.; Egger, R.; Walbot, V.; Cande, W.Z. Maize multiple archesporial cells 1 (mac1), an ortholog of rice TDL1A, modulates cell proliferation and identity in early anther development. Development 2012, 139, 2594–2603. [Google Scholar] [CrossRef] [Green Version]
- Kelliher, T.; Walbot, V. Hypoxia triggers meiotic fate acquisition in maize. Science 2012, 337, 345–348. [Google Scholar] [CrossRef] [Green Version]
- Hong, L.; Tang, D.; Zhu, K.; Wang, K.; Li, M.; Cheng, Z. Somatic and reproductive cell development in rice anther is regulated by a putative glutaredoxin. Plant Cell 2012, 24, 577–588. [Google Scholar] [CrossRef] [Green Version]
- Chaubal, R.; Zanella, C.; Trimnell, M.R.; Fox, T.W.; Albertsen, M.C.; Bedinger, P. Two male-sterile mutants of zea mays (Poaceae) with an extra cell division in the anther wall. Am. J. Bot. 2000, 87, 1193–1201. [Google Scholar] [CrossRef]
- Lou, Y.; Zhou, H.S.; Han, Y.; Zeng, Q.Y.; Zhu, J.; Yang, Z.N. Positive regulation of AMS by TDF1 and the formation of a TDF1-AMS complex are required for anther development in Arabidopsis thaliana. New Phytol. 2018, 217, 378–391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, J.N.; Zhu, J.; Yu, Y.; Teng, X.D.; Lou, Y.; Xu, X.F.; Liu, J.L.; Yang, Z.N. DYT1 directly regulates the expression of TDF1 for tapetum development and pollen wall formation in Arabidopsis. Plant J. 2014, 80, 1005–1013. [Google Scholar] [CrossRef] [PubMed]
- Zhu, E.; You, C.; Wang, S.; Cui, J.; Niu, B.; Wang, Y.; Qi, J.; Ma, H.; Chang, F. The DYT1-interacting proteins bHLH010, bHLH089 and bHLH091 are redundantly required for Arabidopsis anther development and transcriptome. Plant J. 2015, 83, 976–990. [Google Scholar] [CrossRef]
- Cui, J.; You, C.; Zhu, E.; Huang, Q.; Ma, H.; Chang, F. Feedback regulation of DYT1 by interactions with downstream bHLH factors promotes DYT1 nuclear localization and anther development. Plant Cell 2016, 28, 1078–1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.B.; Zhu, J.; Gao, J.F.; Wang, C.; Li, H.; Li, H.; Zhang, H.Q.; Zhang, S.; Wang, D.M.; Wang, Q.X.; et al. Transcription factor AtMYB103 is required for anther development by regulating tapetum development, callose dissolution and exine formation in Arabidopsis. Plant J. 2007, 52, 528–538. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.; Moon, S.; Lee, Y.S.; Zhu, L.; Liang, W.; Zhang, D.; Jung, K.H.; An, G. Defective tapetum cell death 1 (DTC1) regulates ROS levels by binding to metallothionein during tapetum degeneration. Plant Physiol. 2016, 170, 1611–1623. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Shi, J.; Yang, X. Role of Lipid Metabolism in Plant Pollen Exine Development; Springer: Cham, Switzerland, 2016. [Google Scholar]
- Li, Z.; Zhu, T.; Liu, S.; Jiang, Y.; Liu, H.; Zhang, Y.; Xie, K.; Li, J.; An, X.; Wan, X. Genome-wide analyses on transcription factors and their potential microRNA regulators involved in maize male fertility. Crop J. 2021, 9, 1248–1262. [Google Scholar] [CrossRef]
- Wei, X.; Pu, A.; Liu, Q.; Leng, Y.; Fu, Z.; Wu, F.; An, X.; Long, Y. Commercialization and supervision policies of gene edited crops in china and other main countries. ACS Agric. Sci. Technol. 2022, 2, 167–173. [Google Scholar] [CrossRef]
- Sarić, R.; Nguyen, V.D.; Burge, T.; Berkowitz, O.; Trtílek, M.; Whelan, J.; Lewsey, M.G.; Čustović, E. Applications of hyperspectral imaging in plant phenotyping. Trends Plant Sci. 2022, 27, 301–315. [Google Scholar] [CrossRef]
- Shi, Y.; Thomasson, J.A.; Murray, S.C.; Pugh, N.A.; Rooney, W.L.; Shafian, S.; Rajan, N.; Rouze, G.; Morgan, C.L.; Neely, H.L.; et al. Unmanned aerial vehicles for high-throughput phenotyping and agronomic research. PLoS ONE 2016, 11, e0159781. [Google Scholar] [CrossRef] [Green Version]
- Gupta, P.K.; Kulwal, P.L.; Jaiswal, V. Association mapping in plants in the post-GWAS genomics era. Adv. Genet. 2019, 104, 75–154. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Sun, K.; An, X. CRISPR/Cas system: Applications and prospects for maize improvement. ACS Agric. Sci. Technol. 2022, 2, 174–183. [Google Scholar] [CrossRef]



| Types | Gene Names | Gene IDs (Zm00001d) | Positions | Annotations | References |
|---|---|---|---|---|---|
| Genes regulating meristem activity | CT2 | 027886 | Chr1: 16721214–16732176 | Subunit (Gα) of a heterotrimeric GTP binding protein | [70] |
| FEA2 | 051012 | Chr4: 136764371–136769212 | LRR receptor-like protein | [71] | |
| FEA3 | 040130 | Chr3: 28709631–28715222 | LRR receptor | [72] | |
| FEA4 | 037317 | Chr6: 120722612–120728273 | bZIP transcription factor | [73] | |
| TD1 | 014793 | Chr5: 63455339–63461620 | LRR receptor-like kinase | [74] | |
| TVT1-R | 045192 | Chr9: 15719302–15730427 | Ribonucleotide reductase | [75] | |
| ZmGB1 | 033422 | Chr1: 262590898–262600232 | Subunit (Gβ) of a heterotrimeric GTP binding protein | [76] | |
| ZmWUS1 | 001948 | Chr2: 3416796–3418004 | Homeodomain transcription factor | [77] | |
| ZmWUS2 | 026537 | Chr10: 147855536–147856873 | Homeodomain transcription factor | [77] | |
| Genes controlling BM development | BA1 | 042989 | Chr3: 186013129–186016764 | bHLH transcription factor | [78,79] |
| BA2 | 003897 | Chr2: 65741213–65756966 | Nucleoprotein | [64] | |
| BIF1 | 008749 | Chr8: 18950258–18955333 | Aux/IAA protein | [80,81] | |
| BIF2 | 031068 | Chr1: 175806351–175810932 | Serine/threonine protein kinase | [81,82,83,84] | |
| BIF4 | 037691 | Chr6: 134087331–134094170 | Aux/IAA protein | [81] | |
| KN1 | 033859 | Chr1: 276071835–276082742 | KNOTTED1-like homeobox (KNOX) transcription factor | [85,86,87] | |
| LG2 | 042777 | Chr3: 179386227–179397947 | bZIP transcription factor | [88] | |
| QDtbn1 | 053358 | Chr4: 227482692–227483909 | Kelch repeat-containing F-box protein | [89] | |
| RTE | 030656 | Chr1: 151219729–151227321 | Membrane-localized boron efflux transporter | [90] | |
| SPI1 | 044069 | Chr3: 218285795–218290961 | Flavin monooxygenase | [91] | |
| TLS1 | 032461 | Chr1: 227447816–227454579 | Boron channel protein | [92] | |
| TSH4 | 020941 | Chr7: 137272100–137278639 | SBP-box transcription factor | [93] | |
| TU1 | 052180 | Chr4: 181855808–181865542 | MADS box transcription factor | [94] | |
| UB2 | 031451 | Chr1: 190381366–190388089 | SBP-box transcription factor | [95] | |
| UB3 | 052890 | Chr4: 203609847–203617018 | SBP-box transcription factor | [95,96] | |
| VT2 | 008700 | Chr8: 17393223–17400365 | Grass-specific tryptophan aminotransferase | [97] | |
| ZFL1 | 026231 | Chr10: 141560362–141566267 | FLO/LFY homolog | [98] | |
| ZFL2 | 002449 | Chr2: 12912591–12918568 | FLO/LFY homolog | [98] | |
| Genes controlling meristem fate | BD1 | 022488 | Chr7: 178604458–178608405 | ERF transcription factor | [99] |
| IDS1 | 034629 | Chr1: 298421359–298428550 | AP2 transcription factor | [100,101] | |
| NA1 | 042843 | Chr3: 181819965–181824489 | 5α-steroid reductase | [102] | |
| RA1 | 020430 | Chr7: 113570910–113574437 | Cys2-His2 zinc-finger transcription factor | [103] | |
| RA2 | 039694 | Chr3: 12156780–12160565 | LOB-domain transcription factor | [104] | |
| RA3 | 022193 | Chr7: 172483459–172490694 | Trehalose-6phosphate phosphatase | [105] | |
| REL2 | 024523 | Chr10: 75992328–76004412 | Transcriptional co-repressor similar to TOPLESS protein | [106] | |
| Si3 | 004130 | Chr2: 85360621–85369941 | Putative transcriptional regulator | [107] | |
| SID1 | 019230 | Chr7: 23052961–23067089 | AP2 transcription factor | [101] | |
| TS1 | 003533 | Chr2: 47103687–47110872 | Lipoxygenase | [6] | |
| TS2 | 028806 | Chr1: 46953827–46958171 | Short-chain alcohol dehydrogenase | [108] | |
| MicroRNAs mediated inflorescence development | CG1 | - * | Chr3: 7773052–7777625 | MiRNA156 | [109] |
| FZT | 027412 | Chr1: 4722956–4738332 | DICER-LIKE1 | [110] | |
| TS4 | - * | Chr3: 144916511–144920220 | MiRNA172 | [111] | |
| Genes controlling tassel branch angel | BAD1 | 005737 | Chr2: 185420859–185424689 | TCP transcription factor | [112] |
| BAF1 | 045427 | Chr9: 21784850–21788875 | AT hook transcription factor | [113] | |
| LG1 | 002005 | Chr2: 4229354–4236035 | SBP-box transcription factor | [114] | |
| TSH1 | 039113 | Chr6: 170246513–170250985 | GATA zinc-finger protein | [115] |
| BMS System | Gene Names | Elements | References | ||||
|---|---|---|---|---|---|---|---|
| SPT | ZmMs45 | pZmMs5126::ZmMs45 | pPG47::Bt1:ZmAA | 35SEN-pLTP2::DsRed2 | [42] | ||
| MCS | ZmMs7 | pZmMs7::ZmMs7 | pPG47::Bt1:ZmAA | pLTP2::mCherry | p35S::Bar | pZm13::Dam | [169,174,178] |
| MCS | ZmMs30 | PZmMs30::ZmMs30 | pPG47::Bt1:ZmAA | pLTP2::mCherry | p35S::Bar | [169,174,178] | |
| MCS | ZmMs33 | PZmMs33::ZmMs33 | pPG47::Bt1:ZmAA | pLTP2::mCherry | p35S::Bar | pZm13::Dam | [169,174,178] |
| DGMS | ZmMs7 | PZmMs5126::ZmMs7 | pLTP2::mCherry | p35S::Bar | [170] | ||
| DGMS | ZmMs44 | pZmMs44::ZmMs44 miRNA | pPG47::Bt1:ZmAA | 35SEN-pLTP2::DsRed2 | [179] | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Bao, J.; Wei, X.; Wu, S.; Fang, C.; Li, Z.; Qi, Y.; Gao, Y.; Dong, Z.; Wan, X. Genetic Structure and Molecular Mechanisms Underlying the Formation of Tassel, Anther, and Pollen in the Male Inflorescence of Maize (Zea mays L.). Cells 2022, 11, 1753. https://doi.org/10.3390/cells11111753
Wang Y, Bao J, Wei X, Wu S, Fang C, Li Z, Qi Y, Gao Y, Dong Z, Wan X. Genetic Structure and Molecular Mechanisms Underlying the Formation of Tassel, Anther, and Pollen in the Male Inflorescence of Maize (Zea mays L.). Cells. 2022; 11(11):1753. https://doi.org/10.3390/cells11111753
Chicago/Turabian StyleWang, Yanbo, Jianxi Bao, Xun Wei, Suowei Wu, Chaowei Fang, Ziwen Li, Yuchen Qi, Yuexin Gao, Zhenying Dong, and Xiangyuan Wan. 2022. "Genetic Structure and Molecular Mechanisms Underlying the Formation of Tassel, Anther, and Pollen in the Male Inflorescence of Maize (Zea mays L.)" Cells 11, no. 11: 1753. https://doi.org/10.3390/cells11111753
APA StyleWang, Y., Bao, J., Wei, X., Wu, S., Fang, C., Li, Z., Qi, Y., Gao, Y., Dong, Z., & Wan, X. (2022). Genetic Structure and Molecular Mechanisms Underlying the Formation of Tassel, Anther, and Pollen in the Male Inflorescence of Maize (Zea mays L.). Cells, 11(11), 1753. https://doi.org/10.3390/cells11111753







